
A Knowledge Based Framework for Case-specific Diagnosis
Ganesh Ram Santhanam, Gopalakrishnan Sivaprakasam, Giora Slutzki and Samik Basu
Iowa State University, Ames, IA 50014, U.S.A.
Keywords:
Diagnosis, Knowledge Bases, Description Logic, Qualitative Reasoning.
Abstract:
We present a framework whereby the expert knowledge of a domain is represented as a description logic
knowledge base. Based on this framework, we present an approach that uses a knowledge based system for
diagnosis that allows users to key in findings for a case, and obtain the corresponding differential diagnosis
for a case. The framework also prompts hypothetical findings that can effectively guide the user towards a
targeted diagnosis. The framework allows iterative and interactive updates of the case specific knowledge.
Computing the differential diagnosis and hypotheses can be formulated directly as conjunctive queries on the
original knowledge base using the case specific knowledge. We illustrate the applicability of our framework
in the context of medical diagnosis, although the approach is equally applicable in a broad range of diagnosis
problems such as network forensics and criminal investigation.
1 INTRODUCTION
In many diagnostic applications, expert domain
knowledge can be represented by a knowledge base
(KB) that consists of a set of findings, a set of con-
sequences, evidential relationships relating each find-
ing to the likely consequences it indicates, and possi-
bly other relationships between findings and diseases.
Given a subset of findings that hold true in a partic-
ular ‘case’, case-specific diagnosis is the process of
identifying the most likely consequences, called the
differential diagnosis (Long et al., 1988; Miller et al.,
1982). For example, in clinical diagnostic decision
support (Szolovits et al., 1988; Ely et al., 2011), a
doctor looks to narrow down the disease(s) that are
most likely diagnoses given a set of symptoms (case)
(Barnett et al., 1987; Ramnarayan et al., 2003). Simi-
larly, the police may want to identify a set of suspects
that are likely to have committed a crime given a set of
supporting evidences (case) relating to the crime; and
computer network administrators may want to dis-
cover suspected host(s) and the potential role(s) they
could have played in launching a coordinated attack
(Wang and Daniels, 2008; Pilli et al., 2010), given a
set of relevant detected network events.
In this paper, we present a knowledge representa-
tion and reasoning system for case-specific diagnosis
that guides a user progressively towards the most sat-
isfactory (differential) diagnosis by iteratively query-
ing the KB with respect to the evolving case. The
contributions of this paper with respect to past work
on diagnosis (Szolovits et al., 1988; Sohrabi et al.,
2010; Baral et al., 2000) are as follows.
1. We provide a knowledge based framework for dif-
ferential diagnosis where conjunctive queries are
used to reason about domain knowledge that is
relevant to a given case, taking into consideration
case-specific evolution of findings.
2. We allow case-specific knowledge to evolve by
the addition of findings (e.g., a patient may
present new symptoms), and treat case-specific di-
agnosis as an iterative process of recomputing and
refining the differential diagnosis based on new
test results relevant to the case.
3. Moreover, we treat case-specific diagnosis as an
interactive process, i.e., a user can change the
case-specific knowledge by adding different sets
of findings during the course of diagnosis, lead-
ing to the corresponding changes in the differen-
tial diagnoses in the subsequent iteration.
Organization. The rest of the paper is organized as
follows. Section 2 describes a medical diagnosis tool
called Dxplain used to demonstrate our framework,
and how domain knowledge can be represented as a
Description Logic (DL) knowledge base. Section 3
describes how we reason with domain knowledge us-
ing DL conjunctive queries. Section 4 discusses how
conjunctive queries can be extended to compute sug-
gested hypotheses during diagnosis. Section 5 sum-
marizes and discusses directions for future research.
482
Ram Santhanam G., Sivaprakasam G., Slutzki G. and Basu S..
A Knowledge Based Framework for Case-specific Diagnosis.
DOI: 10.5220/0005272104820488
In Proceedings of the International Conference on Agents and Artificial Intelligence (ICAART-2015), pages 482-488
ISBN: 978-989-758-074-1
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

2 BACKGROUND
We present our framework in the context of medical
diagnosis, where the basic knowledge includes facts
about the findings/symptoms, the diseases and their
relationships.
2.1 Case Study: Medical Diagnosis
Dxplain (Barnett et al., 1987; Hoffer et al., 2005) is an
expert system for clinical diagnostic decision support
developed and enhanced by the Massachusetts Gen-
eral Hospital since 1984. The Dxplain KB consists of
5000 clinical findings and 2400 diseases and their re-
lationships (averaging about 50 findings per disease).
In the ‘case analysis mode,’ DXplain uses a graphical
interface to collect findings of a case from a user (doc-
tor) and produces a list of diseases that are likely to be
the consequences of the given findings. DXplain also
provides for each disease a set of findings that sup-
port that disease at various levels of intensity. Dxplain
uses Bayesian probabilistic estimates of likelihood in
order to quantify the strength of evidence of a finding
to a disease and rank diseases. In contrast, the pro-
posed framework will allow users to address different
types of questions using the DL query constructs, thus
broadening the scope of differential diagnosis beyond
what is achievable using Dxplain.
2.2 Semantic Information in Dxplain
Findings. A finding is a clinical manifestation (clin-
ical symptoms, signs, epidemiological data, personal
biodata) or the result of laboratory tests. Each finding
has an attribute ‘Finding Importance’ that can hold
values on a totally ordered (ordinal) scale. This at-
tribute indicates how important it is to explain the
presence of the finding (considering its significance).
For example, a runny nose is less important than acute
right abdominal pain.
Diseases. Each disease is described by three at-
tributes.
1. Disease Importance, indicates the rating on a to-
tally ordered value scale from 1 to 5 (higher values
signifying higher importance).
2. Disease Prevalence, indicates how common the
disease is on a totally ordered (ordinal) scale:
Very Common, Common, Rare and Very Rare.
3. Disease Urgency, indicates whether urgent ac-
tion is needed or not, if the disease was actually
present.
Table 1: Attributes and their Ordinal Scales.
Attribute Label Range
Finding Importance η
f
{η
f
1
, η
f
2
, η
f
3
, η
f
4
, η
f
5
}
Disease Importance η
d
{η
d
1
, η
d
2
, η
d
3
, η
d
4
, η
d
5
}
Disease Prevalence φ
d
{φ
d
1
, φ
d
2
, φ
d
3
, φ
d
4
}
Disease Urgency υ
d
{Urgent, Normal}
Finding Frequency µ
f d
{Indicates,
Eliminates}
Strength of Support σ
f d
{σ
−1
, σ
0
, σ
1
, σ
2
}
Finding - Disease Relationships. A relationship be-
tween a finding and a disease signifies that the finding
either increases or diminishes the support (evidence)
for the existence of that disease. This relationship is
described by two attributes.
1. Finding Frequency/Correlation, indicates how
frequently the finding is seen in the disease, or
whether it rules out the disease. This attribute is
evaluated on a qualitative scale ‘indicates’ to de-
note that the disease should always be considered
given the finding; and ‘eliminates’ to denote that
the presence of the finding excludes the disease.
2. Strength of Support, indicates how strongly the
finding suggests or supports the disease. This is
evaluated on a qualitative (ordinal) scale (Sup-
ports / Strongly supports / Very strongly supports;
or Supports Absence
1
).
Disease - Disease Relationship: Similarity. For
each disease d, Dxplain maintains a set of similar dis-
eases that should be considered if d is being consid-
ered in the diagnosis.
The attributes of the findings, diseases and their
relationships, and their values on an ordinal (rather
than cardinal) scale are defined in Table 1.
2.3 Knowledge Base Example
We illustrate and motivate representation and reason-
ing about domain and case-specific knowledge in the
proposed system using a simplified example of a med-
ical KB
2
with 5 findings and 4 diseases. The graph
representation of the KB as well as the evolution of
the case in terms of the findings of a patient entered
by a doctor is shown in Figure 1. Note that f
1
sup-
ports, and f
3
supports the absence of d
1
; f
2
strongly
supports, f
3
very strongly supports, and f
4
supports
the absence of d
2
; f
4
supports d
3
; f
5
very strongly
1
Supports the absence of a disease.
2
The KB in this example is simplified compared to the
actual medical KB in Dxplain or other tools such as Isabel.
AKnowledgeBasedFrameworkforCase-specificDiagnosis
483
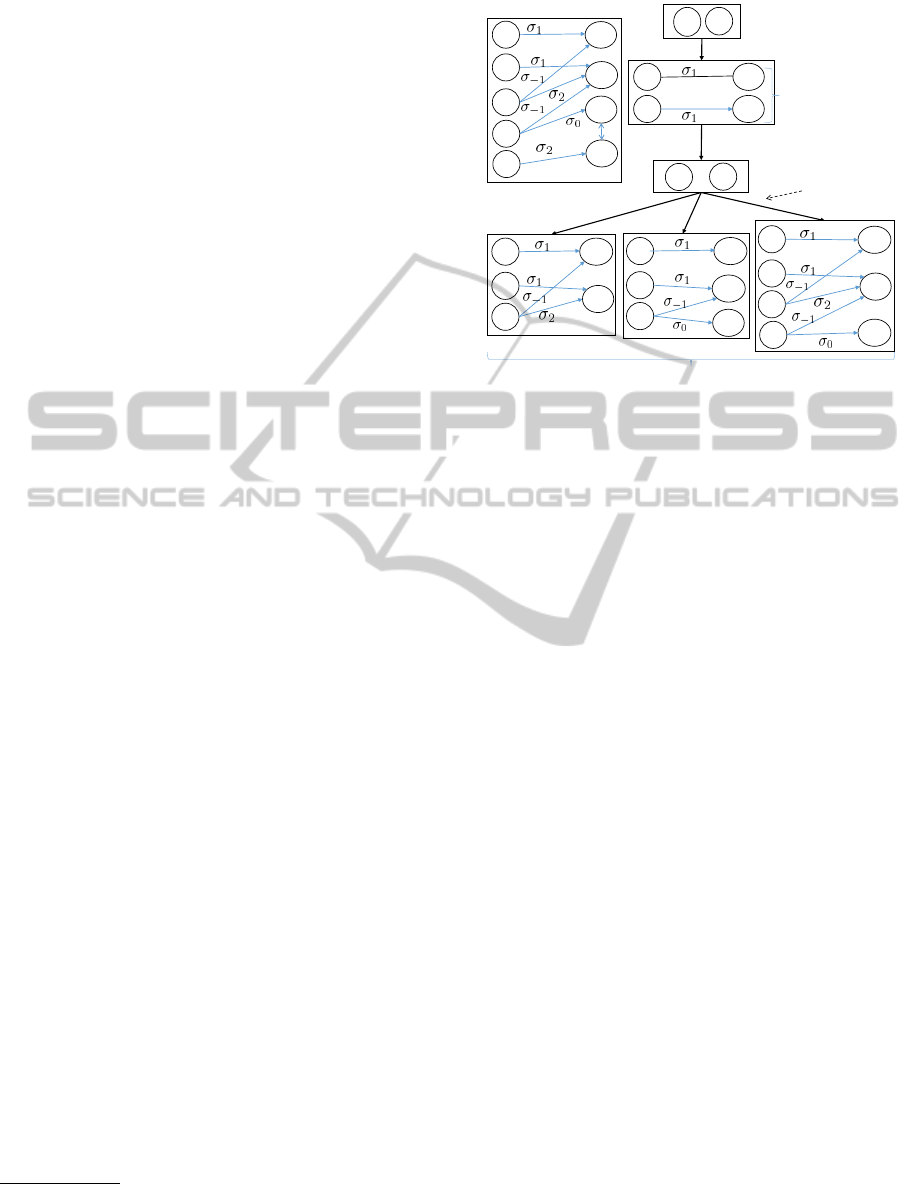
supports d
4
; and d
3
and d
4
are similar to each other.
The sequence of steps taken by the doctor (D) and the
system (S) in tandem during diagnosis is as follows.
• [D:] Provide Initial Case Findings. Suppose the
doctor provides the initial case findings for a pa-
tient, say {f
1
, f
2
}.
• [S:] The system then performs two computations:
– Differential Diagnosis. The system computes
the differential diagnosis {d
1
, d
2
} because f
1
and f
2
support d
1
and d
2
respectively. Note
that d
1
and d
2
cannot be distinguished based on
the available evidence, as the level of support
offered by the respective findings in the case is
σ
1
.
– Hypothesis Generation. Further, the system
suggests a set {f
3
, f
4
} of hypothetical findings
back to the doctor. The finding f
3
is suggested
because it enhances the support for d
2
while di-
minishing support for d
1
and f
4
is suggested
because it diminishes the support for d
2
, leav-
ing the support for d
1
unchanged.
• [D:] Update of Findings. On being presented
with the differential diagnosis and the correspond-
ing hypotheses, the doctor may choose to test (by
a physical exam, ordering a laboratory procedure
such as a blood test, etc.) the presence or absence
of some of them based on his own expertise or ex-
perience. When the results are known, the doctor
may update
3
the case findings by adding one or
more of the suggested hypothesis that tested posi-
tive for the patient and remove the ones for which
the patient tested negative.
• [S:] Recomputation of Differential Diagnosis.
Consider the scenario, where the doctor adds f
3
(as its presence is validated). This results in the
case scenario presented in the bottom-left of Fig-
ure 1. Note that the addition of f
3
adds no new
diseases, but changes the level of support to d
1
and d
2
in a way that allows the doctor to infer that
d
2
is more likely than d
1
. On the other hand, if f
4
were added to the case, then it can lead to a dif-
ferential diagnosis by which d
1
may be inferred
to be more likely than d
2
, in addition to the inclu-
sion of d
3
in the differential diagnosis. Finally, if
both f
3
and f
4
are added to the case, then the re-
sulting differential diagnosis includes d
3
, and the
the doctor may infer that d
2
is more likely than d
1
given the evidence.
3
While the hypotheses only provide suggestions for tests
that the doctor may order, updates are based on results of
such tests.
Add f
3
,f
4
Add f
4
Add f
3
f
1
f
2
f
4
d
1
d
2
d
3
f
1
f
2
d
1
d
2
f
1
f
2
f
3
f
5
d
1
d
2
d
3
d
4
f
4
f
1
f
2
f
3
d
1
d
2
f
4
d
3
f
1
f
2
f
3
d
1
d
2
Original Domain Knowledge
Compute Differential Diagnosis
Possible Evolutions of Case-Specific Knowledge
Differential
Diagnosis
f
1
f
2
(Initial Case)
f
3
f
4
Suggested Hypotheses
Compute Hypothetical Findings
Interactive Step
Figure 1: Original KB and Case Analysis Example.
In the above example, at each step the addition
of new findings results in computation of (a) the new
differential diagnosis; and (b) a set of hypothetical
findings relevant to the case. Note that the doctor
can choose various subsets of the set of hypotheses
presented to him in the case, each leading to possi-
bly different differential diagnoses in the successive
steps. In the following section, we present the DL KB
for capturing the domain-specific and case-specific
knowledge followed by the queries to the knowledge
base that computes the differential diagnosis and hy-
potheses.
3 REASONING WITH KB USING
DLs
We assume basic knowledge of DLs (Baader et al.,
2003) for the rest of the paper. A DL KB is a tuple
K = hA, T , Ri where A is an ABox, T is a TBox
and R is an RBox. The TBox and RBox in K con-
stitute the schema or structure underlying the domain
knowledge and the ABox specifically contains knowl-
edge about individuals in K. A is a set of concept and
role assertions of the form C(a) and r(a, b) (unary
and binary predicates) respectively, where C is a con-
cept, r is a role, and a and b are individuals. T and R
are sets of terminological axioms that state how con-
cepts and roles are related to each other (respectively).
3.1 Diagnostic Knowledge as a DL KB
We now illustrate how to model the domain knowl-
edge in terms of a DL KB K = hA, T , Ri for our
ICAART2015-InternationalConferenceonAgentsandArtificialIntelligence
484

running example from the domain of medical diagno-
sis. The concept and role assertions in the ABox A
and the list of concept and role inclusions in T and R
are given in Figure 2.
Concepts and Roles for Domain Knowledge. The
concept names ϕ and δ in K correspond to individual
findings and diseases respectively. The role names
σ
−1
, σ
0
, σ
1
and σ
2
correspond to relationships be-
tween findings and diseases in the original domain
knowledge with the respective evidential strengths in-
dicated by the corresponding subscripts. These con-
stitute generic domain knowledge common to all the
conceivable cases to be analyzed.
Concepts to Encode Attributes of Findings and
Diseases. The attributes of findings and diseases, in-
cluding finding and disease importance, finding fre-
quency, disease urgency, etc. are encoded as general
concepts in K. For example, K will have concepts
η
d
1
, . . . , η
d
5
, η
f
1
, . . . , η
f
5
, µ
f
, υ
d
, etc., corresponding to
the labeling functions (see Table 1).
Concept Hierarchy. Attributes such as finding and
disease importance have different levels of valuations
in Dxplain that are totally ordered. We model one
general concept for each valuation of that attribute
and whenever appropriate, impose the total ordering
over these valuations using concept inclusions. For
example, disease importance is an attribute that has
valuations from 1 to 5, and we model this with inclu-
sions η
d
i
v η
d
i+1
for i = 1, . . . , 4 in T .
Role Hierarchy. Roles such as σ
1
, σ
2
, etc. indicate
various degrees of relationship, e.g., strength of sup-
port of a disease by a finding. We model the fact that
one role represents a higher degree of support than an-
other by role hierarchies. For example, to model the
fact that a finding f which very strongly supports a
disease d also strongly supports it, we include a role
inclusion axiom σ
2
v σ
1
in the RBox R. Note that
σ
−1
is a role that denotes that the finding offers evi-
dence in support of the absence of the disease.
We assume that assertions in K pertaining to the
above concepts and roles are not updated during the
course of case analysis in Dxplain, as they constitute
domain knowledge that is not expected to change dur-
ing the course of the diagnosis.
Concepts for Case-specific Knowledge. As men-
tioned above, a case is a set of findings, that may in-
clude (a) Positive Findings that are marked as present
in the case; (b) Negative Findings that are marked
to be absent in the case; and (c) Unknown Findings
that are marked as being of no interest to the case.
The concept names ϕ
+
, ϕ
−
and ϕ
u
correspond to
Figure 2: DL Encoding of Original Domain KB K.
the positive, negative and unknown case-specific find-
ings. For example, during case analysis, when a doc-
tor inputs a finding as present (absent or unknown)
for the current case, the corresponding finding in K is
labeled with the concept ϕ
+
(ϕ
−
or ϕ
u
respectively).
We also define a concept ϕ
c
= ϕ
+
tϕ
−
tϕ
u
that cor-
responds to the set of all findings (positive, negative
or unknown) in the case.
3.2 Case-Specific Diagnosis
A new case is initiated by the doctor by keying in a set
C of positive and negative findings. The system then
queries the KB to obtain (a) the differential diagnosis
D(C)and (b) a set of suggested findings or hypotheses
denoted by H(C) that are not already in the case (and
possibly could help refine the diagnosis by increasing
or decreasing the evidential support of, or eliminating
one or more diseases in the differential).
Conjunctive Queries for DL Reasoning. In order to
compute D(C) and H(C) in each iteration, we con-
struct appropriate conjunctive queries on K. A con-
junctive query (CQ) (Glimm et al., 2008) is an ex-
istentially quantified conjunction of atoms on a set
of variables V such that each atom asserts a con-
cept membership or role relationship between one or
more of the variables (or individuals). A CQ can op-
tionally have a free variable (not in V ), certain val-
uations of which may satisfy the CQ. For example,
∃y, z.Q(x) ∧ P (x, y) ∧ P (y, z) ∧ P (z, x) is a CQ
where V = {y, z}, x is a free variable, and Q and P
correspond to a concept and a role respectively. The
CQ
P (d) = ∃f. ϕ
c
(f) ∧ δ(d) ∧ σ
0
(f, d)
states that there exists a finding in the case C that sup-
ports some disease d in the domain. The variable d is
free in the query, resulting in a set D(C) representing
AKnowledgeBasedFrameworkforCase-specificDiagnosis
485
the differential diagnosis. Note that, in view of the
role hierarchy specified in R, the last conjunct σ
0
ac-
counts for support offered by f to d at any level. Once
P (d) is computed, the system adds each such disease
d to the case-specific knowledge using the concept δ
c
,
i.e., an assertion δ
c
(d) is added to the case-specific
ABox. We next formulate the generation of hypothe-
ses as a CQ that obtains all findings in K that provide
either positive support of support of the absence of
any of the existing diseases in the differential diagno-
sis D(C).
Q(f) = ∃d. ¬ϕ
c
(f) ∧ δ
c
(d) ∧ σ(f, d)
The above CQ states that there exists a disease d
in the differential diagnosis D(C) that is offered some
evidential support by a finding f that is not present
in the case findings C. Note that the role σ is de-
fined in R as one that includes all types of evidential
support (including support of the absence of disease)
from findings and diseases. Since f is a free variable,
Q(f) obtains a set of findings exactly corresponding
to those in H(C). Also, the negation in the first term
(checking that f is not in the case-specific knwoledge
so far) can be viewed as a syntactic construct, and
hence will not make query answering undecidable.
Once a disease differential D(C) and the set
H(C) of suggested findings are presented to the user,
the user may chose to proceed by changing the case in
one of two ways: (a) by marking one or more of the
suggested findings as Positive, Negative or Unknown,
or (b) by including findings that are not in H(C) as
Positive or Negative for the case. This triggers a new
iteration for computing D(C
0
) and H(C
0
) with re-
spect to new case specific findings C
0
. The user con-
tinues to change the case iteratively until he/she is sat-
isfied with the obtained disease differential (i.e., the
diagnosis is satisfactory).
We represent the initial set of findings for a case
by the ABox C
0
. Further, we view iterative addition
of findings to the case as a sequence of ABoxes begin-
ning with C
0
, namely C
1
, C
2
, . . . , C
n
. C
n
is an ABox
that provides a differential diagnosis that satisfies a
target criteria defined by the user. For instance, the
user may want to include at least one serious disease
with very high level of evidence from case findings.
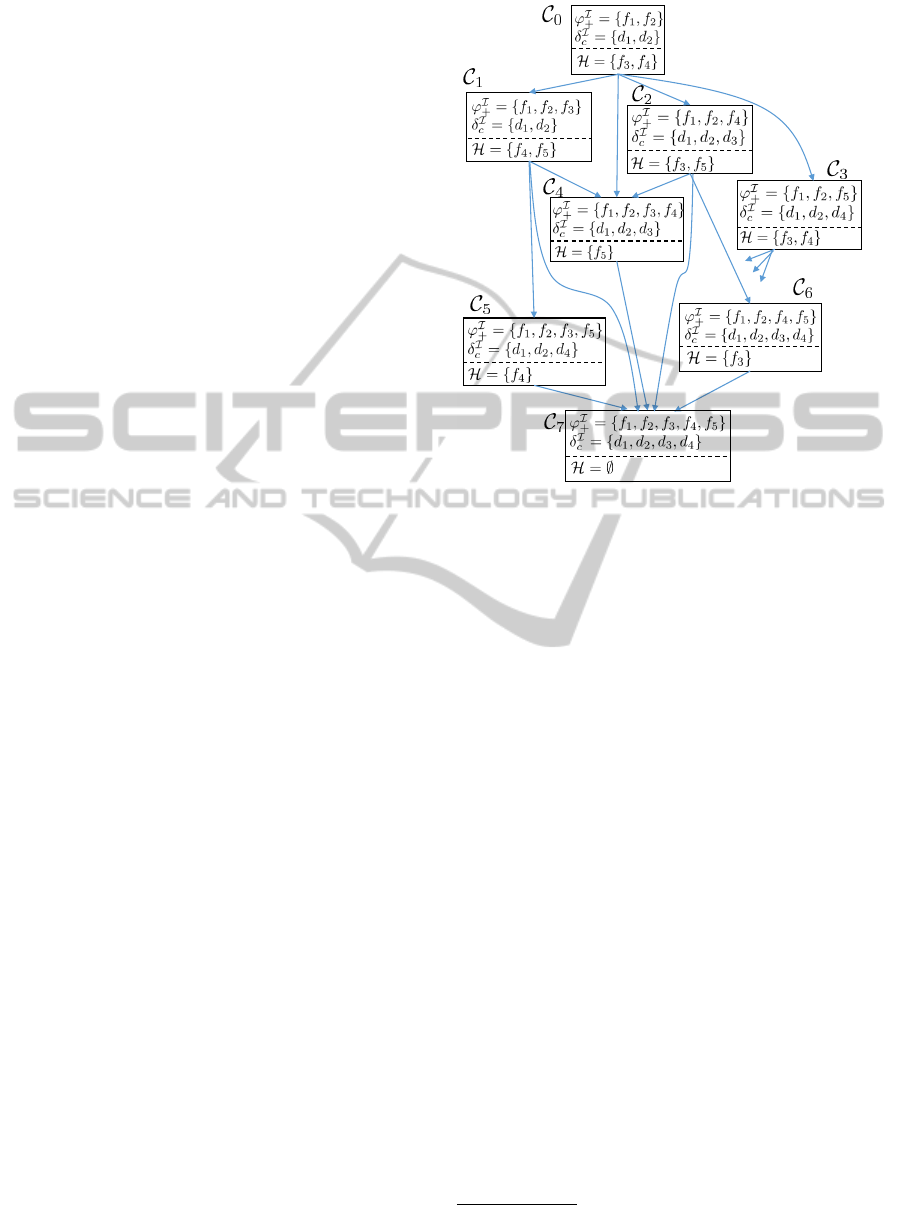
Figure 3 illustrates a possible evolution of ABoxes
for the running example in Figure 1. The evolution
corresponds to the successive addition of a set of pos-
itive findings to the initial case. Each ABox contains
assertions (δ
c
) corresponding to the diseases that are
included in the differential diagnosis for the current
set of case findings (ϕ
+
). For readability, each ABox
in Figure 3 shows only assertions that correspond to
case-specific knowledge during the iteration.
Figure 3: Evolution of ABoxes during Case Analysis for
the example in Figure 1. The contents of the ABoxes are
shown in terms of the interpretation of concepts rather than
the assertions (e.g., δ
c
(d
1
), δ
c
(d
2
)) themselves. Each ABox
is also annotated with a set H of hypotheses.
4 FINDING TARGETED
HYPOTHESIS
The goal of providing suggested hypotheses to the
doctor is to prompt the doctor about case information
(findings), whose inclusion into the case may alter the
differential diagnosis in desired ways. In the previous
section, we presented one way to compute H(C) as
the set of findings that either increase or decrease the
evidential support for one or more diseases in D(C).
There may be other potentially useful hypotheses that
help the doctor navigate the space of differential di-
agnoses as desired, during case evolution; here we
present three such examples.
a. A doctor may have a strong suspicion that one of
the diseases, d ∈ D(C) is actually present in the
patient. In such a case, the doctor may find it use-
ful if prompted (only) with the findings that are
known to increase the evidence for d (based on
the original domain knowledge). Given a disease
d, the following CQ obtains findings f (free vari-
able) that are not already in the case C, which also
very strongly
4
support disease d.
4
Note that this is only one possible way to obtain a set
of findings that strongly support a specific disease d.
ICAART2015-InternationalConferenceonAgentsandArtificialIntelligence
486

Q
0
d
(f) = ¬ϕ
c
(f) ∧ δ
c
(d) ∧ σ
2
(f, d)
b. A doctor may choose to first expand the differen-
tial diagnosis to include diseases that are of high
importance level (e.g., require immediate atten-
tion) and are possible given the case. Recall that
such diseases are labeled with the concept η
d
5
in
K. We compute the desired hypotheses in this
case by looking for findings that support both a
d ∈ D(C) as well as a d
0
/∈ D(C) such that
η
d
(d
0
) = η
d
5
. The corresponding CQ is as fol-
lows.
Q
0
(f) = ∃d, d
0
.
¬ϕ
c
(f) ∧ δ
c
(d) ∧ ¬δ
c
(d
0
)
∧ η
d
5
(d
0
) ∧ σ
0
(f, d
0
)
c. A doctor may want to include diseases of impor-
tance level at least 4 and are similar to any of
the diseases presently in the differential diagnosis.
The corresponding set of findings is then obtained
by the following CQ, assuming ψ represents the
similarity relation between diseases.
Q
0
(f) = ∃d, d
0
.
¬ϕ
c
(f) ∧ δ
c
(d) ∧ ¬δ
c
(d
0
)
∧ η
d
4
(d
0
) ∧ ψ(d, d
0
) ∧ σ
0
(f, d
0
)
Note that in the above CQ, the diseases of impor-
tance level 4 and 5 will be included because of the
inclusion axiom η
d
5
v η
d
4
in the TBox.
5 SUMMARY & FUTURE WORK
In this paper, we present a knowledge based frame-
work for addressing the case-specific diagnosis prob-
lem. The framework allows users to obtain the dif-
ferential diagnosis for a case, and prompts hypothet-
ical findings that can effectively guide the user to-
wards a diagnosis that is supported by evidences from
case findings. The framework facilitates iterative and
interactive updates of case specific knowledge as an
evolution of sequences of ABoxes. We show that the
queries relevant to differential diagnosis and hypothe-
ses generation can be formulated directly as conjunc-
tive queries on the original knowledge base using the
case specific knowledge. We present the applicability
of our framework in the context of medical diagnosis.
We note that however, the generality of our frame-
work makes it applicable to other diagnosis problems
such as network forensics and criminal investigation.
In addition to its generality, the proposed frame-
work provides an approach for addressing relevant
and interesting problems in diagnosis. One of the im-
portant requirements of any diagnostic system is the
justification of diagnosis, i.e., what portions of the
domain knowledge and which findings can be used to
explain a conclusion for a particular instance of case-
specific diagnosis. We can facilitate justification by
computing a proof of correctness of the results ob-
tained from the conjunctive queries executed.
Finally, we conjecture that the interactive and iter-
ative nature of our framework allows for effective di-
agnosis and discovery of consequences that were pre-
viously unknown; this is likely to have significant im-
pact in medical domain resulting in discovery of new
relationships between findings and diseases, as also
in identifying new traits of diseases. In this sense,
this paper provides a road map for addressing various
challenging problems in diagnosis.
ACKNOWLEDGMENTS
We are grateful to Edward P. Hoffer, M.D., Associate
Clinical Professor of Medicine at the Harvard Med-
ical School for clarifying to us the working of Dx-
plain and Jason Maude, CEO and Co-founder, Isabel
Healthcare for his insightful comments on the diag-
nostic tool Isabel. The first three authors were par-
tially supported by NSF grant 1116050.
REFERENCES
Baader, F., Calvanese, D., McGuinness, D. L., Nardi, D.,
and Patel-Schneider, P. F., editors (2003). The De-
scription Logic Handbook: Theory, Implementation,
and Applications. Cambridge University Press.
Baral, C., McIlraith, S., and Son, T. (2000). Formulating
diagnostic problem solving using an action language
with narratives and sensing. In Proceedings of the 7th
International Conference on Principles of Knowledge
Representation and Reasoning, pages 311–322.
Barnett, G. O., Cimino, J. J., Hupp, J. A., and Hoffer, E. P.
(1987). Dxplain: an evolving diagnostic decision-
support system. Journal of the American Medical As-
sociation, 258(1):67–74.
Ely, J. W., Graber, M. L., and Croskerry, P. (2011). Check-
lists to reduce diagnostic errors. Academic Medicine,
86:1040–2446.
Glimm, B., Horrocks, I., Lutz, C., and Sattler, U. (2008).
Conjunctive query answering for the description logic.
Journal of artificial intelligence research, 31:157–
204.
Hoffer, E. P., Feldman, M. J., Kim, R. J., Famiglietti, K. T.,
and Barnett, G. O. (2005). Dxplain: patterns of use
of a mature expert system. In Proceedings of the An-
nual Symposium of the American Medical Informatics
Association, volume 2005, pages 321–325.
Long, W. J., Naimi, S., Criscitiello, M., and Larsen, G.
(1988). Differential diagnosis generation from a
causal network with probabilities. In Proceedings of
AKnowledgeBasedFrameworkforCase-specificDiagnosis
487

the International Conference on Computers in Cardi-
ology, pages 185–188. IEEE.
Miller, R. A., Pople Jr, H. E., and Myers, J. D. (1982).
Internist-I, an experimental computer-based diagnos-
tic consultant for general internal medicine. New Eng-
land Journal of Medicine, 307:468–476.
Pilli, E. S., Joshi, R. C., and Niyogi, R. (2010). Network
forensic frameworks: Survey and research challenges.
Digital Investigation, 7(1):14–27.
Ramnarayan, P., Tomlinson, A., Rao, A., Coren, M., Win-
row, A., and Britto, J. (2003). Isabel: a web-based
differential diagnostic aid for paediatrics: results from
an initial performance evaluation. Archives of disease
in childhood, 88(5):408–413.
Sohrabi, S., Baier, J. A., and McIlraith, S. A. (2010). Diag-
nosis as planning revisited. In Proceedings of the 12th
International Conference on the Principles of Knowl-
edge Representation and Reasoning, pages 26–36.
Szolovits, P., Patil, R. s., and Schwartz, W. b. (1988). Ar-
tificial intelligence in medical diagnosis. Annals of
Internal Medicine, 108(1):80–87.
Wang, W. and Daniels, T. E. (2008). A graph based
approach toward network forensics analysis. ACM
Transactions on Information and System Security,
12(1):4.
ICAART2015-InternationalConferenceonAgentsandArtificialIntelligence
488
