
Machine Reading of Biological Texts
Bacteria-Biotope Extraction
Wouter Massa
1
, Parisa Kordjamshidi
1,2
, Thomas Provoost
1
and Marie-Francine Moens
1
1
Department of Computer Science, KU Leuven, Celestijnenlaan 200A, 3001, Heverlee, Belgium
2
Department of Computer Science, University of Illinois at Urbana-Champaign,
201 North Goodwin Avenue, 61801-2302, Urbana, IL, U.S.A.
Keywords:
Natural Language Processing, Text Mining, Relation Extraction, BioNLP, Bioinformatics, Bacteria, Bacteria
Biotopes.
Abstract:
The tremendous amount of scientific literature available about bacteria and their biotopes underlines the need
for efficient mechanisms to automatically extract this information. This paper presents a system to extract
the bacteria and their habitats, as well as the relations between them. We investigate to what extent current
techniques are suited for this task and test a variety of models in this regard. To detect entities in a biological
text we use a linear chain Conditional Random Field (CRF). For the prediction of relations between the entities,
a model based on logistic regression is built. Designing a system upon these techniques, we explore several
improvements for both the generation and selection of good candidates. One contribution to this lies in the
extended flexibility of our ontology mapper, allowing for a more advanced boundary detection. Furthermore,
we discover value in the combination of several distinct candidate generation rules. Using these techniques,
we show results that are significantly improving upon the state of art for the BioNLP Bacteria Biotopes task.
1 INTRODUCTION
A vast amount of scientific literature is available about
bacteria biotopes and their properties (Bossy et al.,
2013). Processing this literature can be very time-
consuming for biologists, as efficient mechanisms
to automatically extract information from these texts
are still limited. Biologists need information about
ecosystems where certain bacteria live in. Hence,
having methods that rapidly summarize texts and list
properties and relations of bacteria in a formal way
becomes a necessity. Automatic normalization of the
bacteria and biotope mentions in the text against cer-
tain ontologies facilitates extending the information in
ontologies and databases of bacteria. Biologists can
then easily query for specific properties or relations,
e.g. which bacteria live in the gut of a human or in
which habitat Bifidobacterium Longum lives.
The Bacteria Biotopes subtask (BB-Task) of the
BioNLP Shared Task (ST) 2013 is the basis of this
study. It is the third event in this series, following the
same general outline and goals of the previous events
(N
´
edellec et al., 2013). BioNLP-ST 2013 featured six
event extraction tasks all related to “Knowledge base
construction”. It attracted wide attention, as a total of
38 submissions from 22 teams were received.
The BB-Task consists of three subtasks. In the
first subtask habitat entities need to be detected in a
given biological text and the entities must be mapped
onto a given ontology. The habitat entities vary from
very specific concepts like ‘formula fed infants’ to
very general concepts like ‘human’. The second sub-
task is focused on the extraction of two relations: a
Localization and a PartOf relation. These relations
need to be predicted between a given set of entities
(bacteria, habitats and geographical locations). Lo-
calization relations occur between a bacterium and
a habitat or geographical location, PartOf relations
only occur between habitats. The third subtask is an
extended combination of the two other subtasks: enti-
ties need to be detected in a text and relations between
these entities need to be extracted. In this paper we fo-
cus on the first two subtasks.
We first describe related work done in context of
the BioNLP-ST (Section 2). We then discuss our
methodology for the two subtasks (Section 3). Next,
we discuss our experiments and compare our results
with the official submissions to BioNLP-ST 2013
(Section 4). We end with a conclusion (Section 5).
55
Massa W., Kordjamshidi P., Provoost T. and Moens M..
Machine Reading of Biological Texts - Bacteria-Biotope Extraction.
DOI: 10.5220/0005214700550064
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2015), pages 55-64
ISBN: 978-989-758-070-3
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

2 RELATED WORK
The BB-task along with the experimental dataset has
been initiated for the first time in the BioNLP Shared
Task 2011 (Bossy et al., 2011). Three systems were
developed in 2011 and five systems for its extended
version proposed in the 2013 shared task (Bossy et al.,
2013). In 2011 the following systems participated in
this task. TEES (Bjorne and Salakoski, 2011) was
proposed by UTurku as a generic system which uses
a multi-class Support Vector Machine classifier with
linear kernel. It made use of Named Entity Recogni-
tion patterns and external resources for the BB model.
The second system was JAIST (Nguyen and Tsu-
ruoka, 2011), specifically designed for the BB-task.
It uses CRFs for entity recognition and typing and
classifiers for coreference resolution and event extrac-
tion. The third system was Bibliome (Ratkovic et al.,
2011), also specifically designed for this task. This
system is rule-based, and exploits patterns and do-
main lexical resources.
The three systems used different resources for
Bacteria name detection which are the List of
Prokaryotic Names with Standing in Nomenclature
(LPNSN), names in the genomic BLAST page of
NCBI and the NCBI Taxonomy, respectively. The
Bibliome system was the winner for detecting the
Bacteria names as well as for the coreference reso-
lution and event extraction. The important factor in
their outperformance was exploiting the resources and
ontologies. They found useful matching patterns for
the detection of entities, types and events. Using their
manually drawn patterns and rules performed better
than other task participant systems, in which learning
models apply more general features.
In the 2013 edition of this task, the event extrac-
tion is defined in a similar way but an extension to
the 2011 edition considered biotope normalization us-
ing a large ontology of biotopes called OntoBiotope.
The task was proposed in three subtasks to which we
pointed in Section 1. Five teams participated in these
subtasks. In the first subtask all entities have to be
predicted, even if they are not involved in any rela-
tion. The participated systems performed reasonably
well. However, the difficulty of this task has been
boundary detection.
The participating systems obtained a very low re-
call for the relation extraction even when the entities
and their boundaries are given (subtask 2 and 3). The
difficulty of the relation extraction is partially due to
the high diversity of bacteria and locations. The many
mentions of different bacteria and localization in the
same paragraph makes it difficult to select the right
links between them. The second difficulty lies in the
high frequency of anaphora. This makes the extrac-
tion of the relations beyond sentence level difficult.
The strict results of the third task were very poor,
due to struggling with the difficulties of both previ-
ous tasks i.e, boundary detection and link extraction.
For detecting entities (subtask 1), one submission
(Bannour et al., 2013) worked with generated syntac-
tical rules. Three other submissions (Claveau, 2013),
(Karadeniz and
¨
Ozg
¨
ur, 2013) and (Grouin, 2013) used
an approach similar to ours. They generated candi-
dates in an initial phase from texts. These candidates
were subsequently selected by trying to map them
onto the ontology. Two submissions (Claveau, 2013)
and (Karadeniz and
¨
Ozg
¨
ur, 2013) generated candi-
dates by extracting noun phrases. One submission
(Grouin, 2013) used a CRF model to generate candi-
dates, as we do in this work. However, we test candi-
dates more thoroughly and consider every continuous
subspan of tokens in each candidate instead of just the
candidate itself, which explains our improved results.
For the relation extraction with given entities (sub-
task 2), there were four submissions. One system
from LIMSI (Grouin, 2013) relied solely on the fact
that the relation was seen in the training set which
fails to yield a reasonable accuracy. A second sys-
tem BOUN (Karadeniz and
¨
Ozg
¨
ur, 2013) extracted
relations using only simple rules, e.g. in a specific
paragraph they created relations between all locations
and the first bacterium in that paragraph. A third sys-
tem IRISA (Claveau, 2013) used a nearest neighbor
approach. Another system was TEES (Bj
¨
orne and
Salakoski, 2013) (an improved version of the UTurku
participation in 2011) which provided the best results.
However, the results were still poor.
One reason for this lies in the limited scope of
candidates that the submitted systems considered,
e.g. TEES (Bj
¨
orne and Salakoski, 2013) and IRISA
(Claveau, 2013) only examined relations between a
habitat and location that occur in the same sentence.
One of our contributions lies in considering more pos-
sible relations, including relations across sentences.
This is confirmed by a much better recall, as can be
seen in Section 4.3.2.
3 METHODOLOGY
In this section we lay out our developed system. For
each of subtasks 1 and 2, we first discuss the goal of
the subtask, followed by an explanation of our used
methodology. The performance of our model is dis-
cussed in the next section (Section 4).
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
56

3.1 Subtask 1: Entity Detection and
Ontology Mapping
The goal of this subtask is to detect habitat entities
in texts and map them onto concepts defined by the
OntoBiotope-Habitat ontology. For each entity the
name, the location in the text and the corresponding
ontology entry need to be predicted. E.g. the ex-
pected output for a text consisting of the single sen-
tence “This organism is found in adult humans and
formula fed infants as a normal component of gut
flora.” is:
T1 Habitat 27 33 adult humans
T2 Habitat 44 63 formula fed infants
T3 Habitat 44 51 formula
T4 Habitat 89 92 gut
N1 OntoBiotope Annotation:T1 Ref:MBTO:00001522
N2 OntoBiotope Annotation:T2 Ref:MBTO:00000308
N3 OntoBiotope Annotation:T3 Ref:MBTO:00000798
N4 OntoBiotope Annotation:T4 Ref:MBTO:00001828
Four habitat entities are found in this sentence and
they are mapped onto four different ontology entries.
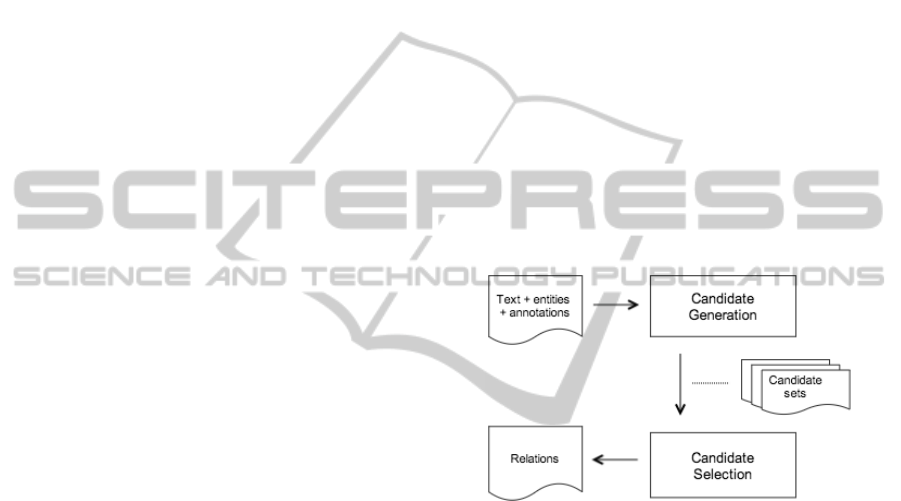
Figure 1 gives an overview of the followed ap-
proach. We first search in the text for token spans
(candidates) that might contain one or more enti-
ties (Section 3.1.1). These generated candidates are
given to a Candidate Selection module, that searches
substrings within the candidate for entities (Section
3.1.3). This Candidate Selection module uses an On-
tology Mapper (Section 3.1.2), finding the ontology
entry that matches closest to a given substring. Ad-
ditionally it returns a dissimilarity value to give an
indication of how close the match is. Based on this
dissimilarity, we can decide to classify part of a can-
didate as the given entry or not.
Figure 1: Overview of the followed approach in subtask 1.
3.1.1 Candidate Generation
The Candidate Generation module generates token
spans from a given input text. The goal of the Gener-
ation module is to quickly reduce a large text to a can-
didate set that can be analysed more efficiently. First
the text is split into sentences and tokens, then ev-
ery sentence is mapped onto a set of candidates. We
use the given annotation files of the Stanford Parser
(Klein and Manning, 2003) to split the texts and to-
kenize the sentences. Sentences are assumed to be
independent in the model, i.e. we do not use informa-
tion from one sentence in another sentence.
Conditional Random Fields. To generate candi-
dates, we use Conditional Random Fields (CRF) (Sut-
ton and McCallum, 2006). In particular, we choose a
linear chain CRF; previous research shows that these
perform well for various natural language process-
ing tasks, especially Named Entity Recognition (Lei
et al., 2014). In contrast to general purpose noun
phrase extractors used by some other existing models
for this task, a CRF can easily exploit the information
of the given annotated files as features.
A CRF model is an undirected probabilistic graph-
ical model G = (V, E) with vertices V and edges E.
The vertices represent a set of random variables with
the edges showing the dependencies between them.
The set of observed random variables is denoted by
X and the unknown/output random variables are de-
noted by Y . This model represents a probability dis-
tribution over a large number of random variables by a
product of local functions that each depend on a small
subset of variables, called factors.
A CRF generally defines a probability distribution
p(y|x), where x, y are specific assignments of respec-
tive variables X and Y as follows:
p(y|x) =
1
Z(x)
∏
Ψ
F
∈G
Ψ
F
(x
F
, y
F
) (1)
where x
F
are those observed variables that are part of
factor F and similarly for y
F
and Y . Ψ
F
: V
n
→ R is
the potential function associated with factor F and is
defined in terms of the features f
Fk
(x
F
, y
F
) as:
Ψ
F
(x
F
, y
F
) = exp
(
∑
k
λ
Fk
f
Fk
(x
F
, y
F
)
)
(2)
The parameters of the conditional distribution λ
Fk
are
trained with labelled examples. Afterwards, using the
trained model the most probable output variables can
be calculated for a given set of observed variables.
Z(x) is a normalization constant and is computed as:
Z(x) =
∑
y
∏
Ψ
F
∈G
Ψ
F
(x
F
, y
F
) (3)
CRFs can represent any kind of dependencies, but
the most commonly used model, particularly in the
NLP tasks such as Named Entity Recognition is the
Linear-chain model. In this work, we use the linear
chain implementation in Factorie (McCallum et al.,
MachineReadingofBiologicalTexts-Bacteria-BiotopeExtraction
57

2009). Linear chain CRFs consider the dependency
between the labels of the adjacent words. In other
words, each local function f
k
(y
t
, y
t−1
, x
t
) represents
the dependency of each output variable y
t
in location
t in the chain to its previous output variable y
t−1
and
the observed variable x
t
at that location. The global
conditional probability then is computed as the prod-
uct of these local functions (Sutton and McCallum,
2006). With the usual assumption that all local func-
tions share parameters and feature functions, its log-
linear form is now written as:
p(y|x) =
1
Z(x)
exp
(
∑
t
∑
k
λ
k
f
k
(y
t
, y
t−1
, x
t
)
)
(4)
Where the normalization constant is derived in an
analogous manner to equation (3) for the case of se-
quential dependencies.
In our model, every token is an observed variable. The
biological entity labels (e.g. ‘Bacterium’) of the to-
kens are the output variables (or labels).
We now discuss the features that we use, along
with the label set into which the tokens are classified.
CRF Features. The following features are used for
each token:
• Token string
• Stem
• Length token
• Is capitalized (binary)
• Token is present in the ontology (binary)
• Stem is present in the ontology (binary)
• Category of the token in the Cocoa annotations
• Part-of-speech tag
• Dependency relation to the head of the token
The stem is calculated using an online available
Scala implementation
1
of Porter’s stemming algo-
rithm (Porter, 1980). The part-of-speech tag and the
dependency relation to the head are added using the
available annotation files from the Stanford Parser
(Klein and Manning, 2003). Cocoa
2
is a dense anno-
tator for biological text. The Cocoa annotations cover
over 20 different semantic categories like ‘Processes’
and ‘Organisms’.
CRF Labels: Extended Boundary Detection Tags.
We use five different labels for the tokens. The used
labels are:
• Start: The token is the first token of an entity.
1
https://github.com/aztek/porterstemmer
2
http://npjoint.com
• Center: The token is in the middle of an entity.
• End: The token is the last token of an entity.
• Whole: The token itself is an entity.
• None: The token does not belong to an entity.
The most immediate alternative to this is the tra-
ditional IOB labeling (Ramshaw and Marcus, 1995).
An even more simple possibility is a binary labeling
that just indicates if a token belongs to an entity men-
tion or not. The more elaborated proposed labeling
generally performs better in our tests.
3.1.2 Ontology Mapper
The Ontology Mapper maps a string onto the ontol-
ogy entry with the lowest dissimilarity. The dissim-
ilarity between an ontology entry and string is cal-
culated by comparing the string with the name, syn-
onyms and plural of the name and synonyms of the
entry with respect to a certain comparison function.
The plurals are calculated simply by just adding ‘s’ or
‘es’ to the end of the singular form.
To compare two strings they are split into tokens.
The tokens from the two strings are matched to mini-
mize the sum of the relative edit distance between the
matched tokens. If not all tokens can be matched i.e.
the number of tokens in the two strings are different,
1.0 is added to the sum for each remaining token. As
a measure for relative edit distance, we use the Lev-
enshtein distance (Levenshtein, 1966) divided by the
sum of the lengths of the strings to get a number be-
tween 0.0 and 1.0.
3.1.3 Candidate Selection
The Candidate Selection module receives spans of to-
kens as input, it searches within these spans for on-
tology entries. For each span every continuous sub-
span of tokens is tested with the Ontology Mapper.
This means that we select
n(n+1)
2
subspans for every
token span with n tokens. If for a subspan a dissimi-
larity lower than a specific bound is reached, we clas-
sify this subspan as an entity. E.g. for the token span
‘formula fed infants’, six subspans are selected: ‘for-
mula’, ‘fed’, ‘infants’, ‘formula fed’, ‘fed infants’ and
‘formula fed infants’. ‘formula’ and ‘formula fed in-
fants’ are found in the ontology and we classify these
as entities.
Based on cross-validation experiments on the
training and development set, we decided to take as
maximal dissimilarity 0.1, i.e. the subspan must be
very close to an ontology entry. This very strict pa-
rameter allows us to be less strict in the Candidate
Generation module: every entity that has a minimum
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
58
probability of 0.1 to contain entities will be tested.
The sensitivity of our results with respect to this mea-
sure is further discussed in Subsection 4.3.1 and Table
2.
3.1.4 Additional Improvements
Dashed Words. Not all entities in the texts consist
of one or more tokens, some entities are only a part of
a token. E.g. in the token ‘tick-born’, ‘tick’ is an en-
tity. To handle these cases we search for all the words
that contain one or more dashes. These words are split
and every part is matched against the ontology. These
parts are easy to match because they are usually just
nouns in singular form.
Extending the Ontology. Mappings from phrases
onto ontology entries are given in the training and de-
velopment set. These phrases are usually similar to
the name or a synonym of the ontology entry. How-
ever in rare cases the phrase is not similar to the name
or a synonym. Based on the assumption that the given
mapping is correct we can extend the ontology. We do
this by adding the phrase as a new synonym to the on-
tology entry. Some submissions to the BioNLP-ST
2013 task used this approach as well (Grouin, 2013),
(Karadeniz and
¨
Ozg
¨
ur, 2013).
Correcting Boundaries. An important part of the
task is to predict the correct boundaries of the enti-
ties. E.g. for the noun phrase ‘blood-sucking tsetse
fly’, it is not sufficient to predict ‘fly’ or ‘tsetse fly’.
The whole noun phrase is the correct entity in this
case. This particular example is hard because ‘blood-
sucking tsetse fly’ does not occur in the ontology. To
handle this case we add to each found entity the de-
pendent words that precede the entity. These depen-
dent words can be extracted by using the given parser
annotations. E.g. from the phrase ‘blood-sucking
tsetse fly’ the entity ‘tsetse fly’ is selected, ‘blood-
sucking’ is added to it because its headword is ‘fly’.
Filter out Parents. Many generated candidates re-
fer to the same entity, it is required that we predict
every entity only once. E.g. in the phrase ‘person
with untreated TB’ the entities ‘person’ and ‘person
with untreated TB’ are detected. They refer both to
the same habitat, ‘person’ is just a more general term
to describe ‘person with untreated TB’. That is why
we filter ‘person’ out. We can do this by using the
parent/child relations given in the ontology. The on-
tology entry ‘person’ is a parent (a more general term)
of the entry ‘person with untreated TB’, so we only
predict the phrase ‘person with untreated TB’ in this
case. Because the ontology is a deep graph of entities,
we test this parent/child relationship recursively.
3.2 Subtask 2: Relation Extraction
In this subtask relations need to be extracted from a
text based on annotated entities in the text. There are
three types of entities: habitats, geographical entities
and bacteria. Two types of relations exists: Local-
ization and PartOf. Localization relations are always
between a bacterium and a habitat or geographical lo-
cation. PartOf relations occur between two habitats.
We handle these two relations independently. In the
training and development set combined, Localization
and PartOf relations are responsible for respectively
81% and 19% of the relations.
We used a similar approach for both relation types.
We will describe our approach for Localization rela-
tions. Our model consists of two modules. A first
module generates sets of relation candidates from the
text using simple rules (Section 3.2.1). These sets are
then forwarded to a second module that trains for each
set a separate model (Section 3.2.2). Figure 2 shows
a visualization of our approach.
Figure 2: Overview of the followed approach in subtask 2.
3.2.1 Candidate Generation
The Candidate Generation module reduces the set of
all possible relations, i.e. all combinations of bacte-
ria and locations, to multiple smaller sets of candidate
relations. Every set is created by using a generation
rule. These smaller sets are then forwarded to the
Candidate Selection module that will try to identify
if a candidate relation is really a relation or not.
We use generation rules for two reasons. On one
side we decrease the overall number of candidates by
a significant amount. On the other side we group sim-
ilar types of relations to build more specific models.
For every set of candidate relations defined by a gen-
eration rule, we build a separate model to test these
relations. A good candidate generation method gen-
erates a relatively large number of correct relations
while keeping the number of wrong relations to a min-
imum.
We tested 5 different candidate generation rules
for Localization relations:
MachineReadingofBiologicalTexts-Bacteria-BiotopeExtraction
59

• All Possible: All combinations of bacteria and lo-
cations are possible.
• Same Sentence: The bacterium and location oc-
cur in the same sentence. This assumption is
used by two submissions: (Bj
¨
orne and Salakoski,
2013) and (Claveau, 2013).
• Previous Bacteria: The bacterium is the first bac-
terium that occurs before the location in the text.
• Next Bacteria: The bacterium is the first bac-
terium that occurs after the location in the text.
• Paragraph Subject: The text is split into para-
graphs. The bacterium is the first bacterium that
occurs in the paragraph of the location. This is
used by one submission: (Karadeniz and
¨
Ozg
¨
ur,
2013).
The results from section 4.3.2 are achieved by
combining the ‘Same sentence’ and ‘Previous bac-
teria’ generation rule, which yields the best perfor-
mance.
3.2.2 Candidate Selection
The Candidate Generation module forwards different
sets of candidate relations to the Candidate Selection
module. This Candidate Selection module builds for
every set a separate logistic regression model (using
the Factorie toolkit (McCallum et al., 2009)). We use
these logistic regression models as binary classifiers
(is a relation or not). In the training phase, the models
are trained based on positive and negative relations
extracted from example texts. In the testing phase,
each set of candidate relations is tested by their sepa-
rate model.
The model uses the following features based on
the two involved entities in a relation:
• The type of the entity
• Surface form
• Is capitalized (binary)
• Stem of each entity token
• Category of each entity token in the Cocoa anno-
tations
• Part-of-speech tag of each entity token
• Dependency relation to the head of each token
None of the above features combine information
from both the bacterium and location. We tested some
features that do this, but without a significant influ-
ence on F1, as we saw a slightly better precision with
a small drop in recall. The tested features are:
• Token distance between bacterium and location
• Length of syntactic path between bacterium and
location
• The depth of the tree that contains the syntactic
path
• Whether the bacterium or location occurs first
Alternative Models. Besides this model we also
tested a nearest neighbor model. In this, we compare a
candidate with a seen example based on the sequence
of part-of-speech tags that occur on the syntactic path
between the bacterium and location. Between these
two sequences of tags the edit distance is calculated.
Finally, the candidate is classified as a relation if the
closest seen example with respect to this distance en-
codes a real relation.
In another approach we used two language models
based on the tokens between the bacterium and loca-
tion, where a separate model for positive and negative
relations was built. Here, a candidate is classified as
a relation if the probability that the candidate is gen-
erated by the positive model is higher than the proba-
bility for the negative model.
Both alternative models failed to achieve reason-
able performance.
4 EXPERIMENTS
In the first subsection we describe the data set and the
resources. The subsections thereafter then present the
results and discussions.
4.1 Data Set
The data set consists of public available documents
from web pages from bacteria sequencing projects
and from the MicrobeWiki encyclopedia (Bossy et al.,
2013). The data is divided into a training, a develop-
ment and a test set. The solution files of the training
and development set are provided. The solution files
of the test set are not available, but it is possible to
test a solution with an online evaluation service
3
with
a minimal time of 15 minutes between two submis-
sions. During the contest the minimal time between
two submissions has been 24 hours. We limited our
use of the online evaluation service to keep our results
comparable with the contest submissions.
The data consists of 5,183 annotated entities and
2,260 annotated relations. The data was manually an-
notated twice followed by a conflict resolution phase
3
http://genome.jouy.inra.fr/∼rbossy/cgi-bin/bionlp-eval
/BB.cgi
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
60

(Bossy et al., 2013). Table 1 gives an overview of the
data distribution. The training and development set is
the same for both subtasks, but the test set is different.
Table 1: Summary statistics of the data set.
Training/Dev Testset 1 Testset 2
Documents 78 27 26
Words 25,828 7,670 10,353
Entities 3,060 877 1,246
Relations 1,265 328 667
4.2 Used Ontology
In the first subtask the OntoBiotope-Habitat ontol-
ogy
4
is used. This ontology contains 1,756 habitat
concepts. For each concept an id, the name and ex-
act and related synonyms are given. Additionally if a
concept can be described by a more general concept,
an is a relation is given. The ontology entry ‘dental
caries’ is for example:
id: MBTO:00001830
name: dental caries
related_synonym: "tooth decay" [TyDI:30379]
exact_synonym: "dental cavity" [TyDI:30380]
is_a: MBTO:00002063 ! caries
4.3 Results
The results are presented separately for the two sub-
tasks of entity detection and relation extraction.
4.3.1 Entity Detection and Ontology Mapping
The score is calculated by mapping the predicted enti-
ties onto the entities of the reference solution. Entities
are paired in a way that the sum of the dissimilarities
are minimized. The dissimilarity between a predicted
entity and a reference entity is based on boundary ac-
curacy and the semantic similarity between the ontol-
ogy concepts. Based on this optimal mapping of enti-
ties the Slot Error Rate (SER) is calculated. A perfect
solution has a SER score of 0, if no entities are pre-
dicted a score of 1 is obtained. The SER is calculated
as follows:
SER =
S + I + D
N
(5)
• S: number of substitutions, based on the dissimi-
larity between the matched entities.
• I: number of insertions, the number of predicted
entities that could not be paired.
4
http://bibliome.jouy.inra.fr/MEM-OntoBiotope/Onto
Biotope BioNLP-ST13.obo
• D: number of deletions, the number of reference
entities that could not be paired.
• N: number of entities in the reference solution.
Improvement Effects. We implemented four vari-
ations to improve our model (see Section 3.1.4). The
highest improvement is achieved by correcting the
boundaries and filtering out redundant parents. Al-
though handling dashed words gives only a slight im-
provement, it is definitely worth to use it because it
increases the number of found entities without creat-
ing much incorrect entities. Extending the ontology
improves our solution only by a very small margin.
Influence of the Maximal Dissimilarity. As ex-
plained in section 3.1.3, the Candidate Selection mod-
ule receives spans of tokens as input and searches
within these spans for ontology entries. For a spe-
cific subspan of tokens, the Ontology Mapper returns
the ontology entry that best matches, together with a
dissimilarity measure. Based on cross-validation ex-
periments we picked 0.1 as maximal dissimilarity, i.e.
we classify all subspans with a lower dissimilarity as
0.1 as a found entity.
Table 2 shows the SER score together with the
number of Substitutions, Insertions and Deletions (us-
ing 10 fold cross validation on the training and devel-
opment set) for several values of maximal dissimilar-
ity. For a range of low thresholds, only a very small
variation in the number of Substitutions and Deletions
is observed. However, the number of Insertions in-
creases steadily with an increasing maximal dissimi-
larity. This is because we allow subspans to be less
and less similar to the ontology entries, causing an in-
creasing number of wrongly extracted entities.
Table 2: Influence of the maximal dissimilarity on entity
detection performance.
Dissimilarity Sub Ins Del SER
0.05 212 195 181 0.38
0.10 210 197 180 0.38
0.15 212 208 180 0.39
0.20 211 212 180 0.39
0.25 227 236 173 0.41
0.30 230 249 169 0.41
0.35 319 497 141 0.61
Comparison with Contest Submissions. Testing
our model with the online evaluation service, we ob-
tained a SER score of 0.36 which is significantly bet-
ter than all submissions to BioNLP-ST 2013. The best
result of the contest is a SER score of 0.46 (IRISA).
MachineReadingofBiologicalTexts-Bacteria-BiotopeExtraction
61
We also improved the precision and F1 compared
to all submissions. Recall, precision and F1 were re-
spectively 0.68, 0.73 and 0.70. The IRISA submission
scored a higher recall but a lower precision than our
model. Table 3 shows our scores together with the
scores of the submissions to BioNLP-ST 2013.
Table 3: Subtask 1 results compared to contest submissions.
Participant SER Recall Precision F1
IRISA 0.46 0.72 0.48 0.57
Boun 0.48 0.60 0.59 0.59
LIPN 0.49 0.61 0.61 0.61
LIMSI 0.66 0.35 0.62 0.44
Ours 0.36 0.68 0.73 0.70
Some reasons why we outperform the others are:
• With a CRF model it is easy to consider any infor-
mation through the addition of features. However,
many systems that use a CRF to generate candi-
dates are based on a general purpose noun phrase
extractor, and do not use the biological annota-
tions that are supplied.
• We search within each candidate for matches,
which makes it possible that a candidate contains
multiple entities.
• We redefine the boundaries of an entity by us-
ing the head annotations from the given Stanford
parser annotated data.
The main weakness of our model is that an entity
needs to be very close to a name or synonym of an
ontology entry to be detected. We picked a value of
0.1 as maximal dissimilarity. This means that entities
that do not occur in the ontology or are described by
an unknown synonym can not be found. We imple-
mented an improvement by correcting the boundaries
to lower the impact of this weakness. In this way,
words that are not seen in the ontology can be part of
an entity if its head word occurs in the ontology.
4.3.2 Relation Extraction
Baseline Model. To better analyse the performance
of our approach, we have first built a baseline model.
This model predicts Localization relations between
all bacteria and locations that occur in the same sen-
tence and no PartOf relations. The results are pre-
sented in Table 5. Considering the achieved scores in
BioNLP-ST 2013, this model performs dramatically
better. It outperforms all submissions based on F1
due to a much higher recall. But the precision of one
submission (TEES) is clearly better (0.82).
This baseline model predicts 53% of the Local-
ization relations. Based on the fact that this baseline
model only predicts relations within the same sen-
tence, we know that about half of the Localization re-
lations occur in the same sentence, for the other half
multiple sentences need to be examined.
Performance on Different Relation Types. We
use a similar approach for PartOf relations as for Lo-
calization relations. Table 4 shows the performance
of our model for the prediction of one relation type
separately and the prediction of both types jointly. We
see a very low precision if we only predict PartOf re-
lations, this is due to the fact that we recall many re-
lations wrongly and there are only few true PartOf
relations in the texts. When we combine our Local-
ization and PartOf model the result is worse than the
Localization model on itself. The PartOf model de-
creases the overall precision of our model much more
compared to the gain in recall.
Table 4: Relation extraction results for the different relation
types.
Model Recall Precision F1
Localization 0.59 0.50 0.54
PartOf 0.09 0.15 0.12
Combined 0.68 0.35 0.46
Comparison with Contest Submissions. We
tested our solution with the available online eval-
uation service and receive a F1 of 0.67 which is
significantly better than all submissions to BioNLP-
ST 2013. The best result of the contest achieved
a F1 of 0.42 (TEES). Our recall and precision are
respectively 0.71 and 0.63. This recall is much higher
than all the contest submissions, one submission
(TEES) scored a better precision (0.82). Table 5
shows our achieved results together with the scores
of the official submissions to BioNLP-ST 2013.
Table 5: Subtask 2 results compared to contest submissions.
Participant Recall Precision F1
TEES-2.1 0.28 0.82 0.42
IRISA 0.36 0.46 0.40
Boun 0.21 0.38 0.27
LIMSI 0.04 0.19 0.06
Baseline 0.43 0.47 0.45
Ours 0.71 0.63 0.67
Some reasons why we outperform the others are:
• We use a combination of generation rules, the
contest submissions were mainly limited to one
specific generation rule.
• We do not predict PartOf relations in our final
model due to low accuracy and overall negative
impact.
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
62

Bacterium Model. The logistic regression model
achieves significantly better results than the baseline
model and all contest submissions. However, many
of the used features have only very little influence.
We remark that almost comparable results can be
achieved by a model that always predicts true unless
the bacterium name starts with ‘bacteri’. This sort of
model is of course not generic and largely overfits the
data. It works well because it succeeds in excluding a
significant amount of false relations. Labeled entities
occur in surface forms ‘bacterium’, ‘bacterial infec-
tions’, . . . These forms occur relatively often in texts,
but they rarely appear in Localization relations. The
reason for this is that when the word ‘bacterium’ ap-
pears in a text, it usually does not refer to the general
concept but to a specific bacterium discussed previ-
ously in the text. However, to avoid overfitting it is
preferred to use such patterns in the data by including
relevant features, rather than implementing strict de-
cision rules based on them. In the case of the above
characteristic, the name of the specific bacterium en-
tity is added as a feature in our system.
5 CONCLUSION
In this paper we discussed an approach for the
first two subtasks of the Bacteria Biotopes task of
BioNLP-ST 2013. For the first subtask (entity detec-
tion and ontology mapping) we implemented a model
based on Conditional Random Fields. In this sys-
tem, candidates are generated from the text and thor-
oughly inspected to find matches within the ontology.
We also devised several improvements for the bound-
ary detection of entities. Our model achieved signif-
icantly better results than all official submissions to
BioNLP-ST 2013.
For the second subtask (relation extraction) we
generated candidates with multiple generation rules
(e.g. all bacteria and locations that occur in the same
sentence). To select a candidate we used a logistic
regression model. Because we used a combination of
generation rules we achieved a much higher recall and
therefore a much better score than all official submis-
sions to BioNLP-ST 2013.
In spite of these pronounced gains, we think there
is still room for improvement, especially for the sec-
ond subtask. One potential improvement of our
model will be to consider long distance dependen-
cies between the bacterium and location, more con-
textual features and additional background knowl-
edge from external resources. In this direction, us-
ing structured output prediction and joint learning
frameworks will help us to consider these kind of
knowledge for an end-to-end entity and relation ex-
traction model (Kordjamshidi and Moens, 2013; Ko-
rdjamshidi and Moens, 2014).
ACKNOWLEDGEMENTS
The authors would like to thank the Research Founda-
tion Flanders (FWO) for funding this research (grant
G.0356.12), as well as the bilateral project of KU
Leuven and Tsinghua University DISK (DIScovery of
Knowledge on Chinese Medicinal Plants in Biomedi-
cal Texts, grant BIL/012/008). Also we would like to
thank the reviewers for their insightful comments and
remarks.
REFERENCES
Bannour, S., Audibert, L., and Soldano, H. (2013).
Ontology-based semantic annotation: an automatic
hybrid rule-based method. In Proceedings of the
BioNLP Shared Task 2013 Workshop, pages 139–143,
Sofia, Bulgaria. ACL.
Bjorne, J. and Salakoski, T. (2011). Generalizing biomedi-
cal event extraction. In Proceedings of BioNLP Shared
Task 2011 Workshop. ACL.
Bj
¨
orne, J. and Salakoski, T. (2013). TEES 2.1: Automated
Annotation Scheme Learning in the BioNLP 2013
Shared Task. In Proceedings of the BioNLP Shared
Task 2013 Workshop, pages 16–25, Sofia, Bulgaria.
ACL.
Bossy, R., Golik, W., Ratkovic, Z., Bessi
`
eres, P., and
N
´
edellec, C. (2013). BioNLP shared Task 2013 –
An Overview of the Bacteria Biotope Task. In Pro-
ceedings of the BioNLP Shared Task 2013 Workshop,
pages 161–169, Sofia, Bulgaria. ACL.
Bossy, R., Jourde, J., Bessieres, P., van de Guchte, M., and
Nedellec, C. (2011). BioNLP shared task 2011 - Bac-
teria Biotope. In Proceedings of BioNLP Shared Task
2011 Workshop. ACL, pages 56–64.
Claveau, V. (2013). IRISA participation to BioNLP-ST
2013: lazy-learning and information retrieval for in-
formation extraction tasks. In Proceedings of the
BioNLP Shared Task 2013 Workshop, pages 188–196,
Sofia, Bulgaria. ACL.
Grouin, C. (2013). Building a contrasting taxa extractor for
relation identification from assertions: Biological tax-
onomy & ontology phrase extraction system. In Pro-
ceedings of the BioNLP Shared Task 2013 Workshop,
pages 144–152, Sofia, Bulgaria. ACL.
Karadeniz, I. and
¨
Ozg
¨
ur, A. (2013). Bacteria biotope detec-
tion, ontology-based normalization, and relation ex-
traction using syntactic rules. In Proceedings of the
BioNLP Shared Task 2013 Workshop, pages 170–177,
Sofia, Bulgaria. ACL.
Klein, D. and Manning, C. D. (2003). Fast exact inference
with a factored model for natural language parsing. In
MachineReadingofBiologicalTexts-Bacteria-BiotopeExtraction
63

Advances in Neural Information Processing Systems
15 (NIPS), pages 3–10. MIT Press.
Kordjamshidi, P. and Moens, M.-F. (2013). Designing con-
structive machine learning models based on general-
ized linear learning techniques. In NIPS Workshop on
Constructive Machine Learning.
Kordjamshidi, P. and Moens, M.-F. (2014). Global machine
learning for spatial ontology population. Journal of
Web Semantics: Special issue on Semantic Search.
Lei, J., Tang, B., Lu, X., Gao, K., Jiang, M., and Xu,
H. (2014). A comprehensive study of named en-
tity recognition in chinese clinical text. Journal
of the American Medical Informatics Association,
21(5):808–814.
Levenshtein, V. (1966). Binary Codes Capable of Cor-
recting Deletions, Insertions and Reversals. Soviet
Physics Doklady, 10:707.
McCallum, A., Schultz, K., and Singh, S. (2009). FACTO-
RIE: Probabilistic programming via imperatively de-
fined factor graphs. In Neural Information Processing
Systems (NIPS).
N
´
edellec, C., Bossy, R., Kim, J.-D., Kim, J.-J., Ohta, T.,
Pyysalo, S., and Zweigenbaum, P. (2013). Overview
of BioNLP shared task 2013. In Proceedings of
the BioNLP Shared Task 2013 Workshop, pages 1–7,
Sofia, Bulgaria. ACL.
Nguyen, N. T. H. and Tsuruoka, Y. (2011). Extracting
bacteria biotopes with semi-supervised named entity
recognition and coreference resolution. In Proceed-
ings of BioNLP Shared Task 2011 Workshop. ACL.
Porter, M. (1980). An algorithm for suffix stripping. Pro-
gram, 14(3):130–137.
Ramshaw, L. A. and Marcus, M. P. (1995). Text chunk-
ing using transformation-based learning. In Proceed-
ings of the 3rd ACL Workshop on Very Large Corpora,
pages 82–94. Cambridge MA, USA.
Ratkovic, Z., Golik, W., Warnier, P., Veber, P., and Nedel-
lec, C. (2011). Task Bacteria Biotope-The Alvis Sys-
tem. In Proceedings of BioNLP Shared Task 2011
Workshop. ACL.
Sutton, C. and McCallum, A. (2006). Introduction to Con-
ditional Random Fields for Relational Learning. MIT
Press.
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
64
