
Spotting Differences Among Observations
Marko Rak, Tim K
¨
onig and Klaus-Dietz T
¨
onnies
Department of Simulation and Graphics, Otto-von-Guericke University, Magdeburg, Germany
Keywords:
Density Difference, Kernel Density Estimation, Scale Space, Blob Detection, Affine Shape Adaption.
Abstract:
Identifying differences among the sample distributions of different observations is an important issue in many
fields ranging from medicine over biology and chemistry to physics. We address this issue, providing a gen-
eral framework to detect difference spots of interest in feature space. Such spots occur not only at various
locations, they may also come in various shapes and multiple sizes, even at the same location. We deal with
these challenges in a scale-space detection framework based on the density function difference of the obser-
vations. Our framework is intended for semi-automatic processing, providing human-interpretable interest
spots for further investigation of some kind, e.g., for generating hypotheses about the observations. Such in-
terest spots carry valuable information, which we outline at a number of classification scenarios from UCI
Machine Learning Repository; namely, classification of benign/malign breast cancer, genuine/forged money
and normal/spondylolisthetic/disc-herniated vertebral columns. To this end, we establish a simple decision
rule on top of our framework, which bases on the detected spots. Results indicate state-of-the-art classification
performance, which underpins the importance of the information that is carried by these interest spots.
1 INTRODUCTION
Sooner or later a large portion of pattern recognition
tasks come down to the question What makes X dif-
ferent from Y ? Some scenarios of that kind are:
Detection of forged money based on image-
derived features: What makes some sort of
forgery different from genuine money?
Comparison of medical data of healthy and
non-healthy subjects for disease detection:
What makes data of the healthy different from
that of the non-healthy?
Comparison of document data sets for text re-
trieval purposes: What makes this set of docu-
ments different from another set?
Apart from this, spotting differences in two or more
observations is of interest in fields of computational
biology, chemistry or physics. Looking at it from a
general perspective, such questions generalize to
What makes the samples of group X different
from the samples of group Y ?
This question usually arises when we deal with
grouped samples in some feature space. For humans,
answering such questions tends to become more chal-
lenging with increasing number of groups, samples
and feature space dimensions, up to the point where
we miss the forest for the trees. This complexity is
not an issue to automatic approaches, which, on the
other hand, tend to either overfit or underfit patterns
in the data. Therefore, semi-automatic approaches are
needed to generate a number of interest spots which
are to be looked at in more detail.
We address this issue by a scale-space difference
detection framework. Our approach relies on the den-
sity difference of group samples in feature space. This
enables us to identify spots where one group domi-
nates the other. We draw on kernel density estimators
to represent arbitrary density functions. Embedding
this into a scale-space representation, we are able to
detect spots of different sizes and shapes in feature
space in an efficient manner. Our framework:
• applies to d-dimensional feature spaces¿
• is able to reflect arbitrary density functions
• selects optimal spot locations, sizes and shapes
• is robust to outliers and measurement errors
• produces human-interpretable results
Our presentation is structured as follows. We out-
line the key idea of our framework in Section 2. The
specific parts of our framework are detailed in Sec-
tion 3, while Section 4 comprises our results on sev-
eral data sets. In Section 5, we close with a summary
of our work, our most important results and an outline
of future work.
5
Rak M., König T. and Tönnies K..
Spotting Differences Among Observations.
DOI: 10.5220/0005165300050013
In Proceedings of the International Conference on Pattern Recognition Applications and Methods (ICPRAM-2015), pages 5-13
ISBN: 978-989-758-076-5
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

2 FOUNDATIONS
Searching for differences between the sample distri-
bution of two groups of observations g and h, we,
quite naturally, seek for spots where the density func-
tion f
g
(x) of group g dominates the density function
f
h
(x) of group h, or vice versa. Hence, we try to find
positive-/negative-valued spots of the density differ-
ence
f
g−h
(x) = f
g
(x) − f
h
(x) (1)
w.r.t. the underlying feature space R
d
with x ∈ R
d
.
Such spots may come in various shapes and sizes. A
difference detection framework should be able to deal
with these degrees of freedom. Additionally, it must
be robust to various sources of error, e.g., from mea-
surement, quantization and outliers.
We propose to superimpose a scale-space repre-
sentation to the density difference f
g−h
(x) to achieve
the above-mentioned properties. Scale-space frame-
works have been shown to robustly handle a wide
range of detection tasks for various types of struc-
tures, e.g., text strings (Yi and Tian, 2011), per-
sons and animals (Felzenszwalb et al., 2010) in
natural scenes, neuron membranes in electron mi-
croscopy imaging (Seyedhosseini et al., 2011) or mi-
croaneurysms in digital fundus images (Adal et al.,
2014). In each of these tasks the function of interest
is represented through a grid of values, allowing for
an explicit evaluation of the scale-space. However, an
explicit grid-based approach becomes intractable for
higher-dimensional feature spaces.
In what follows, we show how a scale-space rep-
resenation of f
g−h
(x) can be obtained from kernel
density estimates of f
g
(x) and f
h
(x) in an implicit
fashion, expressing the problem by scale-space kernel
density estimators. Note that by the usage of kernel
density estimates our work is limited to feature spaces
with dense filling. We close with a brief discussion on
how this can be used to compare observations among
more than two groups.
2.1 Scale Space Representation
First, we establish a family l
g−h
(x;t) of smoothed ver-
sions of the densitiy difference l
g−h
(x). Scale param-
eter t ≥0 defines the amount of smoothing that is ap-
plied to l
g−h
(x) via convolution with kernel k
t
(x) of
bandwidth t as stated in
l
g−h
(x;t) = k
t
(x) ∗ f
g−h
(x). (2)
For a given scale t, spots having a size of about
2
√
t will be highlighted, while smaller ones will be
smoothed out. This leads to an efficient spot detection
scheme, which will be discussed in Section 3. Let
l
g
(x;t) = k
t
(x) ∗ f
g
(x) (3)
l
h
(x;t) = k
t
(x) ∗ f
h
(x) (4)
be the scale-space representations of the group den-
sities f
g
(x) and f
h
(x). Looking at Equation 2 more
closely, we can rewrite l
g−h
(x;t) equivalently in terms
of l
g
(x;t) and l
h
(x;t) via Equation 3 and 4. This reads
l
g−h
(x;t) = k
t
(x) ∗ f
g−h
(x) (5)
= k
t
(x) ∗
h
f
g
(x) − f
h
(x)
i
(6)
= k
t
(x) ∗ f
g
(x) −k
t
(x) ∗ f
h
(x) (7)
= l
g
(x;t) −l
h
(x;t). (8)
The simple yet powerful relation between the left
and the right-hand side of Equation 8 will allow us
to evaluate the scale-space representation l
g−h
(x) im-
plicitly, i.e., using only kernel functions. Of major im-
portance is the choice of the smoothing kernel k
t
(x).
According to scale-space axioms, k
t
(x) should suf-
fice a number of properties, resulting in the uniform
Gaussian kernel of Equation 9 as the unique choice,
cf. (Babaud et al., 1986; Yuille and Poggio, 1986).
φ
t
(x) =
1
p
(2πt)
d
exp
−
1
2t
x
T
x
(9)
2.2 Kernel Density Estimation
In kernel density estimation, the group density f
g
(x)
is estimated from its n
g
samples by means of a ker-
nel function K
B
g
(x). Let x
g
i
∈ R
d×1
with i = 1, . . . , n
g
being the group samples. Then, the group density es-
timate is given by
ˆ
f
g
(x) =
1
n
g
n
g
∑
i=1
K
B
g
x −x
g
i
. (10)
Parameter B
g
∈ R
d×d
is a symmetric positive definite
matrix, which controls the influences of samples to
the density estimate. Informally speaking, K
B
g
(x) ap-
plies a smoothing with bandwidth B
g
to the “spiky
sample relief” in feature space.
Plugging kernel density estimator
ˆ
f
g
(x) into the
scale-space representation l
g
(x;t) defines the scale-
space kernel density estimator
ˆ
l
g
(x;t) to be
ˆ
l
g
(x;t) = k
t
(x) ∗
ˆ
f
g
(x). (11)
Inserting Equation 10 into the above, we can trace
down the definition of the scale-space density estima-
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
6

tor
ˆ
l
g
(x;t) to the sample level via transformation
ˆ
l
g
(x;t) = k
t
(x) ∗
ˆ
f
g
(x) (12)
= k
t
(x) ∗
"
1
n
g
n
g
∑
i=1
K
B
g
x −x
g
i
#
(13)
=
1
n
g
n
g
∑
i=1
(k
t
∗K
B
g
)
x −x
g
i
. (14)
Though arbitrary kernels can be used, we choose
K
B
(x) to be a Gaussian kernel Φ
B
(x) due to its con-
venient algebraic properties. This (potentially non-
uniform) kernel is defined as
Φ
B
(x) =
1
p
det(2πB)
exp
−
1
2
x
T
B
−1
x
. (15)
Using the above, the right-hand side of Equa-
tion 14 simplifies further because of the Gaussian’s
cascade convolution property. Eventually, the scale-
space kernel density estimator
ˆ
l
g
(x;t) is given by
Equation 16, where I ∈ R
d×d
is the identity.
ˆ
l
g
(x;t) =
1
n
g
n
g
∑
i=1
Φ
tI+B
g
x −x
g
i
(16)
Using this estimator, the scale-space representa-
tion l
g
(x;t) of group density f
g
(x) and analogously
that of group h can be estimated for any (x;t) in an
implicit fashion. Consequently, this allows us to es-
timate the scale-space representation l
g−h
(x;t) of the
density difference f
g−h
(x) via Equation 7 by means
of kernel functions only.
2.3 Bandwidth Selection
When regarding bandwidth selection in such a scale-
space representation, we see that the impact of differ-
ent choices for bandwidth matrix B vanishes as scale t
increases. This can be seen when comparing matrices
tI + 0 and tI + B where 0 represents the zero matrix,
i.e., no bandwidth selection at all. We observe that
relative differences between them become neglectable
once ktIk kBk. This is especially true for large
sample sizes, because the bandwidth will then tend
towards zero for any reasonable bandwidth selector
anyway. Hence, we may actually consider setting B
to 0 for certain problems, as we typically search for
differences that fall above some lower bound for t.
Literature bares extensive work on bandwidth ma-
trix selection, for example, based on plug-in es-
timators (Duong and Hazelton, 2003; Wand and
Jones, 1994) or biased, unbiased and smoothed cross-
validation estimators (Duong and Hazelton, 2005;
Sain et al., 1992). All of these integrate well with our
scale-space density difference framework. However,
in view of the argument above, we propose to compro-
mise between a full bandwidth optimization and hav-
ing no bandwidth at all. We define B
g
= b
g
I and use
an unbiased least-squares cross-validation to set up
the bandwidth estimate for group g. For Gaussian ker-
nels, this leads to the optimization of 17, cf. (Duong
and Hazelton, 2005), which we achieved by golden
section search over b
g
.
argmin
B
g
1
n
g
p
det(4πB
g
)
+
1
n
g
(n
g
−1)
n
g
∑
i=1
n
g
∑
j=1
j6=i
(Φ
2B
g
−2Φ
B
g
)(x
g
i
−x
g
j
) (17)
2.4 Multiple Groups
If differences among more than two groups shall be
detected, we can reduce the comparison to a number
of two-group problems. We can consider two typi-
cal use cases, namely one group vs. another and one
group vs. rest. Which of the two is more suitable de-
pends on the specific task at hand. Let us illustrate
this using two medical scenarios. Assume we have
a number of groups which represent patients having
different diseases that are hard to discriminate in dif-
ferential diagnosis. Then we may consider the second
use case, to generate clues on markers that make one
disease different from the others. In contrast, if these
groups represent stages of a disease, potentially in-
cluding a healthy control group, then we may consider
the first use case, comparing only subsequent stages
to give clues on markers of the disease’s progress.
3 METHOD
To identify the positve-/negative-valued spots of a
density difference, we apply the concept of blob de-
tection, which is well-known in computer vision, to
the scale-space representation derived in Section 2.
In scale-space blob detection, some blobness criterion
is applied to the scale-space representation, seeking
for local optima of the function of interest w.r.t. space
and scale. This directly leads to an efficient detection
scheme that identifies a spot’s location and size. The
latter corresponds to the detection scale.
In a grid-representable problem we can evaluate
blobness densely over the scale-space grid and iden-
tify interesting spots directly using the grid neigh-
borhood. This is intractable here, which is why we
rely on a more refined three-stage approach. First, we
trace the local spatial optima of the density difference
SpottingDifferencesAmongObservations
7

(a) “Isometric” View (b) Top View
Figure 1: Detection results for a two-group (red/blue) problem in two-dimensional feature space (xy-plane) with augmented
scale dimension s; Red squares and blue circles visualize the samples of each group; Red/blue paths outline the dendrogram of
scale-space density difference optima for the red/blue group dominating the other group; Interesting spots of each dendrogram
are printed thick; Red/blue ellipses characterize the shape for each of the interest spots.
through scales of the scale-space representation. Sec-
ond, we identify the interesting spots by evaluating
their blobness along the dendrogram of optima that
was obtained during the first stage. Having selected
spots and therefore knowing their locations and sizes,
we finally calculate an elliptical shape estimate for
each spot in a third stage.
Spots obtained in this fashion characterize ellipti-
cal regions in feature space as outlined in Figure 1.
The representation of such regions, i.e., location, size
and shape, as well as its strength, i.e., its scale-space
density difference value, are easily interpretable by
humans, which allows to look at them in more detail
using some other method. This also renders a limita-
tion of our work, because non-elliptical regions may
only be approximated by elliptical ones. We now give
a detailed description of the three stages.
3.1 Scale Tracing
Assume we are given an equidistant scale sampling,
containing non-negative scales t
1
, . . . , t
n
in increasing
order and we search for spots where group g dom-
inates h. More precisely, we search for the non-
negatively valued maxima of l
g−h
(x;t
i−1
). The op-
posite case, i.e., group h dominates g, is equivalent.
Let us further assume that we know the spatial lo-
cal maxima of the density difference l
g−h
(x;t
i−1
) for
a certain scale t
i−1
and we want to estimate those of
the current scale t
i
. This can be done taking the previ-
ous local maxima as initial points and optimizing each
w.r.t. l
g−h
(x;t
i
). In the first scale, we take the sam-
ples of group g themselves. As some maxima may
be converged to the same location, we merge them
together, feeding unique locations as initials into the
next scale t
i+1
only. We also drop any negatively-
valued locations as these are not of interest to our
task. They will not become of interest for any higher
scale either, because local extrema will not enhance as
scale increases, cf. (Lindeberg, 1998). Since deriva-
tives are simple to evaluate for Gaussian kernels, we
can use Newton’s method for spatial optimization.
We can assemble gradient
∂
∂x
l
g−h
(x;t) and Hessian
∂
2
∂x∂x
T
l
g−h
(x;t) sample-wise using
∂
∂x
Φ
B
(x) = −Φ
B
(x)B
−1
x and (18)
∂
2
∂x∂x
T
Φ
B
(x) = Φ
B
(x)
B
−1
xx
T
B
−1
−B
−1
. (19)
Iterating this process through all scales, we form
a discret dendrogram of the maxima over scales. A
dendrogram branching means that a maxima formed
from two (or more) maxima from the preceding scale.
3.2 Spot Detection
The maxima of interest are derived from a scale-
normalized blobness criterion c
γ
(x;t). Two main cri-
teria, namely the determinant of the Hessian (Bret-
zner and Lindeberg, 1998) and the trace of the Hes-
sian (Lindeberg, 1998) have been discussed in litera-
ture. We focus on the former, which is given in Equa-
tion 20
1
, as it has been shown to provide better scale
selection properties under affine transformation of the
feature space, cf. (Lindeberg, 1998).
c
γ
(x;t) = t
γd
(−1)
d
det
∂
2
∂x∂x
T
l
g−h
(x;t)
| {z }
(20)
= t
γd
c(x;t) (21)
1
(−1)
d
handles even and odd dimensions consistently.
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
8

Because the maxima are already spatially optimal, we
can search for spots that maximize c
γ
(x;t) w.r.t. the
dendrogram neighborhood only. Parameter γ ≥ 0 can
be used to introduce a size bias, shifting the detected
spot towards smaller or larger scales. The definition
of γ highly depends on the type of spot that we are
looking for, cf. (Lindeberg, 1996). This is impractical
when we seek for spots of, for example, small and
large skewness or extreme kurtosis at the same time.
Addressing the parameter issue, we search for
all spots that maximize c
γ
(x;t) locally w.r.t. some
γ ∈ [0, ∞). Some dendrogram spot s with scale-space
coordinates (x
s
;t
s
) is locally maximal if there exists
a γ-interval such that its blobness c
γ
(x
s
;t
s
) is larger
than that of every spot in its dendrogram neighbor-
hood N (s). This leads to a number of inequalities,
which can be written as
t
γd
s
c(x
s
;t
s
) >
∀n∈N (s)
t
γd
n
c(x
n
;t
n
) or (22)
γd log
t
s
t
n
>
∀n∈N (s)
log
c(x
n
;t
n
)
c(x
s
;t
s
)
. (23)
The latter can be solved easily for the γ-interval, if
any. We can now identify our interest spots by look-
ing for the maxima along the dendrogram that locally
maximize the width of the γ-interval. More precisely,
let w
γ
(x
s
;t
s
) be the width of the γ-interval for dendro-
gram spot s, then s is of interest if the dendrogram
Laplacian of w
γ
(x;t) is negative at (x
s
;t
s
), or equiva-
lently, if
w
γ
(x
s
;t
s
) >
1
N (s)
∑
n∈N (s)
w
γ
(x
n
;t
n
). (24)
Intuitively, a spot is of interest if its γ-interval width
is above neighborhood average. This is the only as-
sumption we can make without imposing limitations
on the results. Interest spots indentified in this way
will be dendrogram segments, each ranging over a
number of consecutive scales.
3.3 Shape Adaption
Shape estimation can be done in an iterative manner
for each interest spot. The iteration alternatingly up-
dates the current shape estimate based on a measure
of anisotropy around the spot and then corrects the
bandwidth of the scale-space smoothing kernel ac-
cording to this estimate, eventually reaching a fixed
point. The second moment matrix of the function of
interest is typically used as an anisotropy measure,
e.g., in (Lindeberg and Garding, 1994) and (Mikola-
jczyk and Schmid, 2004). Since it requires spatial in-
tegration of the scale-space representation around the
interest spot, this measure is not feasible here.
We adapted the Hessian-based approach of (Lake-
mond et al., 2012) to d-dimensional problems.
The aim is to make the scale-space representation
isotropic around the interest spot, iteratively mov-
ing any anisotropy into the symmetric positive defi-
nite shape matrix S ∈R
d×d
of the smoothing kernel’s
bandwidth tS. Thus, we lift the problem into a gen-
eralized scale-space representation l
g−h
(x;tS) of non-
uniform scale-space kernels, which requires us to re-
place the definition of φ
t
(x) by that of Φ
B
(x).
Starting with the isotropic S
1
= I, we decompose
the current Hessian via
∂
2
∂x∂x
T
l
g−h
(·;tS
i
) = VD
2
V
T
(25)
into its eigenvectors in columns of V and eigenvalues
on the diagonal of D
2
. We then normalize the latter to
unit determinant via
D =
d
p
det(D)D (26)
to get a relative measure of anisotropy for each of
the eigenvector directions. Finally, we move the
anisotropy into the shape estimate via
S
i+1
=
V
T
D
−
1
2
V
S
i
VD
−
1
2
V
T
(27)
and start all over again. Iteration terminates when
isotropy is reached. More precisely: when the ratio
of minimal and maximal eigenvalue of the Hessian
approaches one, which usually happens within a few
iterations.
4 EXPERIMENTS
We next demonstrate that interest spots carry valuable
information about a data set. Due to the lack of data
sets that match our particular detection task a ground
truth comparison is impossible. Certainly, artificially
constructed problems are an exception. However, the
generalizability of results is at least questionable for
such problems. Therefore, we chose to benchmark
our approach indirectly via a number of classification
tasks. The rational is that results that are comparable
to those of well-established classifiers should under-
pin the importance of the identified interest spots.
We next show how to use these interest spots for
classification using a simple decision rule and detail
the data sets that were used. We then investigate pa-
rameters of our approach and discuss the results of the
classification tasks in comparison to decision trees,
Fisher’s linear discriminant analysis, k-nearest neigh-
bors with optimized k and support vector machines
with linear and cubic kernels. All experiments were
performed via leave-one-out cross-validation.
SpottingDifferencesAmongObservations
9

(a) “Isometric” View (b) Top View
Figure 2: Feature space decision boundaries (black plane curves) obtained from group likelihood criterion for the two-
dimensional two-group problem of Figure 1; Red squares and blue circles visualize the samples of each group; Red/blue
paths outline the dendrogram of scale-space density difference optima for the red/blue group dominating the other group;
Interesting spots of each dendrogram are printed thick; Red/blue ellipses characterize the shape for each of the interest spots.
10
−6
10
−4
10
−2
10
0
10
−6
10
−4
10
−2
10
0
p
red
p
blue
Figure 3: Sample group likelihoods and decision boundary
(black diagonal line) for the two-group problem of Figure 1.
4.1 Decision Rule
To perform classification we establish a simple deci-
sion rule based on interest spots that were detected us-
ing the one group vs. rest use case. Therefore, we de-
fine a group likelihood criterion as follows. For each
group g, having the set of interest spots I
g
, we define
p
g
(x) = max
s∈I
g
l
g−h
(x
s
;t
s
S
s
)
·exp
−
1
2
(x −x
s
)
T
(t
s
S
s
)
−1
(x −x
s
)
. (28)
This is a quite natural trade-off, where the first factor
favors spots s with high density difference, while the
latter factor favors spots with small Mahalanobis dis-
tance to the location x that is investigated. We may
also think of p
g
(x) as an exponential approximation
of the scale-space density difference using interesting
spots only. Given this, our decision rule simply takes
the group that maximizes the group likelihood for the
location of interest x. Figure 2 and Figure 3 illustrate
the decision boundary obtained from this rule.
4.2 Data Sets
We carried out our experiments on three classifi-
cation data sets taken from UCI Machine Learn-
ing Repository. A brief summary of them is given
in Table 1. In the first task, we distinguish be-
tween benign and malign breast cancer based on
manually graded cytological charateristics, cf. (Wol-
berg and Mangasarian, 1990). In the second task,
we distinguish between genuine and forged money
based on wavelet-transform-derived features from
photographs of banknote-like specimen, cf. (Glock
et al., 2009). In the third task, we differentiate among
normal, spondylolisthetic and disc-herniated vertebral
columns based on biomechanical attributes derived
from shape and orientation of the pelvis and the lum-
bar vertebral column, cf. (Berthonnaud et al., 2005).
4.3 Parameter Investigation
Before detailing classification results, we investigate
two aspects of our approach. Firstly, we inspect
the importance of bandwidth selection, benchmarking
no kernel density bandwidth against the least-squares
cross-validation technique that we use. Secondly, we
determine the influence of the scale sampling rate.
For the latter we space n + 1 scales for various n
equidistantly from zero to
t
n
= F
−1
χ
2
(1 −ε|d) max
g
d
q
det(Σ
g
)
, (29)
where F
−1
χ
2
(·|d) is the cumulative inverse-χ
2
distribu-
tion with d degrees of freedom and Σ
g
is the covari-
ance matrix of group g. Intuitively, t
n
captures the
extent of the group with largest variance up to a small
ε, i.e., here 1.5 ·10
−8
.
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
10
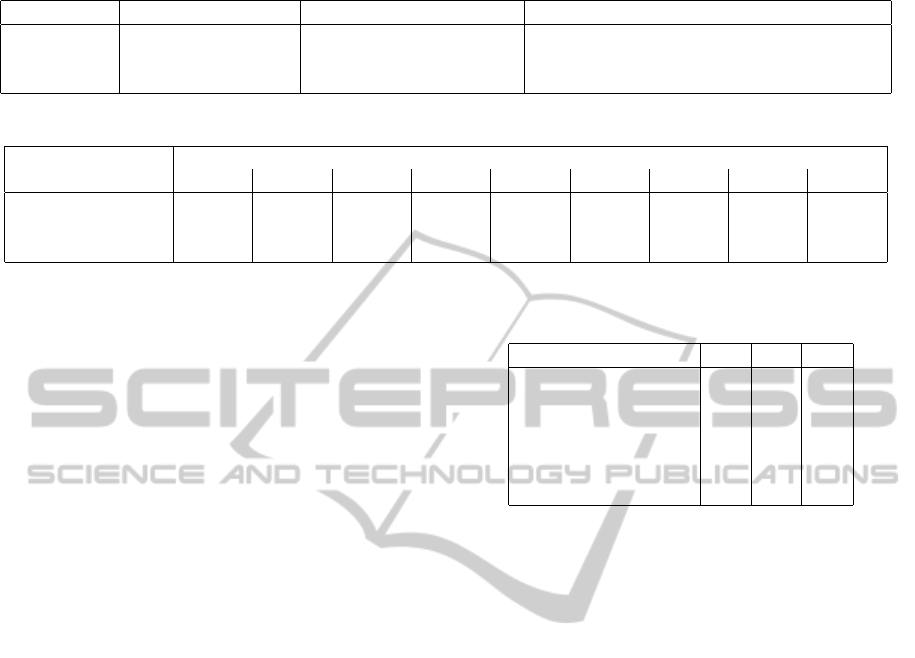
Table 1: Data sets from UCI Machine Learning Repository.
Breast Cancer (BC) Banknote Authentication Vertebral Column
Groups benign / malign genuine / forged normal / spondylolisthetic / herniated discs
Samples 444 / 239 762 / 610 100 / 150 / 60
Dimensions 10 4 6
Table 2: Classification accuracy of our decision rule in b%c for data sets of Table 1 with/without bandwidth selection.
Scale sampling rate n
100 125 150 175 200 225 250 270 300
Breast Cancer 65 / 65 97 / 97 97 / 97 95 / 95 97 / 97 95 / 95 97 / 97 96 / 96 97 / 97
Banknote Authen. 96 / 94 96 / 96 96 / 96 98 / 98 98 / 98 98 / 98 98 / 98 98 / 98 99 / 99
Vertebral Column 87 / 82 88 / 83 88 / 84 88 / 83 88 / 85 88 / 85 88 / 86 88 / 86 88 / 87
To investigate the two aspects, we compare classi-
fication accuracies with and without bandwidth selec-
tion as well as sampling rates ranging from n = 100
to n = 300 in steps of 25. From the results, which are
given in Table 2, we observe that bandwidth selection
is almost neglectable for the Breast Cancer (BC) and
the Banknote Authentication (BA) data set. However,
the impact is substantial throughout all scale sampling
rates for the Vertebral Column (VC) data set. This
may be due to the comparably small number of sam-
ples per group for this data set. Regarding the sec-
ond aspect, we observe that for the BA and VC data
set the classification accuracy slightly increases when
the scale sampling rate rises. Regarding the BC data
set, accuracy remains relatively stable, except for the
lower rates. From the results we conclude that band-
width selection is a necessary part for interest spot
detection. We further recommend n ≥ 200, because
accuracy starts to saturate at this point for all data sets.
For the remaining experiments we used bandwidth se-
lection and a sampling rate of n = 200.
4.4 Classification Results
A comparison of classification accuracies of our de-
cision rule against the aforementioned classifiers is
given in Table 3. For the BC data set we observe that
except for the support vector machine (SVM) with cu-
bic kernel all approaches were highly accurate, scor-
ing between 94% and 97% with our decision rule be-
ing topmost. Even more similar to each other are re-
sults for the BA data set, where all approaches score
between 97% and 99%, with ours lying in the middle
of this range. Results are most diverse for the VC data
sets. Here, the SVM with cubic kernel again performs
significantly worse than the rest, which all score be-
tween 80% and 85%, while our decision rule peaks
at 88%. Other research showed similar scores on the
given data sets. For example the artificial neural net-
works based on pareto-differential evolution in (Ab-
Table 3: Classification accuracies of different classifiers in
b%c for data sets of Table 1.
BC BA VC
decision tree 94 98 82
k-nearest neighbors 97 99 80
Fisher’s discriminant 96 97 80
linear kernel SVM 96 99 85
cubic kernel SVM 90 98 74
our decision rule 97 98 88
bass, 2002) obtained 98% accuracy for the BC data
set, while (Rocha Neto et al., 2011) achieved 83% to
85% accuracy on the VC data set with SVMs with dif-
ferent kernels. These results suggest that our interest
points carry information about a data set that are sim-
ilarly important than the information carried by the
well-established classifiers.
Confusion tables for our decision rule are given
in Table 4 for all data sets. As can be seen, our ap-
proach gave balanced inter-group results for the BC
and the BA data set. We obtained only small inac-
curacies for the recall of the benign (96%) and gen-
uine (97%) groups as well as for the precision of the
malign (94%) and forged (96%) groups. Results for
the VC data set were more diverse. Here, a num-
ber of samples with disc herniation were mistaken
for being normal, lowering the recall of the herniated
group (86%) noticeably. However, more severe inter-
group imbalances were caused by the normal sam-
ples, which were relatively often mistaken for being
spondylolisthetic or herniated discs. Thus, recall for
the normal group (76%) and precision for the herni-
ated group (74%) decreased significantly. The latter is
to some degree caused by a handful of strong outliers
from the normal group that fall into either of the other
groups, which can already be seen from the group
likelihood plot in Figure 4. This finding was made by
others as well, cf. (Rocha Neto and Barreto, 2009).
The other classifiers performed similarly balanced
on the BA and BC data set. Major differences occured
SpottingDifferencesAmongObservations
11

Table 4: Confusion table for predicted/actual groups of our decision rule for data sets of Table 1.
(a) Breast Cancer
H
H
H
H
P
A
ben. mal.
ben. 429 4 99
mal. 15 235 94
96 98 b%c
(b) Banknote Authentication
H
H
H
H
P
A
gen. for.
gen. 742 0 100
for. 20 610 96
97 100 b%c
(c) Vertebral Column
H
H
H
H
P
A
norm. spon. hern.
norm. 76 1 6 91
spon. 10 145 2 92
hern. 14 4 52 74
76 96 86 b%c
10
−30
10
−20
10
−10
10
0
10
−30
10
−20
10
−10
10
0
max
1
p
spond.
, p
hern.
2
p
normal
Figure 4: Sample group likelihoods and decision boundary
(black diagonal line) for the Vertebral Column data set of
Table 1; Normal, spondylolisthetic and herniated discs in
blue, magenta and red, respectively.
on the VC data set only. A precision/recall compar-
ison of all classifiers on the VC data set is given in
Table 5. We observe that the precision of the normal
and the herniated group are significantly lower (gap >
12%) than that of the spondylolisthetic group for all
classifiers except for our decision rule, for which at
least the normal group is predicted with a similar pre-
cision. Regarding the recall we note an even more
unbalanced behavior. Here, a strict ordering from
spondylolisthetic over normal to herniated disks oc-
curs. The differences of the recall of spondylolisthetic
and normal are significant (gap > 16 %) and those
between normal and herniated are even larger (gap >
18 %) among all classifiers that we compared against.
The recalls for our decision rule are distributed differ-
ently, ordering the herniated before the normal group.
Also the magnitude of differences is less significant
(gaps ≈ 10%) for our decision rule. Results of this
comparison indicate that the information that is car-
ried by our interest points tends to be more balanced
among groups than the information carried by the
well-established classifiers that we compared against.
5 CONCLUSION
We proposed a detection framework that is able to
identify differences among the sample distributions
of different observations. Potential applications are
Table 5: Classification precision/recall of different classi-
fiers in b%c for the Vertebral Column data set of Table 1.
norm. spon. hern.
decision tree 69 / 83 97 / 95 68 / 50
k-nearest neighbors 70 / 74 96 / 96 58 / 55
Fisher’s discriminant 70 / 80 87 / 92 74 / 48
linear kernel SVM 76 / 85 97 / 96 72 / 61
cubic kernel SVM 59 / 82 90 / 91 52 / 18
our decision rule 91 / 76 92 / 96 74 / 86
manifold, touching fields such as medicine, biology,
chemistry and physics. Our approach bases on the
density function difference of the observations in fea-
ture space, seeking to identify spots where one obser-
vation dominates the other. Superimposing a scale-
space framework to the density difference, we are able
to detect interest spots of various locations, size and
shapes in an efficient manner.
Our framework is intended for semi-automatic
processing, providing human-interpretable interest
spots for potential further investigation of some kind.
We outlined that these interest spots carry valuable
information about a data set at a number of classifi-
cation tasks from the UCI Machine Learning Repos-
itory. To this end, we established a simple decision
rule on top of our framework. Results indicate state-
of-the-art performance of our approach, which under-
pins the importance of the information that is carried
by these interest spots.
In the future, we plan to extend our work to sup-
port repetitive features such as angles, which cur-
rently is a limitation of our approach. Modifying our
notion of distance, we would then be able to cope with
problems defined on, e.g., a sphere or torus. Future
work may also include the migration of other types of
scale-space detectors to density difference problems.
This includes the notion of ridges, valleys and zero-
crossings, leading to richer sources of information.
ACKNOWLEDGEMENTS
This research was partially funded by the project “Vi-
sual Analytics in Public Health” (TO 166/13-2) of the
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
12

German Research Foundation.
REFERENCES
Abbass, H. A. (2002). An evolutionary artificial neural net-
works approach for breast cancer diagnosis. Artificial
Intelligence in Medicine, 25:265–281.
Adal, K. M., Sidibe, D., Ali, S., Chaum, E., Karnowski,
T. P., and Meriaudeau, F. (2014). Automated detection
of microaneurysms using scale-adapted blob analysis
and semi-supervised learning. Computer Methods and
Programs in Biomedicine, 114:1–10.
Babaud, J., Witkin, A. P., Baudin, M., and Duda, R. O.
(1986). Uniqueness of the Gaussian kernel for scale-
space filtering. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 8:26–33.
Berthonnaud, E., Dimnet, J., Roussouly, P., and Labelle,
H. (2005). Analysis of the sagittal balance of the
spine and pelvis using shape and orientation param-
eters. Journal of Spinal Disorders and Techniques,
18:40–47.
Bretzner, L. and Lindeberg, T. (1998). Feature tracking with
automatic selection of spatial scales. Computer Vision
and Image Understanding, 71:385–392.
Duong, T. and Hazelton, M. L. (2003). Plug-in bandwidth
matrices for bivariate kernel density estimation. Jour-
nal of Nonparametric Statistics, 15:17–30.
Duong, T. and Hazelton, M. L. (2005). Cross-validation
bandwidth matrices for multivariate kernel density es-
timation. Scandinavian Journal of Statistics, 32:485–
506.
Felzenszwalb, P. F., Girshick, R. B., McAllester, D., and
Ramanan, D. (2010). Object detection with discrim-
inatively trained part-based models. IEEE Transac-
tions on Pattern Analysis and Machine Intelligence,
32:1627–1645.
Glock, S., Gillich, E., Schaede, J., and Lohweg, V. (2009).
Feature extraction algorithm for banknote textures
based on incomplete shift invariant wavelet packet
transform. In Proceedings of the Annual Pattern
Recognition Symposium, volume 5748, pages 422–
431.
Lakemond, R., Sridharan, S., and Fookes, C. (2012).
Hessian-based affine adaptation of salient local image
features. Journal of Mathematical Imaging and Vi-
sion, 44:150–167.
Lindeberg, T. (1996). Edge detection and ridge detection
with automatic scale selection. In Proceedings of the
IEEE Computer Society Conference on Computer Vi-
sion and Pattern Recognition, pages 465–470.
Lindeberg, T. (1998). Feature detection with automatic
scale selection. International Journal of Computer Vi-
sion, 30:79–116.
Lindeberg, T. and Garding, J. (1994). Shape-adapted
smoothing in estimation of 3-d depth cues from affine
distortions of local 2-d brightness structure. In Pro-
ceedings of the European Conference on Computer
Vision, pages 389–400.
Mikolajczyk, K. and Schmid, C. (2004). Scale & affine in-
variant interest point detectors. International Journal
of Computer Vision, 60:63–86.
Rocha Neto, A. R. and Barreto, G. A. (2009). On the appli-
cation of ensembles of classifiers to the diagnosis of
pathologies of the vertebral column: A comparative
analysis. IEEE Latin America Transactions, 7:487–
496.
Rocha Neto, A. R., Sousa, R., Barreto, G. A., and Cardoso,
J. S. (2011). Diagnostic of pathology on the verte-
bral column with embedded reject option. In Pattern
Recognition and Image Analysis, volume 6669, pages
588–595. Springer Berlin Heidelberg.
Sain, S. R., Baggerly, K. A., and Scott, D. W. (1992). Cross-
validation of multivariate densities. Journal of the
American Statistical Association, 89:807–817.
Seyedhosseini, M., Kumar, R., Jurrus, E., Giuly, R., Ellis-
man, M., Pfister, H., and Tasdizen, T. (2011). De-
tection of neuron membranes in electron microscopy
images using multi-scale context and Radon-like fea-
tures. In Proceedings of the International Conference
on Medical Image Computing and Computer-assisted
Intervention, pages 670–677.
Wand, M. P. and Jones, M. C. (1994). Multivariate plug-in
bandwidth selection. Computational Statistics, 9:97–
116.
Wolberg, W. and Mangasarian, O. (1990). Multisurface
method of pattern separation for medical diagnosis ap-
plied to breast cytology. In Proceedings of the Na-
tional Academy of Sciences, pages 9193–9196.
Yi, C. and Tian, Y. (2011). Text string detection from natu-
ral scenes by structure-based partition and grouping.
IEEE Transactions on Image Processing, 20:2594–
2605.
Yuille, A. L. and Poggio, T. A. (1986). Scaling theorems for
zero crossings. IEEE Transactions on Pattern Analy-
sis and Machine Intelligence, 8:15–25.
SpottingDifferencesAmongObservations
13
