
Alignment of Cyclically Ordered Trees
∗
Takuya Yoshino
1
and Kouichi Hirata
2
1
Graduate School of Computer Science and Systems Engineering, Kyushu Institute of Technology,
Kawazu 680-4, Iizuka 820-8502, Japan
2
Department of Artificial Intelligence, Kyushu Institute of Technology, Kawazu 680-4, Iizuka 820-8502, Japan
Keywords:
Alignment, Alignment Distance, Segmental Alignment Distance, Cyclically Ordered Tree, Tai Mapping.
Abstract:
In this paper, as unordered trees preserving the adjacency among siblings, we introduce the following three
kinds of a cyclically ordered tree, that is, a biordered tree that allows both a left-to-right and a right-to-left order
among siblings, a cyclic-ordered tree that allows cyclic order among siblings in a left-to-right direction and a
cyclic-biordered tree that allows cyclic order among siblings in both left-to-right and right-to-left directions.
Then, we design the algorithms to compute the alignment distance and the segmental alignment distance
between biordered trees in O(n
2
D
2
) time and ones between cyclic-ordered trees and cyclic-biordered trees in
O(n
2
D
4
) time, where n is the maximum number of nodes and D is the maximum degree in two given trees.
1 INTRODUCTION
Comparing tree-structured data is one of the impor-
tant tasks for many research areas such as pattern
recognition, natural language processing, machine
learning, data mining, bioinformatics, and so on. In
these researches, the tree-structured data are well re-
garded as rooted labeled trees (trees, for short). Also
a tree is ordered if the left-to-right order among sib-
lings is fixed and unordered otherwise.
An edit distance (Tai, 1979) is one of the standard
distance measures between trees. The edit distance is
formulated as the minimum cost to transform from a
tree to another tree by applying edit operations of a
substitution, a deletion and an insertion to trees.
It is known that the edit distance is closely related
to a Tai mapping (Tai, 1979). The minimum cost of
Tai mappings coincides with the edit distance (Tai,
1979). Then, whereas the problem of computing the
edit distance between ordered trees is tractable (De-
maine et al., 2009), one between unordered trees
is MAX SNP-hard (Zhang and Jiang, 1994). This
MAX SNP-hardness holds even if both trees are bi-
nary (Hirata et al., 2011).
An alignment distance is an alternative distance
measure between trees introduced by (Jiang et al.,
∗
This work is partially supported by Grant-in-Aid
for Scientific Research 24240021, 24300060, 25540137,
26280085 and 26370281 from the Ministry of Education,
Culture, Sports, Science and Technology, Japan.
1995) and applied to comparing RNA secondary
structures in bioinformatics (H¨ochsmann et al., 2003;
Schiermer and Giegerich, 2013; Shapiro and Zhang,
1990; Zhang, 1998). The alignment distance is for-
mulated as the minimum cost of possible alignments
(as trees) obtained by first inserting nodes labeled
with spaces into two trees such that the resulting trees
have the same structure and then overlaying them. In
operational, the alignment distance is an edit distance
such that every insertion precedes to deletions.
Kuboyama (Kuboyama, 2007) has first formulated
an alignable mapping as a variation of the Tai map-
ping. Then, he has shown that the alignment dis-
tance coincides with the minimum cost of alignable
mappings and the alignable mapping coincides with a
less-constrained mapping (Lu et al., 2001). As same
as the edit distance, whereas the problem of comput-
ing the alignment distance between ordered trees is
tractable, one between unordered trees is MAX SNP-
hard (Jiang et al., 1995). On the other hand, this prob-
lem becomes tractable if the degrees of unordered
trees are bounded (Jiang et al., 1995).
In the above results of computing distances, we
deal with either ordered or unordered trees. Note
that unordered trees allow all of the permutations
among siblings. On the other hand, several appli-
cations require to allow just some permutations, not
all of the permutations, among siblings. For exam-
ple, when representing graphs with cyclic compounds
such as monosaccharides in glycans (Hizukuri et al.,
263
Yoshino T. and Hirata K..
Alignment of Cyclically Ordered Trees.
DOI: 10.5220/0005207802630270
In Proceedings of the International Conference on Pattern Recognition Applications and Methods (ICPRAM-2015), pages 263-270
ISBN: 978-989-758-076-5
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

2005) and molecules in molecular graphs (Horv´ath
et al., 2010) as trees, the adjacency of nodes in the
compounds is represented as the adjacency among
siblings in the tree representation. Also, when
comparing or modeling RNA secondary structures
as trees (H¨ochsmann et al., 2003; Schiermer and
Giegerich, 2013; Shapiro and Zhang, 1990; Zhang,
1998), the base pairs in nucleotides are connected
with preserving the adjacency among siblings.
Hence, as unordered trees preserving the adja-
cency among siblings, in this paper, we formulate
the following three kinds of a cyclically ordered
tree. Let v
1
, . .. , v
n
be siblings from left to right.
Then, we say that a tree is biordered if it allows
two orders v
1
, . . . , v
n
and v
n
, . . . , v
1
. Also we say
that a tree is cyclic-ordered if it allows a cyclic
order v
i
, . . . , v
n
, v
1
, . . . , v
i−1
for every i (1 ≤ i ≤ n).
Furthermore, we say that a tree is cyclic-biordered
if it allows cyclic orders v
i
, . . . , v
n
, v
1
, . . . , v
i−1
and
v
i
, . . . , v
1
, v
n
, . . . , v
i−1
for every i (1 ≤ i ≤ n).
Since an unordered binary tree is always cycli-
cally ordered, the problem of computing the edit dis-
tance, the segmental distance (Kan et al., 2014) and
the bottom-up distance (Valiente, 2001; Kuboyama,
2007) between cyclically ordered trees is also
MAX SNP-hard (Hirata et al., 2011; Yamamoto
et al., 2014). On the other hand, the problems of
computing the isolated-subtree (or constrained) dis-
tance (Zhang, 1995; Zhang, 1996), the accordant
(or Lu’s) distance (Lu, 1979; Kuboyama, 2007),
the LCA-preserving (or degree-2) distance (Zhang
et al., 1996) and the top-down (or degree-1) dis-
tance (Selkow, 1977; Chawathe, 1999) between un-
ordered trees are tractable, so are the problems of
computing these distances between cyclically ordered
trees.
In this paper, we focus on the alignment dis-
tance and a segmental alignment distance, which is
an alignment distance to preserve the parent-children
relationship as possible (Yoshino and Hirata, 2013),
between cyclically ordered trees, because the prob-
lems of computing both distances are tractable if the
degrees of unordered trees are bounded. Note that the
algorithms to compute all of the above tractable vari-
ations of the edit distance between unordered trees
contain the maximum weighted bipartite matching al-
gorithm (Yamamoto et al., 2014; Zhang et al., 1996)
or originally the minimum cost maximum flow algo-
rithm (Wang et al., 2003; Zhang, 1996).
On the other hand, in this paper, by directly ex-
tending the recurrences to compute the alignment dis-
tance between ordered trees (Jiang et al., 1995), we
first design the algorithms to compute the alignment
distance between biordered trees in O(n
2
D
2
) time,
where n is the maximum number of nodes and D is the
maximum degree in two given trees. This time com-
plexity is same as one between ordered trees (Jiang
et al., 1995). Also we design the algorithms to com-
pute the alignment distance between cyclic-ordered
and cyclic-biordered trees in O(n
2
D
4
) time.
Next, by using the same strategy of (Kan et al.,
2014) to compute a top-down distance for every pair
of nodes in given two cyclically ordered trees in ad-
vance, we design the algorithm to compute the seg-
mental alignment distance between cyclically ordered
trees with the same time complexity as above.
Finally, we give experimental results for the
alignment distance between biordered trees com-
paring with the edit distance between ordered
trees, by using N-glycan data provided from
KEGG (Kyoto Encyclopedia of Genes and Genomes,
http://www.kegg.jp/
).
2 PRELIMINARIES
A tree is a connected graph without cycles. For a tree
T = (V, E), we denote V and E by V(T) and E(T),
respectively. Also the size of T is |V| and denoted by
|T|. We sometime denote v ∈ V(T) by v ∈ T. We
denote an empty tree by
/
0.
A rooted tree is a tree with one node r chosen as
its root. We denote the root of a rooted tree T by
r(T). For each node v in a rooted tree with the root
r, let UP
r
(v) be the unique path from v to r. The
parent of v(6= r), which we denote by par(v), is its
adjacent node on UP
r
(v) and the ancestors of v(6= r)
are the nodes on UP
r
(v) − {v}. We denote the set of
all ancestors of v by anc(v). We say that u is a child
of v if v is the parent of u. The set of children of v is
denoted by ch(v). We call the number of children of
v the degree of v and denote it by d(v), that is, d(v) =
|ch(v)|. Also we define d(T) = max{d(v) | v ∈ T}
and call it the degree of T.
In this paper, we use the ancestor orders < and ≤,
that is, u < v if v is an ancestor of u and u ≤ v if u < v
or u = v. We say that w is the least common ancestor
of u and v, denoted by u⊔ v, if u ≤ w, v ≤ w and there
exists no w
′
such that w
′
≤ w, u ≤ w
′
and v ≤ w
′
. A
(complete) subtree of T = (V, E) rooted by v, denoted
by T[v], is a tree T
′
= (V
′
, E
′
) such that r(T
′
) = v,
V
′
= { u ∈ V | u ≤ v} and E
′
= { (u, w) ∈ E | u, w ∈ V
′
}.
We say that a rooted tree is labeled if each node
is assigned a symbol from a fixed finite alphabet Σ.
For a node v, we denote the label of v by l(v), and
sometimes identify v with l(v). Also let ε 6∈ Σ denote
a special blank symbol and define Σ
ε
= Σ∪ {ε}.
We say that a rooted tree is ordered if a left-to-
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
264

right order among siblings is fixed; unordered oth-
erwise. In particular, for nodes u and v in an or-
dered tree, u is to the left of v, denoted by u v,
if pre(u) ≤ pre(v) and post(u) ≤ post(v) for the pre-
order number pre and the postorder number post.
Furthermore, in this paper, we introduce cyclically
ordered trees by using the following functions σ
+
p,n
(i)
and σ
−
p,n
(i) for 1 ≤ i, p ≤ n.
σ
+
p,n
(i) = ((i+ p− 1) mod n) + 1,
σ
−
p,n
(i) = ((n− i− p+ 1) mod n) + 1.
Definition 1 (Cyclically Ordered Trees). Let T be a
tree and suppose that v
1
, . . . , v
n
are the children of v ∈
T from left to right.
1. We say that T is biordered if T allows the orders
of both v
1
, . . . , v
n
and v
n
, . . . , v
1
.
2. We say that T is cyclic-ordered if T allows the
orders v
σ
+
p,n
(1)
, . . . , v
σ
+
p,n
(n)
for every 1 ≤ p ≤ n.
3. We say that T is cyclic-biordered if T allows the
orders v
σ
+
p,n
(1)
, . . . , v
σ
+
p,n
(n)
and v
σ
−
p,n
(1)
, . . . , v
σ
−
p,n
(n)
for every 1 ≤ p ≤ n.
Sometimes we use the scripts o, b, c, cb, u, and the no-
tation of π ∈ {o, b, c, cb, u} , which we call a π-tree.
It is obvious that the cyclically ordered trees are
an extension of ordered trees and a restriction of un-
ordered trees. The number of orders among siblings
of a node v in ordered trees, biordered trees, cyclic-
ordered trees, cyclic-biordered trees and unordered
trees is 1, 2, d(v), 2d(v) and d(v)!, respectively.
Next, we introduce the alignment distance (Jiang
et al., 1995). Here, for π ∈ {o, b, c, cb, u}, we call an
isomorphism for π-trees a π-isomorphism.
Definition 2 (Alignment (Jiang et al., 1995)). Let T
1
and T
2
be trees and π ∈ {o, b, c, cb, u}. An alignment
between T
1
and T
2
is a tree T obtained by the follow-
ing two steps.
1. Insert new nodes labeled by ε into T
1
and T
2
such
that the resulting trees T
′
1
and T
′
2
are π-isomorphic
with ignoring labels and l(φ(v)) 6= ε whenever
l(v) = ε for a π-isomorphism φ between T
′
1
and
T
′
2
and every node v ∈ T
′
1
.
2. Set T to an obtained tree T
′
1
by relabeling a la-
bel l(v) for every node v ∈ T
′
1
with (l(v), l(φ(v))).
(Note that (ε, ε) 6∈ T .)
Let A
π
(T
1
, T
2
) denote the set of all possible align-
ments between T
1
and T
2
.
We define a cost function γ : (Σ
ε
×Σ
ε
−{(ε, ε)}) 7→
R
+
on pairs of labels. We constrain γ to be a metric,
that is, γ(l
1
, l
2
) ≥ 0, γ(l
1
, l
1
) = 0, γ(l
1
, l
2
) = γ(l
2
, l
1
)
and γ(l
1
, l
3
) ≤ γ(l
1
, l
2
) + γ(l
2
, l
3
). In particular, we
sometimes use a unit cost function such that γ(l
1
, l
2
) =
1 if l
1
6= l
2
. The cost of an alignment T , denoted by
γ(T ), is the sum of the costs of all labels in T .
Definition 3 (Alignment Distance (Jiang et al.,
1995)). Let π ∈ {o, b, c, cb, u}. Then, the alignment
distance between T
1
and T
2
is defined as the mini-
mum cost γ(T ) for every alignment T ∈ A
π
(T
1
, T
2
).
Also we call an alignment with the minimum cost an
optimal alignment.
Example 1. Consider ordered trees T
1
and T
2
in Fig-
ure 1 (left). Then, T in Figure 1 (right) is the opti-
mal alignment between T
1
and T
2
. Under the unit cost
function γ, since γ(T ) = 4, the alignment distance be-
tween T
1
and T
2
is 4.
T
1
T
2
T
Figure 1: Ordered trees T
1
and T
2
(left) and the optimal
alignment T ∈ A
o
(T
1
, T
2
) (right) in Example 1.
3 MAPPING AND DISTANCE
In this section, we introduce a Tai mapping and its
variations, and then the distance as the minimum cost
of all the mappings.
Definition 4 (Tai Mapping (Tai, 1979)). Let T
1
and
T
2
be trees and M ⊆ V(T
1
) ×V(T
2
).
1. We say that a triple (M, T
1
, T
2
) is an ordered
Tai mapping from T
1
to T
2
, denoted by M ∈
M
o
TAI
(T
1
, T
2
), if every pair (u
1
, v
1
) and (u
2
, v
2
) in
M satisfies the following conditions.
(i) u
1
= u
2
iff v
1
= v
2
(one-to-one condition).
(ii) u
1
≤ u
2
iff v
1
≤ v
2
(ancestor condition).
(iii) u
1
u
2
iff v
1
v
2
(sibling condition).
2. We say that a triple (M, T
1
, T
2
) is an unordered
Tai mapping from T
1
to T
2
, denoted by M ∈
M
u
TAI
(T
1
, T
2
), if M satisfies the conditions (i) and
(ii).
In the following, let u
1
, u
2
, u
3
, u
4
∈ ch(u) and
v
1
, v
2
, v
3
, v
4
∈ ch(v).
3. We say that a triple (M, T
1
, T
2
) is a biordered
Tai mapping from T
1
to T
2
, denoted by M ∈
M
b
TAI
(T
1
, T
2
), if M satisfies the above conditions
(i) and (ii) and the following condition (iv).
(iv) For every u ∈ T
1
and v ∈ T
2
such that
(u
1
, v
1
), (u
2
, v
2
), (u
3
, v
3
) ∈ M, one of the fol-
lowing statements holds.
1. u
1
u
2
u
3
iff v
1
v
2
v
3
.
AlignmentofCyclicallyOrderedTrees
265

2. u
1
u
2
u
3
iff v
3
v
2
v
1
.
4. We say that a triple (M, T
1
, T
2
) is a cyclic-ordered
Tai mapping from T
1
to T
2
, denoted by M ∈
M
c
TAI
(T
1
, T
2
), if M satisfies the above conditions
(i) and (ii) and the following condition (v).
(v) For every u ∈ T
1
and v ∈ T
2
such that
(u
1
, v
1
), (u
2
, v
2
), (u
3
, v
3
) ∈ M, one of the fol-
lowing statements holds.
1. u
1
u
2
u
3
iff v
1
v
2
v
3
.
2. u
1
u
2
u
3
iff v
2
v
3
v
1
.
3. u
1
u
2
u
3
iff v
3
v
1
v
2
.
5. We say that a triple (M, T
1
, T
2
) is a cyclic-
biordered Tai mapping from T
1
to T
2
, denoted by
M ∈ M
cb
TAI
(T
1
, T
2
), if M satisfies the above condi-
tions (i) and (ii) and the following condition (vi).
(vi) For every u ∈ T
1
and v ∈ T
2
such that
(u
1
, v
1
), (u
2
, v
2
), (u
3
, v
3
), (u
4
, v
4
) ∈ M, one of
the following statements holds.
1. u
1
u
2
u
3
u
4
iff v
1
v
2
v
3
v
4
.
2. u
1
u
2
u
3
u
4
iff v
2
v
3
v
4
v
1
.
3. u
1
u
2
u
3
u
4
iff v
3
v
4
v
1
v
2
.
4. u
1
u
2
u
3
u
4
iff v
4
v
1
v
2
v
3
.
5. u
1
u
2
u
3
u
4
iff v
4
v
3
v
2
v
1
.
6. u
1
u
2
u
3
u
4
iff v
3
v
2
v
1
v
4
.
7. u
1
u
2
u
3
u
4
iff v
2
v
1
v
4
v
3
.
8. u
1
u
2
u
3
u
4
iff v
1
v
4
v
3
v
2
.
We will use M instead of (M, T
1
, T
2
) simply.
Since a less-constrained mapping (Lu et al., 2001)
coincides with an alignable mapping (Kuboyama,
2007) characterizing the alignment, we formulate the
alignable mapping as the less-constrained mapping.
Definition 5 (Variations of Tai Mapping). Let T
1
and
T
2
be trees, π ∈ {o, b, c, cb, u} and M ∈ M
π
TAI
(T
1
, T
2
).
Here, we denote M− {(r(T
1
), r(T
2
))} by M
−
.
1. We say that M is an alignable map-
ping (Kuboyama, 2007) (or a less-constrained
mapping (Lu et al., 2001)), denoted by
M ∈ M
π
ALN
(T
1
, T
2
), if M satisfies the follow-
ing condition.
∀(u
1
, v
1
), (u
2
, v
2
), (u
3
, v
3
) ∈ M
u
1
⊔ u
2
< u
1
⊔ u
3
=⇒ v
2
⊔ v
3
= v
1
⊔ v
3
.
2. We say that M is a segmental mapping (Kan et al.,
2014), denoted by M ∈ M
π
SG
(T
1
, T
2
), if M satisfies
the following condition.
∀(u, v) ∈ M
−
∃(u
′
, v
′
) ∈ M
(u
′
∈ anc(u)) ∧ (v
′
∈ anc(v))
=⇒
(par(u), par(v)) ∈ M
.
3. We say that M is a segmental alignable map-
ping (Yoshino and Hirata, 2013), denoted by
M ∈ M
π
SGALN
(T
1
, T
2
), if M ∈ M
π
SG
(T
1
, T
2
) ∩
M
π
ALN
(T
1
, T
2
).
4. We say that M is a top-down mapping (Selkow,
1977; Chawathe, 1999) (or a degree-1 mapping),
denoted by M ∈ M
π
TOP
(T
1
, T
2
), if M satisfies the
following condition.
∀(u, v) ∈ M
−
(par(u), par(v)) ∈ M
.
Let M be a mapping from T
1
to T
2
. Let I and J be
the sets of nodes in T
1
and T
2
but not in M. Then, the
cost γ(M) of M is given as follows.
γ(M) =
∑
(u,v)∈M
γ(u, v) +
∑
u∈I
γ(u, ε)+
∑
v∈J
γ(ε, v).
Definition 6 (Variations of Edit Distance). For every
A ∈ {TAI, ALN, SGALN, TOP} and π ∈ { o, b, c, cb, u},
we define the distance τ
π
A
(T
1
, T
2
) as follows.
τ
π
A
(T
1
, T
2
) = min{γ(M) | M ∈ M
π
A
(T
1
, T
2
)}.
Theorem 1. Let T
1
and T
2
be trees and π ∈
{o, b, c, cb, u}.
1. τ
π
TAI
(T
1
, T
2
) coincides with the edit distance (Tai,
1979).
2. τ
π
ALN
(T
1
, T
2
) coincides with the alignment dis-
tance (Kuboyama, 2007).
Theorem 2. Let T
1
and T
2
be trees such that n =
|T
1
| ≥ |T
2
| = m and D = max{d(T
1
), d(T
2
)}.
1. We can compute τ
o
TAI
(T
1
, T
2
) in O(nm
2
(1 +
log
n
m
)) = O(n
3
) time. On the other hand, the
problem of computing τ
u
TAI
(T
1
, T
2
) is MAX SNP-
hard, even if T
1
and T
2
are binary (Demaine
et al., 2009; Zhang and Jiang, 1994; Hirata et al.,
2011).
2. We can compute τ
o
ALN
(T
1
, T
2
) and τ
o
SGALN
(T
1
, T
2
)
in O(nmD
2
) time. On the other hand, the prob-
lem of computing τ
u
ALN
(T
1
, T
2
) and τ
u
SGALN
(T
1
, T
2
)
is MAX SNP-hard, but it is tractable if the de-
grees of T
1
and T
2
are bounded (Jiang et al., 1995;
Yoshino and Hirata, 2013).
Proposition 1 (cf. (Kuboyama, 2007; Yoshino and
Hirata, 2013)). Let T
1
and T
2
be trees and π ∈
{o, b, c, cb, u}. Also suppose that a cost function is
a metric. Then, τ
π
TAI
(T
1
, T
2
) and τ
π
TOP
(T
1
, T
2
) are met-
rics, whereas neither τ
π
ALN
(T
1
, T
2
) nor τ
π
SGALN
(T
1
, T
2
)
is a metric.
Proposition 2. Let T
1
and T
2
be trees. For A ∈
{TAI, ALN, SGALN, TOP} and π ∈ {o, b, c, cb, u} , the
following statements hold.
1. τ
u
A
(T
1
, T
2
) ≤ τ
cb
A
(T
1
, T
2
) ≤ τ
b
A
(T
1
, T
2
) ≤ τ
o
A
(T
1
, T
2
).
2. τ
u
A
(T
1
, T
2
) ≤ τ
cb
A
(T
1
, T
2
) ≤ τ
c
A
(T
1
, T
2
) ≤ τ
o
A
(T
1
, T
2
).
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
266

3. τ
π
TAI
(T
1
, T
2
) ≤ τ
π
ALN
(T
1
, T
2
) ≤ τ
π
SGALN
(T
1
, T
2
) ≤
τ
π
TOP
(T
1
, T
2
).
Proposition 3. For A ∈ {TAI, ALN, SGALN, TOP},
there exist trees T
1
and T
2
satisfying each of the fol-
lowing conditions.
1. τ
c
A
(T
1
, T
2
) < τ
b
A
(T
1
, T
2
).
2. τ
b
A
(T
1
, T
2
) < τ
c
A
(T
1
, T
2
).
Proof. Consider the following trees T
1
, T
2
and T
3
.
T
1
T
2
T
3
Under the unit cost function, Statement 1 follows
that τ
b
TOP
(T
1
, T
2
) = 1 < 3 = τ
c
TOP
(T
1
, T
2
) and Statement
2 followsthat τ
c
TOP
(T
1
, T
3
) = 1 < 3 = τ
b
TOP
(T
1
, T
2
).
Proposition 4. Let T
1
and T
2
be trees and A ∈
{TAI, ALN, SGALN, TOP}.
1. If max{d(T
1
), d(T
2
)} ≤ 1, then it holds
that τ
o
A
(T
1
, T
2
) = τ
b
A
(T
1
, T
2
) = τ
c
A
(T
1
, T
2
) =
τ
cb
A
(T
1
, T
2
) = τ
u
A
(T
1
, T
2
).
2. If max{d(T
1
), d(T
2
)} ≤ 2, then it holds that
τ
b
A
(T
1
, T
2
) = τ
c
A
(T
1
, T
2
) = τ
cb
A
(T
1
, T
2
) = τ
u
A
(T
1
, T
2
).
3. If max{d(T
1
), d(T
2
)} ≤ 3, then it holds that
τ
cb
A
(T
1
, T
2
) = τ
u
A
(T
1
, T
2
).
Proposition 5. For π ∈ {b, c, cb}, there exist trees T
1
and T
2
satisfying each of the following conditions.
1. τ
o
TAI
(T
1
, T
2
) < τ
π
ALN
(T
1
, T
2
).
2. τ
π
ALN
(T
1
, T
2
) < τ
o
TAI
(T
1
, T
2
).
Proof. Consider the following trees T
1
, T
2
and T
3
and
suppose that a cost function is the unit cost function.
T
1
T
2
T
3
T
12
T
13
1. It is obvious that τ
o
TAI
(T
1
, T
2
) = 2. On the other
hand, since the alignment T
12
is an optimal alignment
between T
1
and T
2
for cyclically ordered trees, it holds
that τ
b
ALN
(T
1
, T
2
) = τ
c
ALN
(T
1
, T
2
) = τ
cb
ALN
(T
1
, T
2
) = 3.
Note that τ
o
ALN
(T
1
, T
2
) = 4 (Jiang et al., 1995).
2. Since the alignment T
13
is an optimal
alignment between T
1
and T
3
for cyclically ordered
trees, it holds that τ
b
ALN
(T
1
, T
3
) = τ
c
ALN
(T
1
, T
3
) =
τ
cb
ALN
(T
1
, T
3
) = 1. On the other hand, it is obvious that
τ
o
TAI
(T
1
, T
3
) = 4. Note that τ
o
ALN
(T
1
, T
2
) = 5.
4 ALGORITHMS
In this section, we identify a node with its pos-
torder number. Also let n = |T
1
|, m = |T
2
| (and sup-
pose that n ≥ m), d = min{d(T
1
), d(T
2
)} and D =
max{d(T
1
), d(T
2
)}.
A(n ordered) forest is a sequence [T
1
, . . . , T
n
] of
trees. For a tree T and a node i ∈ T, T(i) is a
forest obtained by deleting the root i in T[i]. For
nodes i ∈ T
1
and j ∈ T
2
, let the children of i and
j be i
1
, . . . , i
s
and j
1
, . . . , j
t
. That is, it holds that
d(i) = s and d( j) = t. Also, for trees T
1
and T
2
,
we denote the forests T
1
(i) = [T
1
[i
1
], . . . , T
1
[i
s
]] and
T
2
( j) = [T
2
[ j
1
], . . . , T
2
[ j
t
]] by F
1
(i
1
, i
s
) and F
2
( j
1
, j
t
).
For A ∈ {ALN, SGALN, TOP} andπ ∈ {o, b, c, cb},
the recurrences in Figure 2 compute the distance τ
π
A
and the forest distance δ
π
A
when containing an empty
tree or forest. Also Figure 3 illustrates the common
recurrences Γ
π
A
and ∆
π
A
to compute τ
π
A
and δ
π
A
.
δ
π
A
(
/
0,
/
0) = 0,
τ
π
A
(T
1
[i],
/
0) = δ
π
A
(T
1
(i),
/
0) + γ(i, ε),
τ
π
A
(
/
0, T
2
[ j]) = δ
π
A
(
/
0, T
2
( j)) + γ(ε, j),
δ
π
A
(T
1
(i),
/
0) =
s
∑
k=1
τ
π
A
(T
1
[i
k
],
/
0),
δ
π
A
(
/
0, T
2
( j)) =
t
∑
k=1
τ
π
A
(
/
0, T
2
[ j
k
]).
Figure 2: The basic recurrences of computing τ
π
A
(T
1
, T
2
).
Γ
π
A
(T
1
[i], T
2
[ j])
= min
τ
π
A
(T
1
[i],
/
0) + min
T∈T
1
(i)
{τ
π
A
(T, T
2
[ j]) − τ
π
A
(T,
/
0)},
τ
π
A
(
/
0, T
2
[ j]) + min
T∈T
2
( j)
{τ
π
A
(T
1
[i], T) − τ
π
A
(
/
0, T)}
,
∆
π
A
(F
1
(i
1
, i
s
), F
2
( j
1
, j
t
))
= min
δ
π
A
(F
1
(i
1
, i
s−1
), F
2
( j
1
, j
t
)) + τ
π
A
(T
1
[i
s
],
/
0),
δ
π
A
(F
1
(i
1
, i
s
), F
2
( j
1
, j
t−1
)) + τ
π
A
(
/
0, T
2
[ j
t
]),
δ
π
A
(F
1
(i
1
, i
s−1
), F
2
( j
1
, j
t−1
)) + τ
π
A
(T
1
[i
s
], T
2
[ j
t
])
.
Figure 3: The common recurrences Γ
π
A
(T
1
, T
2
) and
∆
π
A
(F
1
, F
2
).
Let T
1
(i) = [T
1
[i
1
], . . . , T
1
[i
s
]] and T
2
( j) =
[T
2
[ j
1
], . . . , T
2
[ j
t
]]. Also let 1 ≤ p ≤ s and 1 ≤ q ≤ t.
Then, we denote the forests [T
1
[i
σ
+
p,s
(1)
], . . . , T
1
[i
σ
+
p,s
(s)
]]
and [T
2
[ j
σ
+
q,t
(1)
], . . . , T
2
[ j
σ
+
q,t
(t)
]] by T
p
1
(i) and T
q
2
( j).
Also we denote the forests [T
1
[i
σ
−
p,s
(1)
], . . . , T
1
[i
σ
−
p,s
(s)
]]
and [T
2
[ j
σ
−
q,t
(1)
], . . . , T
2
[ j
σ
−
q,t
(t)
]] by T
−p
1
(i) and T
−q
2
( j).
It is obvious that T
1
(i) = T
1
1
(i) and T
2
( j) = T
1
2
( j).
AlignmentofCyclicallyOrderedTrees
267

Furthermore, the values of p and q in T
p
1
(i),
T
p
1
(i
s
), T
q
2
( j) and T
q
2
( j
t
) are (1) p = q = 1 if π = o,
(2) p = ±1 and q = ±1 if π = b, (3) 1 ≤ p ≤ s and
1 ≤ q ≤ t if π = c and (4) 1 ≤ p ≤ s, −s ≤ p ≤ −1,
1 ≤ q ≤ t and −t ≤ q ≤ −1 if π = cb. Hence, we
prepare the following sets: (1) o(s) = o(t) = {1} , (2)
b(s) = b(t) = {−1, 1}, (3) c(s) = {1, . . . , s}, c(t) =
{1, . . . ,t}, and (4) cb(s) = {−s, . . . , −1, 1, . . . , s},
cb(t) = {−t, . . . , −1, 1, . . .,t}. We refer these sets to
π(s) and π(t) for π ∈ {o, b, c, cb}.
Then, by introducing the sets π(s) and π(t) into
the recurrences in (Jiang et al., 1995), we design the
recurrences of computing τ
π
ALN
(T
1
, T
2
) between cycli-
cally ordered trees T
1
and T
2
as Figure 4.
τ
π
ALN
(T
1
[i], T
2
[ j])
= min
(
min
p∈π(s),q∈π(t)
{δ
π
ALN
(T
p
1
(i), T
q
2
( j)) + γ(i, j)},
Γ
π
ALN
(T
1
[i], T
2
[ j])
)
,
δ
π
ALN
(F
1
(i
1
, i
s
), F
2
( j
1
, j
t
)) = min
∆
π
ALN
(F
1
(i
1
, i
s
), F
2
( j
1
, j
t
)),
γ(i
s
, ε)
+ min
1≤k<t,p∈π(s)
δ
π
ALN
(F
1
(i
1
, i
s−1
), F
2
( j
1
, j
k−1
))
+δ
π
ALN
(T
p
1
(i
s
), F
2
( j
k
, j
t
)))
,
γ(ε, j
t
)
+ min
1≤k<s,q∈π(t)
δ
π
ALN
(F
1
(i
1
, i
k−1
), F
2
( j
1
, j
t−1
))
+δ
π
ALN
(F
1
(i
k
, i
s
), T
q
2
( j
t
)))
.
Figure 4: The recurrences of computing τ
π
ALN
(T
1
, T
2
) be-
tween cyclically ordered trees.
Theorem 3. The recurrences in Figure 4 are correct
to compute τ
π
ALN
(T
1
, T
2
) between cyclically ordered
trees T
1
and T
2
for π ∈ {b, c, cb}.
Proof. In the proof of (Jiang et al., 1995) showing
that the recurrences of computing τ
o
ALN
(T
1
, T
2
) is cor-
rect, the formulas and the cases of an optimal align-
ment tree or forest T are presented as follows.
1. The formula δ
o
ALN
(T
1
(i), T
2
( j)) + γ(i, j) is corre-
sponding to the case that (i, j) is a label in an op-
timal alignment tree T of T
1
[i] and T
2
[ j], and T
contains the alignment of T
1
(i) and T
2
( j).
2. The formula
γ(i
s
, ε) + min
1≤k<t
δ
o
ALN
(F
1
(i
1
, i
s−1
), F
2
( j
1
, j
k−1
))
+δ
o
ALN
(T
1
(i
s
), F
2
( j
k
, j
t
)))
is corresponding to the case that (i
s
, ε) is a label
in an optimal alignment forest T of F
1
(i
1
, i
s
) and
F
2
( j
1
, j
t
), and T contains the alignment of T
1
(i
s
)
and F
2
( j
k
, j
t
) for 1 ≤ k < t.
3. The formula
γ(ε, j
t
) + min
1≤k<s
δ
o
ALN
(F
1
(i
1
, i
k−1
), F
2
( j
1
, j
t−1
))
+δ
o
ALN
(F
1
(i
k
, i
s
), T
2
( j
t
)))
is corresponding to the case that (ε, j
t
) is a la-
bel in an optimal alignment forest T of F
1
(i
1
, i
s
)
and F
2
( j
1
, j
t
), and T contains the alignment of
F
1
(i
k
, i
s
) and T
2
( j
t
) for 1 ≤ k < s.
In just above three formulas, an optimal alignment
T contains and expands the siblings of some node in
T
1
or T
2
(or both). When extending from τ
o
ALN
(T
1
, T
2
)
to τ
π
ALN
(T
1
, T
2
), it is sufficient to deal with more than
two orders in the above three formulas, instead of one
left-to-right order, and then to replace T
1
(i), T
2
( j),
T
1
(i
s
) and T
2
( j
t
) with T
p
1
(i), T
q
2
( j), T
p
1
(i
s
) and T
q
2
( j
t
)
for p ∈ π(s) and q ∈ π(t). Hence, by replacing the for-
mulas in the above statements 1, 2 and 3 with the first
formula in τ
π
ALN
(F
1
(i
1
, i
s
), F
2
( j
1
, j
t
)) and the second
and the third formulas in δ
π
ALN
(F
1
(i
1
, i
s
), F
2
( j
1
, j
t
)) in
Figure 4, we can compute τ
π
ALN
(T
1
, T
2
) correctly.
Theorem 4. We can compute τ
b
ALN
(T
1
, T
2
) in
O(nmD
2
) time. Also we can compute τ
c
ALN
(T
1
, T
2
) and
τ
cb
ALN
(T
1
, T
2
) in O(nmdD
3
) time.
Proof. In Figure 4, the number of recurrences in τ
o
ALN
is 3 and one in δ
o
ALN
is 5; the number of recurrences
in τ
b
ALN
is 6 and one in δ
b
ALN
is 7; the number of re-
currences in τ
c
ALN
is d(i)d( j) + 2 and one in δ
c
ALN
is
d(i) + d( j) + 3; the number of recurrences in τ
cb
ALN
is
4d(i)d( j) + 2 and one in δ
cb
ALN
is 2d(i) + 2d( j) + 3.
According to the proof of (Jiang et al., 1995),
for s = d(i) and t = d( j), we can compute
δ
o
ALN
(F
1
(i
s
′
, i
s
), F
2
( j
t
′
, j
t
)) in O((s − s
′
) × (t − t
′
) ×
((s− s
′
) + (t −t
′
))) = O(d(i)d( j)(d(i) + d( j))) time.
Then, we can compute δ
b
ALN
(F
1
(i
s
′
, i
s
), F
2
( j
t
′
, j
t
)) in
O(d(i)d( j)(d(i) + d( j))) time. So the running time
of computing τ
b
ALN
(T
1
[i], T
2
[ j]) for each (i, j) ∈ T
1
×
T
2
is O(d(i)d( j)(d(i) + d( j))d(i) + d(i)d( j)(d(i) +
d( j))d( j)) = O(d(i)d( j)(d(i) + d( j))
2
). Hence, the
running time of computing τ
b
ALN
(T
1
, T
2
) is:
|T
1
|
∑
i=1
|T
2
|
∑
j=1
O
d(i)d( j)(d(i) + d( j))
2
≤
|T
1
|
∑
i=1
|T
2
|
∑
j=1
O
d(i)d( j)(d(T
1
) + d(T
2
))
2
≤ O
(d(T
1
) + d(T
2
))
2
×
|T
1
|
∑
i=1
d(i) ×
|T
2
|
∑
j=1
d( j)
≤ O(|T
1
| × |T
2
| × (d(T
1
) + d(T
2
))
2
)
= O(nmD
2
).
Also, by focusing on the number of recurrences in
Figure 4, we can compute δ
c
ALN
(F
1
(i
s
′
, i
s
), F
2
( j
t
′
, j
t
))
and δ
cb
ALN
(F
1
(i
s
′
, i
s
), F
2
( j
t
′
, j
t
)) in O((s −
s
′
)d( j) × (t − t
′
)d(i) × ((s − s
′
) + (t − t
′
))) =
O(d(i)
2
d( j)
2
(d(i) + d( j))) time. So the run-
ning time of computing τ
c
ALN
(T
1
[i], T
2
[ j]) and
τ
cb
ALN
(T
1
[i], T
2
[ j]) for each (i, j) ∈ T
1
× T
2
is
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
268
O(d(i)
2
d( j)
2
(d(i) + d( j))d(i) + d(i)
2
d( j)
2
(d(i) +
d( j))d( j)+d(i)d( j)) = O(d(i)
2
d( j)
2
(d(i)+d( j))
2
),
where the last formula d(i)d( j) is corresponding to
the time complexity of computing the first recurrence
in τ
π
ALN
(T
1
[i], T
2
[ j]) in Figure 4. Hence, the running
time of computing τ
c
ALN
(T
1
, T
2
) and τ
cb
ALN
(T
1
, T
2
) is:
|T
1
|
∑
i=1
|T
2
|
∑
j=1
O
d(i)
2
d( j)
2
(d(i) + d( j))
2
≤
|T
1
|
∑
i=1
|T
2
|
∑
j=1
O
d(i)d( j)d(T
1
)d(T
2
)
×(d(T
1
) + d(T
2
)
2
)
≤ O
d(T
1
)d(T
2
)(d(T
1
) + d(T
2
))
2
×
|T
1
|
∑
i=1
d(i) ×
|T
2
|
∑
j=1
d( j)
≤ O(|T
1
| × |T
2
| × d(T
1
)d(T
2
)(d(T
1
) + d(T
2
))
2
)
= O(nmdD
3
).
Next, we design the algorithm to compute the
segmental alignment distance τ
π
SGALN
(T
1
, T
2
) for π ∈
{b, c, cb}. Here, we adopt the same strategy of (Kan
et al., 2014) to compute τ
π
TOP
(T
1
[i], T
2
[ j]) between
T
1
[i] and T
2
[ j] for every pair (i, j) ∈ T
1
× T
2
(1 ≤ i ≤
n, 1 ≤ j ≤ m) in advance.
Then, Figure 5 illustrates the recurrences of com-
puting τ
π
SGALN
(T
1
, T
2
) for cyclically ordered trees.
Here, δ
π
SGALN
(F
1
, F
2
) is same as δ
π
ALN
(F
1
, F
2
) by re-
placing the subscript ALN with SGALN.
τ
π
TOP
(T
1
[i], T
2
[ j])
= min
p∈π(s),q∈π(s)
{δ
π
TOP
(T
p
1
(i), T
q
2
( j)) + γ(i, j)},
δ
π
TOP
(F
1
(i
1
, i
s
), F
2
( j
1
, j
t
)) = ∆
π
TOP
(F
1
(i
1
, i
s
), F
2
( j
1
, j
t
)).
τ
π
SGALN
(T
1
[i], T
2
[ j])
= min
τ
π
TOP
(T
1
[i], T
2
[ j])
·· ·use the value computed in advance,
min
p∈π(s),q∈π(t)
{δ
b
SGALN
(T
p
1
(i), T
q
2
( j)) + γ(i, j)},
Γ
π
SGALN
(T
1
[i], T
2
[ j])
,
δ
π
SGALN
(F
1
(i
1
, i
s
), F
2
( j
1
, j
t
))
·· ·same as δ
π
ALN
with replacing ALN with SGALN.
Figure 5: The recurrence of computing τ
π
SGALN
(T
1
, T
2
) be-
tween cyclically ordered trees.
Theorem 5. The recurrences in Figure 5 are cor-
rect to compute τ
b
SGALN
(T
1
, T
2
) in O(nmD
2
) time, and
τ
c
SGALN
(T
1
, T
2
) and τ
cb
SGALN
(T
1
, T
2
) in O(nmdD
3
) time.
Proof. The correctness follows from Theorem 3 and
(Yoshino and Hirata, 2013). Since the number of re-
currences in τ
b
TOP
in Figure 5 is O(1) and one in τ
c
TOP
and τ
cb
TOP
is O(dD), we can compute τ
b
TOP
(T
1
[i], T
2
[ j])
in O(nm) time and compute τ
c
TOP
(T
1
[i], T
2
[ j]) and
τ
cb
TOP
(T
1
[i], T
2
[ j]) in O(nmdD) time for every pair
(i, j) ∈ T
1
× T
2
. Hence, by Theorem 4, the run-
ning time of computing τ
b
SGALN
(T
1
, T
2
) is O(nm) +
O(nmD
2
) = O(nmD
2
). Also, the running time
of computing τ
c
SGALN
(T
1
, T
2
) and τ
cb
SGALN
(T
1
, T
2
) is
O(nmdD) + O(nmdD
3
) = O(nmdD
3
).
5 EXPERIMENTAL RESULTS
In this section, we give experimental results for τ
b
ALN
comparingwith τ
o
TAI
, by using N-glycan data provided
from KEGG. Here, the number of N-glycan data is
2142, the average number of nodes is 11.09, the av-
erage number of labels is 5.43 and the average depth
and degree are 5.38 and 2.07, respectively.
Figure 6: The correlation diagrams to the edit distance τ
o
TAI
of τ
b
ALN
for N-glycan data.
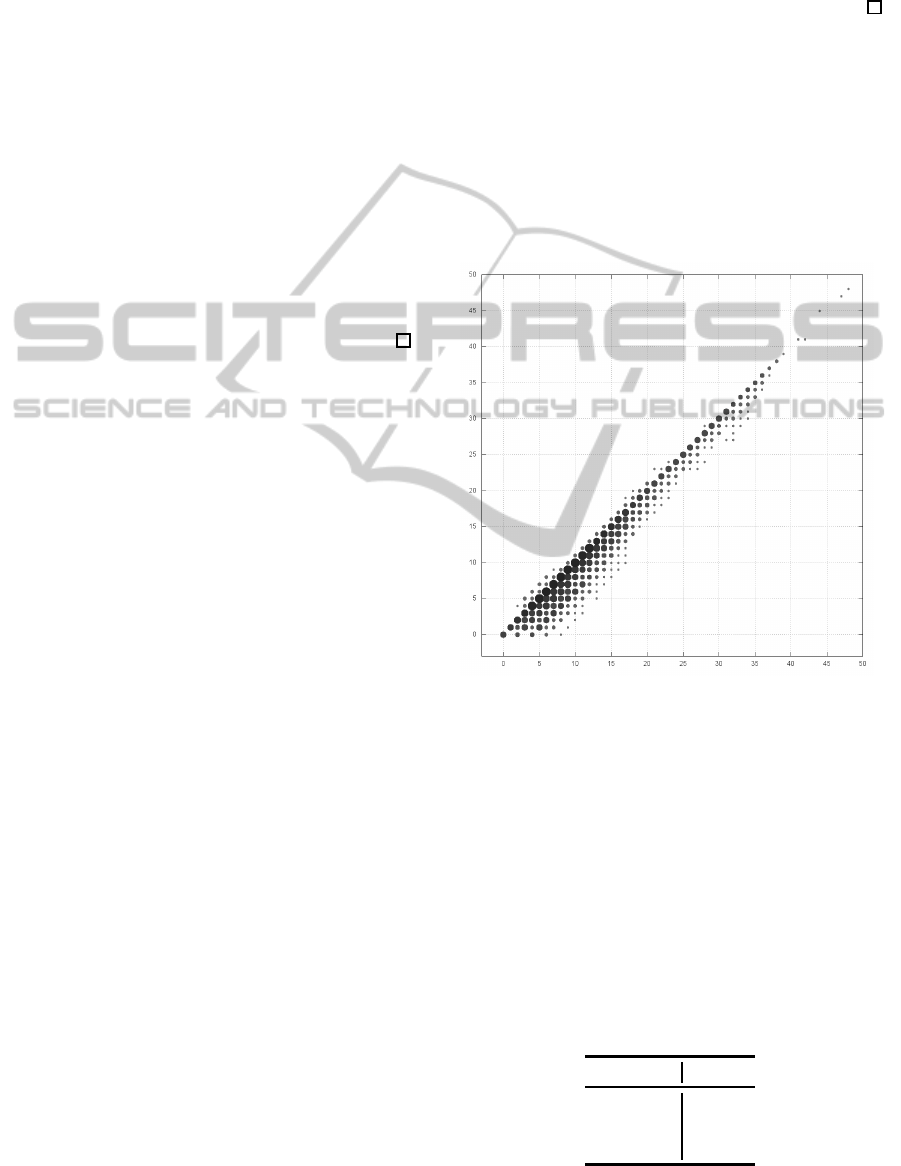
Figures 6 illustrates the correlation diagrams to
τ
o
TAI
of τ
b
ALN
for all the 2293011 pairs of N-glycan
data. The plots in Figures 6 are the ratio (%) of the
pairs of trees whose value of τ
b
ALN
is given as the y-
axis to the value of τ
o
TAI
given in the x-axis.
Figures 6 shows that, for N-glycan data, whereas
τ
b
ALN
tends to be smaller than τ
o
TAI
, we can observe the
pairs that τ
b
ALN
is greater than τ
o
TAI
as Proposition 5.
Table 1 represents the number of pairs comparing
τ
b
ALN
with τ
o
TAI
in all the pairs of N-glycan data.
Table 1: The number of pairs comparing τ
b
ALN
with τ
o
TAI
.
case #pairs
τ
b
ALN
> τ
o
TAI
675
τ
b
ALN
= τ
o
TAI
1193559
τ
b
ALN
< τ
o
TAI
298777
AlignmentofCyclicallyOrderedTrees
269

Hence, we conclude that, τ
b
ALN
≤ τ
o
TAI
for al-
most pairs of N-glycan data; Only 675 pairs (about
0.029%) satisfies that τ
b
ALN
> τ
o
TAI
. This result implies
that τ
b
ALN
(and τ
cb
ALN
) is possible to be a good approxi-
mation of τ
u
TAI
for N-glycan data.
6 CONCLUSION
In this paper, we have formulated biordered, cyclic-
ordered and cyclic-biordered trees as cyclically or-
dered trees, and then designed the algorithms to com-
pute τ
b
ALN
(T
1
, T
2
) and τ
b
SGALN
(T
1
, T
2
) in O(nmD
2
) time
and to compute τ
π
ALN
(T
1
, T
2
) and τ
π
SGALN
(T
1
, T
2
) (π ∈
{c, cb}) in O(nmdD
3
) time. Finally, we have given
the experimental results of computing τ
b
ALN
compar-
ing with τ
o
TAI
by using N-glycan data.
It is a future work to implement the algo-
rithms to compute τ
c
ALN
, τ
cb
ALN
and τ
π
SGALN
(π ∈
{b, c, cb}), and apply τ
π
ALN
and τ
π
SGALN
to real data
such as glycans (Hizukuri et al., 2005) or molecular
graphs (Horv´ath et al., 2010). Also, it is a future work
to apply cyclically ordered trees to compare RNA sec-
ondary structures (H¨ochsmann et al., 2003; Schier-
mer and Giegerich, 2013; Shapiro and Zhang, 1990;
Zhang, 1998).
As the comparison with τ
u
TAI
, it is a future work to
investigate how τ
π
ALN
(π ∈ { b, c, cb}) is a good approx-
imation of τ
u
TAI
and to compare τ
π
ALN
with tractable
variations of τ
u
TAI
such as the isolated-subtree dis-
tance (Zhang, 1996) and the LCA-preserving dis-
tance (Zhang et al., 1996). Also, it is a future work to
solve whether or not the problem of computing τ
u
ALN
is tractable if the number of permutations among sib-
lings is bounded by some polynomial with respect to
degrees.
REFERENCES
Chawathe, S. S. (1999). Comparing hierarchical data in ex-
ternal memory. In Proc. VLDB’99, pages 90–101.
Demaine, E. D., Mozes, S., Rossman, B., and Weimann, O.
(2009). An optimal decomposition algorithm for tree
edit distance. ACM Trans. Algo., 6.
Hirata, K., Yamamoto, Y., and Kuboyama, T. (2011). Im-
proved MAX SNP-hard results for finding an edit dis-
tance between unordered trees. In Proc. CPM’11
(LNCS 6661), pages 402–415.
Hizukuri, Y., Yamanishi, T., Nakamura, O., Yagi, F., Goto,
S., and Kanehisa, M. (2005). Extraction of leukemia
specific glycan motifs in humans by computational
glycomics. Carbohydrate Research, 340:2270–2278.
H¨ochsmann, M., T¨oller, T., Giegerich, R., and Kurtz, S.
(2003). Local similarity in RNA secondary structures.
In Proc. CSB’03, pages 159–168.
Horv´ath, T., Ramon, J., and Wrobel, S. (2010). Frequent
subgraph mining in outerplanar graphs. Data Min.
Knowl. Disc., 21:472–508.
Jiang, T., Wang, L., and Zhang, K. (1995). Alignment of
trees – an alternative to tree edit. Theoret. Comput.
Sci., 143:137–148.
Kan, T., Higuchi, S., and Hirata, K. (2014). Segmental
mapping and distance for rooted ordered labeled trees.
Fundamenta Informaticae, 132:1–23.
Kuboyama, T. (2007). Matching and learning in trees. Ph.D
thesis, University of Tokyo.
Lu, C. L., Su, Z.-Y., and Yang, C. Y. (2001). A new mea-
sure of edit distance between labeled trees. In Proc.
COCOON’01 (LNCS 2108), pages 338–348.
Lu, S.-Y. (1979). A tree-to-tree distance and its application
to cluster analysis. IEEE Trans. Pattern Anal. Mach.
Intell., 1:219–224.
Schiermer, S. and Giegerich, R. (2013). Forest alignment
with affine gaps and anchors, applied in RNA struc-
ture comparison. Theoret. Comput. Sci., 483:51–67.
Selkow, S. M. (1977). The tree-to-tree editing problem. In-
form. Process. Lett., 6:184–186.
Shapiro, B. A. and Zhang, K. (1990). Comparing multi-
ple RNA secondary structures using tree comparision.
Comp. Appl. Biosci., 6:309–318.
Tai, K.-C. (1979). The tree-to-tree correction problem. J.
ACM, 26:422–433.
Valiente, G. (2001). An efficient bottom-up distance be-
tween trees. In Proc. SPIRE’01, pages 212–219.
Wang, Y., DeWitt, D. J., and Cai, J.-Y. (2003). X-Diff: An
effective change detection algorithm for XML docu-
ments. In Proc. ICDE’03, pages 519–530.
Yamamoto, Y., Hirata, K., and Kuboyama, T. (2014).
Tractable and intractable variations of unordered tree
edit distance. Internat. J. Found. Comput. Sci.,
25:307–329.
Yoshino, T. and Hirata, K. (2013). Hierarchy of segmen-
tal and alignable mapping for rooted labeled trees. In
Proc. DDS’13, pages 62–69.
Zhang, K. (1995). Algorithms for the constrained edit-
ing distance between ordered labeled trees and related
problems. Pattern Recog., 28:463–474.
Zhang, K. (1996). A constrained edit distance between un-
ordered labeled trees. Algorithmica, 15:205–222.
Zhang, K. (1998). Computing similarity between RNA sec-
ondary structures. In Proc. IEEE Internat. Joint Symp.
Intell. Sys., pages 126–132.
Zhang, K. and Jiang, T. (1994). Some MAX SNP-hard re-
sults concerning unordered labeled trees. Inform. Pro-
cess. Lett., 49:249–254.
Zhang, K., Wang, J., and Shasha, D. (1996). On the editing
distance between undirected acyclic graphs. Internat.
J. Found. Comput. Sci., 7:43–58.
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
270
