
Supporting Multi-level User-driven Detection of
Guideline Interactions
Luca Piovesan
1
, Gianpaolo Molino
2
and Paolo Terenziani
3
1
Dipartimento di Informatica, Università degli Studi di Torino, Torino, Italy
2
Azienda Ospedaliera San Giovanni Battista, Torino, Italy
3
DISIT, Università del Piemonte Orientale “Amedeo Avogadro”, Alessandria, Italy
Keywords: Computer-interpretable Clinical Guidelines, Comorbidity Treatment, Knowledge Representation,
Ontologies, Guideline Interaction Detection.
Abstract: Clinical practice guidelines are widely used to support physicians, but only on individual pathologies. The
treatment of patients affected by multiple diseases (comorbid patients) requires the development of new
approaches, supporting physicians in the detection of interactions between guidelines. We propose a new
methodology, supporting flexible and physician-driven search and detection. In particular, we provide a
flexible and interactive mechanism to navigate guidelines and our ontology of interactions (between drugs,
or between actions’ goals) at multiple levels of detail, focusing on specific parts of it (e.g., on a specific pair
of actions, or of drugs) to look for interactions. We introduce the notion of “navigation tree”, as the basic
data structure to support multiple-level interaction analysis, and describe navigation and focusing algorithms
operating on it. We also introduce a visualization tool that is based on the “navigation tree”, and further
enhances the user-friendliness of our approach.
1 INTRODUCTION
Clinical practice guidelines (CPGs) are defined as
“systematically developed statements to assist
practitioner and patient decisions about appropriate
healthcare for specific clinical circumstances”
(Committee to Advise the Public Health Service on
Clinical Practice Guidelines, Institute of Medicine
1990).
CPGs exploitation is meant to improve the
quality and to reduce the cost of healthcare, putting
evidence based medicine into practice, and is
progressively spreading in several countries. As a
matter of fact, a lot of national and international
institutions have recently been engaged in
developing and disseminating CPGs. Thousands of
CPGs have been devised in the last years. For
instance, the Guideline International Network
(Guidelines International Network n.d.) groups 100
organizations of 48 countries, and provides a library
of more than 6500 CPGs. CPGs aim to reduce
errors, unjustified practice variation and wasteful
commitment of resources, and encourage best
practices and accountability in medicine.
Moreover, the medical community has started to
recognize that a computer-based management of
CPGs can further increase CPG advantages,
providing relevant benefits (e.g. automatic
connection to the patient databases, and decision
making support) to care providers and patients.
In recent years, the research about computerized
guidelines has reached a relevant role within the
Medical Informatics community, and many different
approaches and projects have been developed to
create domain-independent computer-assisted tools
for managing, acquiring, representing and executing
computer-interpretable clinical guidelines
(henceforth CIGs). See e.g. the collections (Gordon
and Christensen 1995; Fridsma 2001; Ten Teije et
al. 2008; Peleg 2013)).
By definition, clinical guidelines address specific
clinical circumstances (i.e., specific diseases).
However, unfortunately, specific patients may be
affected by more than one disease. The treatment of
patient affected by multiple diseases (comorbid
patients) is one of the main challenges for the
modern healthcare, also due to the aging of
population, and the consequent increase of chronic
diseases. This sets up the urgent need of developing
ways of merging multiple single-disease
interventions to provide professionals’ assistance to
comorbid patients (Riaño and Collado 2013).
413
Piovesan L., Molino G. and Terenziani P..
Supporting Multi-level User-driven Detection of Guideline Interactions.
DOI: 10.5220/0005217404130422
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2015), pages 413-422
ISBN: 978-989-758-068-0
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

However, though some CPGs covering frequently
occurring comorbidities might be devised, there is a
need for formal methodologies to support physicians
in the detection and resolution of interactions
between guidelines, and, ultimately, in the process
of merging two or more guidelines. As a result, in
the very last years some computer-based approaches
have started to face this problem (see the discussion
in Section 5).
In this paper, we focus on one central issue in
the management of multiple CIGs, namely the
development of a methodology addressing
interaction detection. In a recent work (Piovesan et
al. 2014), we have identified three different
knowledge levels at which interactions might occur:
(i) level of the intentions of the CIG actions, (ii)
level of the effects of the drug categories
(recommended by the pharmaceutical actions in the
CIGs), and (iii) level of drugs. We have also pointed
out that, in turn, levels (i) and (ii) may be structured
at different levels of abstraction. Indeed, though a
large variety of representation formalisms exists
(Ten Teije et al. 2008), most CIG formalism support
a hierarchical decomposition of guidelines at
multiple levels of detail, in which composite actions
may be represented, and then refined (possibly at
different levels of abstraction) into their
components. At the finest level of detail therapeutic
pharmaceutical actions in the guideline may
recommend, depending on the accuracy, the use of
drugs or drugs categories, or active principles (thus,
also the interactions between drug categories must
be considered). In turn, drug categories are usually
structured in a hierarchy representing different levels
of detail (see, e.g., ATC (WHO Collaborating Centre
for Drug Statistics Methodology n.d.)). In (Piovesan
et al. 2014), we have also proposed an ontological
representation for the interactions at the different
levels, as well as an algorithm that, given two
actions (or drugs), automatically queries the
ontology to detect interactions between them.
The main goal of this paper is that of extending
the approach in (Piovesan et al. 2014) to provide
user physicians with a flexible support to navigation
and focusing (considering both CIGs and ontology,
at the different levels of detail), in order to
interactively identify actions/drugs on which
interaction analysis should be performed.
2 PROBLEMS AND
METHODOLOGY
The treatment of interactions between CIGs is a very
challenging one, involving difficult problems both at
the knowledge and at the process level.
At the knowledge level, two main limitations
have to be faced:
(K1) defining and acquiring “a priori” a new
guideline for any possible co-morbidity (i.e., for any
possible combination of two or more CIGs) is not
realistic in practice;
(K2) defining and acquiring, for each possible
pair of CIGs G1 and G2, and for each possible pair
of actions (a1G1, a2G2) the interactions between
them is practically unfeasible, too.
At the process level, an automatic process that,
considering two input CIGs G1 and G2, provides as
output the possible interactions between each
possible pair of actions (a1G1, a2G2) is
technically feasible, but practically useless for user
physicians, since the problem is combinatorial, and
too many interactions would be provided to the users
(consider, in particular, the usual dimensions of real-
world CIGs, and the number of alternative paths
they contain).
In this paper, we propose a methodology that
overcomes the above problems.
At the knowledge level, we (K1) consider CIGs
developed for single diseases, and (K2) employ an
ontology in which possible interactions between
actions (or, better, between their goals and
intentions) are modelled independently of the
specific CIGs.
At the process level, we propose a mixed-
initiative algorithm for the detection of interactions
which, taking in input two CIGs, (i) allows user
physicians to integrate their knowledge in order to
focus such detection on relevant sets of actions, and
(ii) exploits the (guideline independent) ontological
knowledge to find and analyse the interactions
between such focused parts.
Notice that the navigation and focusing phase is
interactive and physician-driven, and may be
facilitated by a user-friendly graphical interface. On
the other hand, the interactions between the focused
parts of the CIGs can be automatically provided by
the system (the navigation on the ontology and the
inferences on it are hidden to user-physicians). A
distinguishing feature of our approach is also
flexibility: it supports the navigation and selection of
the focus at different levels of abstraction.
The goal of this paper is to propose a system-
independent methodology. We only assume that (i)
CIGs can be structured at different levels of detail,
as a hierarchical graph, (ii) CIGs contain, besides
composite actions, also actions prescribing the
administration of drug categories (or, possibly, even
HEALTHINF2015-InternationalConferenceonHealthInformatics
414

specific drugs). Specifically, in the following, we
exemplify our approach considering the following
representation (used in GLARE): each action has a
type attribute, which is link to an ontological
concept, and at least one aimsTo attribute, linking it
to its relative intentions in the ontology.
Additionally, pharmaceutical actions have a
substance attribute, linking them to a drug category
or to a specific drug in the ontology.
Our implementation is based on the new version
of GLARE built within the GINSENG project
(Terenziani et al. 2014; Terenziani et al. 2013).
3 ONTOLOGY OF
INTERACTIONS
In (Piovesan et al. 2014), we fully detailed an
ontology of interactions, which is grounded on the
concepts of action goals, called intentions and of
administered drugs, which represent the main
sources of interactions between CIGs.
Before describing the knowledge representation,
it is worth stressing that our long-term goal is to
develop a decision support system highly
collaborative with the user. Following this
desideratum, the knowledge representation and the
inferences made on it should be human-friendly
enough to be understood by the user physician, as a
sort of “second opinion”. Indeed, in the medical
field, physician cannot trust “black box” tools that
simply output suggestions without explain to the
user (in an “understandable” way) how such
conclusions have been reached.
In our approach, action intentions are modelled
as desired variations, with certain modalities
(increasing, decreasing, stability), of some
parameters of the patient status (attributes).
Intentions are also organized along an ISA and
PART-OF taxonomy.
Like intentions, drugs are organized along a
taxonomy of drug categories, exhibiting, at the
bottom level, specific drugs. For such hierarchy, we
exploit existing classifications, such as the
consensus ATC (WHO Collaborating Centre for
Drug Statistics Methodology n.d.). Each drug (or
drug category) is related with effects it causes,
which are modelled as variations of patient’s status
attributes, just as the intentions.
As mentioned in the introduction, interactions
may occur at each level of abstraction (i.e., each
level of the two taxonomies), and the ontology
supports the representation of interactions at all the
levels.
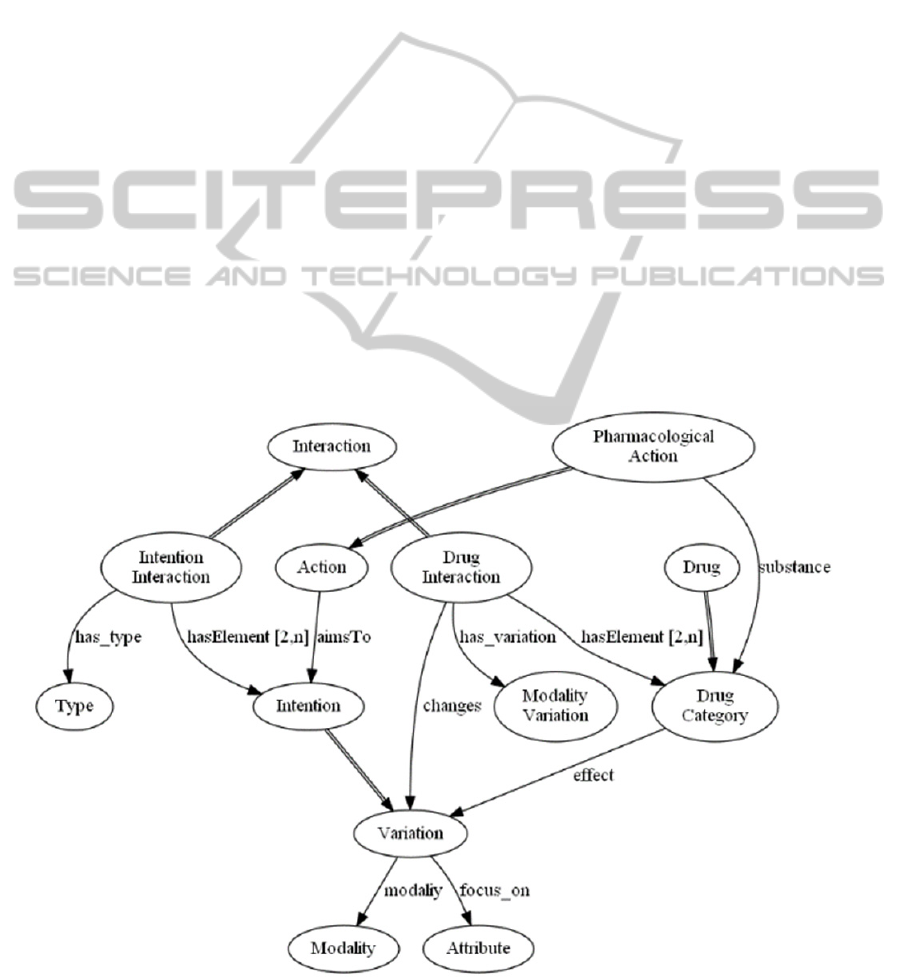
An Intention interaction is an interaction
between two intentions, and it is described by a type.
We identified three basic types: independence
(intentions do not interact), concordance (intentions
reinforce each other), and discordance (intentions
negatively interact with each other). However, more
“sharp” types of interactions can also be added. For
instance, the opposite type (as subtype of
discordance) could be added to cope with intentions
focusing on the same attribute, with opposite
modalities.
Drug interactions occur between two drugs or
drug categories. A drug interaction is characterized
by the modality of the variation that it causes in an
effect of the two drugs it involves.
Both types of interactions can be annotated by
links to the literature showing their evidence.
It is worth stressing that the interactions we
model are action and guideline independent because
they involve (action) intentions and drugs, which are
general concepts. Thus, differently from some other
approaches, when modelling a new CIG it is not
needed to specify all the interactions between the
new CIG and the existing ones because they are
autonomously recognized following the relations
between the actions and their intentions and drugs
prescribed (for pharmaceutical ones).
In the following, Figure 1 shows a glimpse of
part of the interaction ontology, focusing on the
modelling of interactions. At the moment, the
ontology has been validate using parts of guidelines
(see examples in this paper and in (Piovesan et al.
2014)). However, it is scheduled to be integrated in
METAGLARE (Terenziani et al. 2014), recently
developed.
4 A FLEXIBLE USER-DRIVEN
ALGORITHM FOR
INTERACTION DETECTION
AT MULTIPLE LEVELS OF
ABSTRACTION
4.1 Background
In this section, we describe our mixed-initiative
approach to the interaction detection between CIGs.
It is worth stressing that our main goal is to integrate
three fundamental knowledge sources:
(i) Knowledge deriving from the CIGs
structure, such as the decomposition of high
level actions in lower level ones, and the
sequence of actions to be executed
SupportingMulti-levelUser-drivenDetectionofGuidelineInteractions
415
(ii) Knowledge about intentions, drugs, and
their interactions. We detailed how we
organized such a basic medical knowledge
in the previous section
(iii) User physician knowledge about the context
and relevant parts of CIGs for the specific
case
In particular, given the high number of
alternative paths and actions for each CIGs, the
physician’s knowledge regarding the context is of
fundamental importance. Indeed, without
considering such an information, the output of an
autonomous tool could be not useful for the user: it
would contain too many non-organized information,
and most of them would be irrelevant for the specific
case. Exploiting out tool, the user physician can a
priori discard uninteresting parts of CIGs and focus
only on the relevant ones, obtaining, in addition, a
well-structured, easy-to-use and easy-to-analyse
output. In order to accomplish such a result, we aim
at devising a flexible and interactive detection tool
allowing physicians to navigate through the different
abstraction levels, thus supporting the natural
methodology they adopt to cope with CIG analysis.
For instance, at the highest level, a physician may
want to start to consider only the interactions
between the intentions of the high-level actions of
the guidelines. Then, focusing on a specific part of
the guideline, (s)he may want to move down to a
more detailed analysis, considering the
decomposition of composite action into its parts,
and/or the specific drugs category considered in
order to reach the high-level intentions. In general,
our approach will provide physicians with the
possibility of moving in both directions, i.e., going
down from a general to a more specific analysis, or
moving up, from a specific analysis to a higher level
of abstraction.
Another important contribution of our approach
is the possibility of extending CIGs with the
knowledge in the ontology. For instance, many
guidelines only recommend drug categories.
However, at the very end, a specific drug of that
category must be prescribed to the specific patient,
but there are many cases in which, while two drug
categories do not interact, specific drugs of the two
categories do interact. Such interactions can only be
detected in case the CIG knowledge is expanded
with the knowledge in the ontology.
4.2 Data Structures
To provide a flexible support, our algorithm must
rely on suitable data structures. In particular, such
Figure 1: Ontology of Intentions, Drugs and Interactions between them.
HEALTHINF2015-InternationalConferenceonHealthInformatics
416

structures should support the following desiderata:
(i) Maintain the history of the focusing
process, supporting both the addition of
new CIG focuses, and the rollback to upper
focuses
(ii) Maintain the association between CIG
focuses and interactions
As regards specifically the representation of
interactions, the data structure must also
(iii) Support the fact that interactions occur
between pair of actions in the two CIG
focuses
(iv) Support the fact that multiple interactions
may occur between each pair of actions
(since actions may have multiple intentions,
and drugs may have multiple effects)
To store the history, a tree structure is adopted.
Each node of the tree represents a “status” of the
analysis. It consists of three elements: (i) the focused
part of the first CPG (at the chosen abstraction
level), (ii), the focused part of the second CPG (at
the chosen abstraction level), and (iii) the list of
action interactions (found at the chosen level). In
turn, each “focused part” of a CPG simply consists
of a selection of nodes from the CPG itself.
Actions interactions derive from intention or
drug interactions. In particular, the list contains
(when filled by the algorithm) an interaction of two
actions a1 and a2 for each pair of intentions i1
(related to a1) and i2 (related to a2) that interact. In
addition, if a1 and a2 are pharmaceutical actions, the
list contains an interaction for each pair of drugs (or
drug categories) d1 (substance of a1) and d2
(substance of a2) that interact. The list maintains the
link to the ontological concepts, in order to allow the
user to examine the reasons of such conclusions.
Initially, the root of the tree contains the two
input CIGs (at the highest abstraction level) and the
list of interactions is empty. Then, the tree structure
expands to explicitly model (the results of) the
operations performed in a session of work (see, e.g.,
the graphical representation in Figures 2 and 3).
Navigation tree: tree of Views
View: <<CIG
1
,Focus
1
>,<CIG
2
,Focus
2
>,
ActionInteractions>
CIG
i
: a (possibly expanded) CIG, at the
abstraction level chosen by the user-
physician
Focus
i
: a subset of the action nodes in
CIG
i
ActionInteractions: {<action
i
, action
j
,
{Interactions}}
4.3 The Interaction Detection Tool
Our INTERACTION-DETECTION algorithms
support user-driven navigation over a navigation
tree, starting from a given node of the tree (i.e., from
the current view).
At the beginning of a session of work,
considering two guidelines CIG
1
and CIG
2
, the tree
is initialized with just the root node, consisting of
the view <<CIG
1
,{}>,<CIG
2
,{}>, {}> (i.e., at the
beginning, no focuses and no interactions are
identified), which is set as the current view.
At each step, the algorithm allows the user to
choose between four alternative actions:
STOP_ANALYSIS (which simply closes the session
of work), REFINE, ROLL-UP, and DETECT.
REFINE add a new view to the navigation tree (a
child of the current view), which becomes the new
current view. Such a new view is initialized as a
copy of the current view, but it is then refined by
refining actions and/or identifying focuses, through
the ZoomandSelect procedure (see below). On the
other hand, ROLL-UP moves up along the
navigation tree, setting the mother of the current
view as the new current view. Finally, DETECT add
the interactions between the focused actions to the
current view. The DETECT operation exploits the
links between the guideline action description and
the ontology and navigates the ontology in order to
find the modality of the interactions. A full
description of such an operation is reported in
(Piovesan et al. 2014).
Through the ZoomandSelect procedure, users
can iteratively refine a view, by changing the
focuses and/or expanding some of the actions they
contain. Four options are possible.
STOP_FOCUSING simply ends up the procedure.
ADD_TO_FOCUS add some actions in a CIG into
its focus, while REMOVE_FROM_FOCUS remove
actions from the focuses. EXPAND supports the
expansion of some of the actions in the focuses.
First, the user chooses the actions to expand
(variable actions_to_refine) then, while
actions_to_refine is not empty, each action a in it is
independently refined. The REFINEMENT
operation takes in input a view (v) and an action a in
it, and performs one step of refinement. The way in
which such a refinement is obtained depend on the
type of a. If a is a composite action in v, its
expansion is simply the sub-guideline constituted by
the actions composing it. On the other hand, if a is a
pharmaceutical action, the representation of a
contains the attribute substance, whose value is a
link to the ontological entity representing drug
SupportingMulti-levelUser-drivenDetectionofGuidelineInteractions
417

category to be administered (say DrugCat
x
). Thus, a
is expanded as a new piece of guideline, consisting
of several alternative pharmacological actions, one
for each one of the direct descendants of DrugCat
x
in
the ontology.
The REPLACE operation simply substitutes a
with the newly identified refinements into v.
Whenever a new refinement is added, we allow
users to update actions_to_refine, adding actions of
the new refinement to it. In such a way, we provide
users with the possibility of going on with this
process until the desired level of detail is reached.
algorithm INTERACTION-DETECTION
(nt: navigation_tree, current_view:
view)
let current_view be <<CIG
1
, F
1
>,
<CIG
2
, F
2
>, {I}>
begin
user_act the user chooses an
action;
while user_act • STOP_ANALYSIS do
begin
if user_act = “REFINE” then
begin
New_View generate_view();
New_View copy(current_view);
Append_child(New_View,
current_view);
ZoomandSelect(New_View);
current_view New_View;
end
if user_act = “ROLL-UP” then
current_view
mother_of(current_view);
if user_act = “DETECT” then
begin
current_view <<CIG1, F1>,
<CIG2, F2>,
ANALYSE_INTERACTIONS(F1,F2)>;
end
user_act the user chooses an
action;
end while
end
4.4 Graphical Interface
In the following, we explain how we integrated our
algorithms with a graphical interface, in order to
support users in the detection and analysis of
interactions at different levels of detail. The
graphical interface provides a user-friendly
interaction to physicians. In Figures 2 and 3, we
show how the navigation tree is displayed by the
graphical interface, considering as an example a
session of work. In particular, we compare a
simplified part of a CIG for the postoperative
management (PM), with a simplified part of a CIG
for the treatment of acute otitis media (AOM).
Each node in the navigation tree is represented by
three boxes: the first two show the guideline views
at the respective status of expansion, while the third
box contains the interactions detected between
actions contained in the views, when available.
At the beginning of the detection, the interface
only contains the root of the tree, with the guideline
views at the highest level of abstraction.
algorithm ZoomandSelect (v: view)
let v be <<CIG1, F1>, <CIG2, F2>,
{I}>
begin
user_act the user chooses an
action;
while user_act • STOP_FOCUSING do
begin
if user_act = “EXPAND” then
begin
actions_to_refine the user
selects the action(s)to expand
from F1 and F2;
while actions_to_refine {} do
begin
a take an action from
actions_to_refine and
remove it;
refinement REFINE(v, a);
REPLACE(a,refinement, v);
actions_to_refine
actions_to_refine
user selection of actions
from refinement;
end
end
if user_act = “ADD_TO_FOCUS” then
begin
F’ the user selects
action(s) to focus on from
CIG1;
F” the user selects
action(s)
to focus on from CIG2;
F1 F1
F’;
F2 F2
F”;
end
if user_act = “REMOVE_FROM_FOCUS”
then
begin
F’ the user selects
action(s)
to remove from F1;
F” the user selects
action(s)
to remove from F2;
F1 F1 - F’;
F2 F2 - F”;
end
end while
end
For the sake of simplicity, in our example we
suppose that the initial situation (with the guidelines
HEALTHINF2015-InternationalConferenceonHealthInformatics
418

Figure 2: Graphical interface for the INTERACTION DETECTION process. Each node of the tree represents a level of
expansion of the guidelines and may contain the detected interactions. The simulation continues in Figure 3.
at the highest level of abstraction) is represented as
shown in Figure 2.
In such a situation (part (1)), the physician may
want to compare the composite action “thrombosis
prevention” of the PM CIG with the “antibiotic
treatment” of the AOM one. In that case, (s)he
selects to refine the current node. The
ZoomandSelect procedure window, not shown in
this paper for the sake of brevity, allows her/him to
add the two actions to the focus and expand them.
We briefly explain how it works. First, considering
the PM CIG, the physician applies the
“ADD_TO_FOCUS” and “EXPAND” procedures to
the “thrombosis prevention” action. The result is the
expansion of such action. Among the other actions,
the composite action “thrombosis prevention”
recommends the administration of an antithrombotic
agent. Then, the physician decides to focus on such
action, and in particular, among the available
antithrombotic agents, on the administration of a
Vitamin K antagonist (repeating the procedures
“ADD_TO_FOCUS” and “EXPAND”). Now,
supposing that the guideline does not specify the
level of specific drugs, such expansion cannot be
performed using the CIG knowledge only. Then, the
system uses the knowledge in the ATC classification
in order to build the expansion of the selected action,
returning a decision between the administrations of
all the possible Vitamin K antagonists (e.g.,
dicoumarol, warfarin, etc.), which is the expansion
shown in Figure 3. At this point, the physician has
reached the desired level of abstraction and a similar
procedure is performed to expand the “antibiotic
treatment” node for the AOM CIG. When the
refinement is complete, the physician selects the
action STOP_FOCUSING and the node on the left
part of Figure 3 is created.
In the part (2) of Figure 3, we can see a
simplified expansion of the two selected actions.
Notice that the current expansion has been added to
the navigation tree as a child of the first one. At this
time, the third box of the node does not contain any
interaction. If the user performs a DETECT action,
interactions between the focused actions in the two
expansions of the guidelines are automatically
detected navigating the ontology and inserted in the
third box of the node (part (3) of Figure 3). An
interaction is detected between the administrations
of warfarin and erythromycin, which causes an
increase in the anticoagulant effect of the warfarin.
Now, we suppose that the physician is satisfied in
the exploration of this direction, and decide to
explore other possible interactions: (s)he perform a
ROLL-UP action, in order to set the root of the tree
as the current view (the node in 2 is however
maintained; see the part (4) in Figure 3). In the
example, we suppose that the user selects for the
new refinement the same action (“antibiotic
treatment”) for the AOM guideline, but another one,
“antibiotic prophylaxis” for the PM one.
The ZoomandSelect procedure at this time
produces the node in right part (5) of Figure 3, and
the navigation can go on until the desired level of
detail is reached, and the actions to be focused on
are decided. At this point, DETECT can be invoked
again, and so on. Obviously, the navigation tree can
expand at any depth, and there is no limit for the
possible alternative branches at any level, since (in
principle) all alternative expansions, at all levels, can
be explored and maintained in the navigation tree.
SupportingMulti-levelUser-drivenDetectionofGuidelineInteractions
419
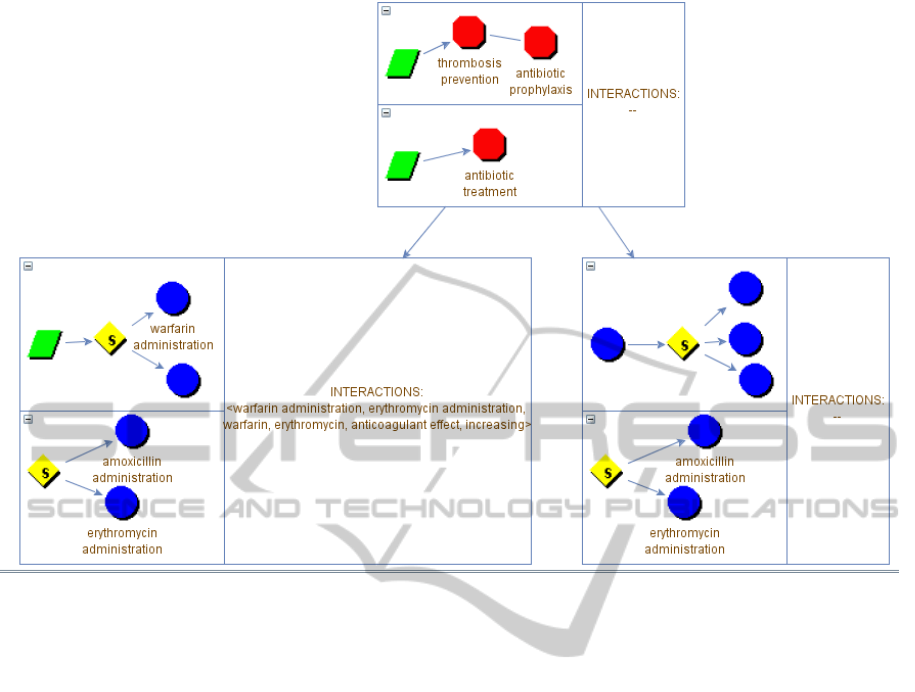
Figure 3: Second part of the simulation.
5 RELATED WORK AND
DISCUSSION
The development of software tools and metho-
dologies to support interaction detection is gaining
an increasing attention in the last years. For instance,
several current tools provide online access to
pharmacological databases (Robert Wood Johnson
Foundation and Partnership for Solutions 2004), or
alerting systems that detect and inform about
interactions (Medscape, Drugs.com, etc.). However,
such tools mostly focus on drug-drug interactions
only. Unfortunately, this is only a very limited
support, when a physician has to detect and analyse
the interactions between two or more guidelines.
To overcome such a limitation, several research
approaches have been proposed in the very last
years. As regards ontologies, GLINDA (Musen et al.
2011) proposes a wide ontology of cross-guideline
interactions.
On the other hand, several other approaches have
focused their attention on methodologies to “merge”
two or more CIGs, “solving” their interactions.
Sánchez-Garzón (Sánchez-Garzón et al. 2013), for
example, attempts to capture the collaborative aspect
of the merging: each guideline is considered as a
physician expert in the treatment of a single disease,
and represented by an agent with hierarchical
planning capabilities. The result is obtained through
the coordination of all the agents, and respects the
recommendations of each guideline. Another
interesting approach, presented in (Michalowski et
al. 2013) and (Wilk et al. 2013), uses constraint
logic programming to identify and address adverse
interactions. In this solution, a constraint logic
programming (CLP) model is derived from the
combination of logical models that represent each
CIG, then a mitigation algorithm is applied to detect
and mitigate interactions. Among rule-based
systems, (López-Vallverdú et al. 2013) represents
guidelines as sets of clinical actions that are
modelled into an ontology. To combine two
treatments, first they are unified in a unique
treatment, then a set of “combination rules” is
applied to detect and avoid possible interactions. A
model-based combination of CIGs is purposed in
(Riaño and Collado 2013), in which guidelines
expressed in a particular formalism can be
automatically merged through a combining operator.
Jafarpour (2013) uses semantic-web rules and an
ontology for the merging criteria. Given these, an
HEALTHINF2015-InternationalConferenceonHealthInformatics
420

Execution Engine dynamically merges several CIGs
according to merge criteria.
On the other hand, the approach in this paper
focuses on interaction detection, rather than on
guideline merge. In this sense, our approach is
largely complementary with respect to the above
approaches in the literature, and can be integrated
with them. However, two main advantages of the
approach in this paper are (i) the fact that it is
flexible, interactive and user-driven: instead or
proposing “black-box solutions” to physicians, we
aim at providing them with user-friendly
investigation and decision supports; (2) the fact that
is allows the analysis at different levels of detail.
The approach in this paper is based on (Piovesan
et al., 2014). Indeed, as discussed in Section 2, an
automatic process that provides as output the
possible interactions between each possible pair of
actions between two CIGs is practically useless for
user-physicians, since the problem is combinatorial,
and too many interactions would be provided as
output. Thus, we suggest to split interaction analysis
into two phases, which can be iteratively repeated in
an interactive and physician-driven process: (1)
focus on specific actions/drugs (at a specific level of
detail), and (2) detect interactions on them. While
the work in (Piovesan et al., 2014) mainly focuses
on the second phase, in this paper we extend it to
cope with the first one. To achieve such a goal, this
paper presents three major original contributions: (1)
analysis of the requirements for the data structures,
and their definition (see the navigation tree, in
Section 4.2); (2) definition of a flexible and
interactive focusing algorithm (Section 4.3); (3)
definition of a user-friendly graphical interface
(Section 4.4). The main limitation of the current
approach is, in our opinion, the fact that it has only
undergone a limited experimental evaluation. Up to
now, it has been tested only on simplified guidelines
or part of them, such as the ones described in the
Section 4, by two physicians of Azienda Ospedaliera
San Giovanni Battista (“Molinette” Hospital) in
Turin. Though the test has been quite successful, a
more systematic and intensive experimental
evaluation should be required, and this is the goal of
our future work.
6 CONCLUSIONS
The treatment of patients affected by multiple
diseases (comorbid patients) is one of the main
challenges for the modern healthcare, also due to the
aging of population, and to the increase of chronic
diseases. Recent studies demonstrates that various
types of interactions must be taken into account
when merging two (or more) CIGs, and propose and
ontology of interactions (Piovesan et al. 2014;
Musen et al. 2011). However, to the best of our
knowledge, our approach is the first one that, having
identified different levels of abstractions in the
analysis of interactions, supports user-driven and
interactive interaction detection over them.
Our flexible approach to interaction detection,
operating at different levels of abstractions, may
support expert physicians to analyse “abstractly”
(i.e., just considering the CIGs, with no reference to
a specific patient) the interactions between two or
more CIGs that are commonly used together (e.g., to
provide some “partial merge” between them). In
such a case, the possibility of reasoning about “high-
level” actions is certainly crucial. Moreover, our
approach can also support a physician treating a
specific comorbid patient. In such a context, though
the abstraction facilities are certainly helpful, the
possibility of moving from the “general” actions in
the guidelines to study of the interactions of specific
drug categories (and drugs) can play a crucial role.
In our short-term future work, we aim at
proposing a more extensive experimental evaluation
of the current approach, and at extending it to cope
also with “patient-guideline action” interactions and
“patient-drug” interactions, and with the temporal
issues (e.g., not all interactions between CIGs are
possible, due to the temporal constraints between
guideline actions). In our long-term future work, we
will support physicians also in the interaction
solving, and, finally, in merging multiple guidelines
in the treatment of a specific patient.
ACKNOWLEDGEMENTS
The work described in paper was partially supported
by Compagnia di San Paolo, in the Ginseng project.
REFERENCES
Committee to Advise the Public Health Service on
Clinical Practice Guidelines, Institute of Medicine,
1990. Clinical practice guidelines directions for a new
program M. J. Field and K. N. Lohr, eds.,
Washington, D.C.: National Academy Press. Available
at: http://site.ebrary.com/id/10068353 [Accessed July
28, 2014].
Fridsma, D.B., 2001. Special Issue on Workflow
Management and Clinical Guidelines. Journal of the
SupportingMulti-levelUser-drivenDetectionofGuidelineInteractions
421

American Medical Informatics Association, 22(1),
pp.1–80.
Gordon, C. and Christensen, J.P. eds., 1995. Health
telematics for clinical guidelines and protocols,
Amsterdam, Netherlands: IOS Press.
Guidelines International Network, Guidelines
International Network website. Available at:
http://www.g-i-n.net/ [Accessed October 14, 2014].
Jafarpour, B. and Abidi, S.S.R., 2013. Merging Disease-
Specific Clinical Guidelines to Handle Comorbidities
in a Clinical Decision Support Setting. In AIME. pp.
28–32.
López-Vallverdú, J.A., Riaño, D. and Collado, A., 2013.
Rule-Based Combination of Comorbid Treatments for
Chronic Diseases Applied to Hypertension, Diabetes
Mellitus and Heart Failure. In R. Lenz et al., eds.
Process Support and Knowledge Representation in
Health Care. Lecture Notes in Computer Science.
Springer Berlin Heidelberg, pp. 30–41.
Michalowski, M. et al., 2013. Using Constraint Logic
Programming to Implement Iterative Actions and
Numerical Measures during Mitigation of
Concurrently Applied Clinical Practice Guidelines. In
N. Peek, R. M. Morales, and M. Peleg, eds. Artificial
Intelligence in Medicine. Lecture Notes in Computer
Science. Springer Berlin Heidelberg, pp. 17–22.
Musen, M.A. et al., 2011. GLINDA Interaction Ontology.
Available at: http://glinda-project.stanford.edu/
guidelineinteractionontology.html [Accessed October
17, 2014].
Peleg, M., 2013. Computer-interpretable clinical
guidelines: A methodological review. Journal of
Biomedical Informatics, 46(4), pp.744–763.
Piovesan, L., Molino, G. and Terenziani, P., 2014. An
ontological knowledge and multiple abstraction level
decision support system in healthcare. Decision
Analytics, 1(1). Available at: http://www.decision
analyticsjournal.com/content/1/1/8.
Riaño, D. and Collado, A., 2013. Model-Based
Combination of Treatments for the Management of
Chronic Comorbid Patients. In Artificial Intelligence
in Medicine. 14th Conference on Artificial Intelligence
in Medicine. Springer, pp. 11–16.
Robert Wood Johnson Foundation and Partnership for
Solutions, 2004. Chronic Conditions: Making the Case
for Ongoing Care. Available at: Chronic Conditions:
Making the Case for Ongoing Care [Accessed October
19, 2014].
Sánchez-Garzón, I. et al., 2013. A Multi-agent Planning
Approach for the Generation of Personalized
Treatment Plans of Comorbid Patients. In N. Peek, R.
Marín Morales, and M. Peleg, eds. Artificial
Intelligence in Medicine. Berlin, Heidelberg: Springer
Berlin Heidelberg, pp. 23–27. Available at:
http://link.springer.com/10.1007/978-3-642-38326-
7_4 [Accessed October 17, 2014].
Ten Teije, A., Miksch, S. and Lucas, P. eds., 2008.
Computer-based medical guidelines and protocols: a
primer and current trends, Amsterdam, Netherlands:
IOS Press.
Terenziani, P. et al., 2013. Towards a Second Generation
of Computer Interpretable Guidelines. In M. Helfert,
C. Francalanci, and J. Filipe, eds. DATA 2013 -
Proceedings of the 2nd International Conference on
Data Technologies and Applications, Reykjavík,
Iceland, 29 - 31 July, 2013. SciTePress, pp. 199–205.
Terenziani, P., Bottrighi, A. and Rubrichi, S., 2014.
META-GLARE: a meta-system for defining your own
CIG system: Architecture and Acquisition. In 6th
International Workshop on Knowledge Representation
for Health Care. pp. 92–107.
WHO Collaborating Centre for Drug Statistics
Methodology, Anatomical Therapeutic Chemical
classification system. Available at: http://www.whocc.
no/atc/ [Accessed October 14, 2014].
Wilk, S. et al., 2013. Mitigation of adverse interactions in
pairs of clinical practice guidelines using constraint
logic programming. Journal of biomedical
informatics, 46(2), pp.341–353.
HEALTHINF2015-InternationalConferenceonHealthInformatics
422
