
Chemo-inspired Genetic Algorithm for Optimizing the Piecewise
Aggregate Approximation
Muhammad Marwan Muhammad Fuad
Forskningsparken 3, Institutt for kjemi, NorStruct
The University of Tromsø - The Arctic University of Norway, NO-9037 Tromsø, Norway
Keywords: Bacterial Foraging, Chemotaxis, Genetic Algorithm, Hybridization, Piecewise Aggregate Approximation,
Time Series.
Abstract: In a previous work we presented DEWPAA: an improved version of the piecewise aggregate approximation
representation method of time series. DEWPAA uses differential evolution to set weights to different
segments of the time series according to their information content. In this paper we use a hybrid of bacterial
foraging and genetic algorithm (CGA) to set the weights of the different segments in our improved
piecewise aggregate approximation. Our experiments show that the new hybrid gives better results in time
series classification.
1 INTRODUCTION
In the last two decades there has been an increasing
interest in temporal data, namely, time series. A time
series is a chronological collection of observations.
This data type is encountered in many scientific and
financial applications. The main feature of time
series is their high-dimensionality. One of the
common approaches to handle the problem of high-
dimensionality of time series is to transform them
into another domain with a lower-dimensionality
followed by an indexing mechanism, called a
dimensionality reduction technique or a
representation method, applied to this lower-
dimensional data.
Several representation methods have been
proposed to reduce the dimensionality of time series
data. Of those we mention Discrete Fourier
Transformation (DFT) (Faloutsos et al., 1994),
Discrete Wavelet Transformation (DWT) (Chan and
Fu 1999), Chebyshev Polynomials (CHEB) (Cai and
Ng, 2004), Symbolic Aggregate approXimation
(SAX) (Lin et al., 2003), Piecewise Linear
Approximation (PLA) (Morinaka et al, 2001).
The Piecewise Aggregate Approximation (PAA)
(Keogh et al., 2000), (Yi and Faloutsos, 2000) is a
time series representation method that has been used
extensively for its simplicity and its low
computational complexity.
In (Muhammad Fuad, M.M., 2012) we applied
differential evolution; a popular bio-inspired
optimization algorithm, to set weights to different
segments of PAA-represented time series, which
reflect the information content of each segment. The
weights were determined through an optimization
process whose output is the weights that maximize
the information content of the PAA representation.
We showed how this modification improves the
performance of PAA.
Differential evolution, however, may suffer from
stagnation; i.e. the inability of progressing towards
global optima. It may also suffer from premature
convergence.
In this paper we propose solving the
aforementioned optimization problem by applying
an alternative optimizer which is a hybrid of two
bio-inspired optimization algorithms: genetic
algorithm and bacterial foraging. We show how this
hybrid yields better results than those obtained when
using differential evolution.
The rest of this paper is organized as follows:
Section 2 presents related work. The hybrid
algorithm is introduced in Section 3, and the
comparison with the previous method is conducted
in Section 4. We conclude this paper in Section 5.
205
Muhammad Fuad M..
Chemo-inspired Genetic Algorithm for Optimizing the Piecewise Aggregate Approximation.
DOI: 10.5220/0005277302050210
In Proceedings of the International Conference on Operations Research and Enterprise Systems (ICORES-2015), pages 205-210
ISBN: 978-989-758-075-8
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

2 RELATED WORK
PAA is a simple, yet efficient, representation method
of time series. PAA reduces the dimensionality of a
time series S from n to N dimensions by segmenting
the time series into equal-sized frames and mapping
each segment to a point that represents the mean of
the points that constitute that segment. The
similarity measure given in the following equation:
N
i
ii
N
ts
N
n
TSd
1
2
,
(1)
is defined on the lower-dimensional space. This
similarity measure is a lower bound of the Euclidean
distance defined on the original space. In Figure 1
we show an example of applying PAA to reduce the
dimensionality of a time series from n=12 to N=3.
The main drawback of PAA is that it is unable to
faithfully represent the original time series due to the
information loss that results from the technique PAA
uses to reduce the dimensionality, as we showed in
(Muhammad Fuad, M.M., 2012). In that paper we
proposed weighting different segments of the PAA
representation differently according to their
information content. We proposed using an
alternative similarity measure for PAA which we
called the Weighted Piecewise Aggregate
Approximation Distance (WPAAD):
N
i
iii
tsw
N
n
TSWPAAD
1
2
,
(2)
1,0
i
w
The challenge here is to find an objective measure to
determine the values of
i
w .
In (Muhammad Fuad, M.M., 2012) we
formulated the above problem as an optimization
problem where the fitness function is the
classification error and the optimization algorithm
seeks to set the values of
i
w that minimize the
classification error.
The optimizer we used in (Muhammad Fuad,
M.M., 2012) was the differential evolution, and we
called our method the Differential Evolutionary
Weighted Piecewise Aggregate Approximation
(DEWPAA).
Figure 1: PAA dimensionality reduction.
Differential Evolution (DE): DE is an evolutionary
optimization algorithm that has the same
computational steps as any other evolutionary
algorithm. Its particularity is in the way the
population members are perturbed, as DE uses a
scaled difference of two other members added to a
third to perform this operation.
DE is particularly adapted to solve continuous
optimization problems, and it has been successfully
used to solve real-life optimization problems.
DE starts by initializing a random population of
vectors. After initialization DE, and for each
member
i
T
(the target vector), creates a donor
vector
D
as a weighted difference of two other
vectors in the population added to a third one. In the
next step DE generates a trial vector
R
through a
crossover operation.
In the next step DE selects which of the trial
vector or the target vector will survive in the next
generation and which will die out. This selection is
based on which of
i
T
and
R
yields a better value of
the fitness function.
The above steps repeat for a number of
generations or until a stopping criterion terminates
the algorithm.
3 CHEMO-INSPIRED GENETIC
ALGORITHM
Hybridization of different optimization algorithms
has been extensively used to solve different
optimization problems. In time series data mining
different hybrid methods have been successfully
used (Muhammad Fuad, 2014a, 2014b, 2014c). The
main advantage that hybridization offers is that the
resulting hybrid method benefits from the strengths
of the two methods, or it avoids their weaknesses.
In this work we use a hybrid of genetic algorithm
and bacterial foraging proposed by (Das and Mishra,
2013) to solve the problem we presented in Section
2.
3.1 The Genetic Algorithm
The Genetic Algorithm (GA) is a famous
evolutionary algorithm that has been applied to solve
a variety of optimization problems. GA is a
population-based global optimization algorithm
which mimics the rules of Darwinian selection in
that weaker individuals have less chance of
surviving the evolution process than stronger ones.
ICORES2015-InternationalConferenceonOperationsResearchandEnterpriseSystems
206

GA captures this concept by adopting a mechanism
that preserves the “good” features during the
optimization process.
In GA a population of candidate solutions (also
called chromosomes) explores the search space and
exploits this by sharing information. These
chromosomes evolve using genetic operations
(selection, crossover, mutation, and replacement).
GA starts by randomly initializing a population
of chromosomes inside the search space. The fitness
function of these chromosomes is evaluated.
According to the values of the fitness function new
offspring chromosomes are generated through the
aforementioned genetic operations. The above steps
repeat for a number of generations or until a
predefined stopping condition terminates the GA.
3.2 Bacterial Foraging
The foraging bahavior of the Escherichia coli (E.
coli) bacteria has inspired a nature-inspired
optimization algorithm called Bacterial Foraging
(BF). The motivation behind this optimizing
algorithm is that in order to perform social foraging,
an animal needs communication capabilities. Over a
period of time this animal gains advantages which
exploit the sensing capabilities of the whole group.
This helps the group to predate on a larger prey, or it
enables the individuals to get better protection
against predators (Kim et al., 2007).
The basis of BF is that animals with poor
foraging strategies tend to be eliminated by natural
selection and they are either replaced by other
individuals with better foraging strategies or they
are shaped into ones which have these desirable
strategies (Passino, 2002). BF formulates this
process as an optimization problem.
The E. coli bacterium moves by means of a set of
flagella, each driven as a biological motor. The two
types of movements the E. coli bacteria perform are
swimming and tumbling. The former takes place
when the flagella rotate in the counterclockwise
direction whereas the latter is achieved by rotating
the flagella in the clockwise direction. Figure 1
shows these two movement types. Together they are
known as chemotaxis (which we will define more
formally later in this section). The aim of
chemotaxis is to help the bacterium approach or
avoid nutrient or noxious substance gradients. This
chemotaxis progress can be destroyed by sudden
environmental changes which cause the elimination
and dispersal of a group of bacteria.
Flagellarotatingcounterclockwise:swimming
Flagellarotatingclockwise:tumbling
Figure 2: The swimming and tumbling movements.
BF finds the minimum of a function
nbp
f
R
;
(nbp is the number of parameters)
by applying four mechanisms; chemotaxis,
swarming, reproduction, and elimination-dispersal.
The position of each member of the population
of N
b
bacteria at the j
th
chemotactic step, k
th
reproduction step, and l
t
h
elimination-dispersal event
is denoted by
b
i
N,...,2,1i| l,k,jk,j,iP
We now describe the four mechanisms we
mentioned earlier in this section:
Chemotaxis: Let
l,k,j
i
be the i
th
bacterium at
the j
th
chemotactic step, k
th
reproduction step, and
l
t
h
elimination-dispersal event, then the
movement of the bacterium can be represented
by:
ii
i
iCl,k,jl,k,1j
T
ii
(3)
where Δ is a vector in the random direction whose
elements lie in the interval [-1, 1].
Swarming: E. coli bacteria demonstrate a
swarming behavior as they travel in rings which
move up the nutrient medium when they are
placed in the center of a semisolid matrix with a
single nutrient chemo-effecter. When simulated
Table 1: The symbols used in the description of bacterial
foraging.
N
b
The number of bacteria in the population
N
c
The number of chemotactic steps
N
s
The swimming length
N
re
The number of reproduction steps
N
ed
The number of elimination-dispersal events
P
ed
The probability of elimination-dispersal
C(i)
The size of the step taken in the random
direction determined by the tumble
Chemo-inspiredGeneticAlgorithmforOptimizingthePiecewiseAggregateApproximation
207

by a high level of succinate the bacteria release
an attractant aspartate which helps them
aggregate into groups and thus move as a swarm.
The cell-to-cell signal in the swam can be
represented by the following function:
b
b
b
N
i
nbp
m
i
mmrepellantrepellant
N
i
nbp
m
i
mm
N
i
i
cccc
h
d
lkjflkjPf
11
2
11
2
attractantattractant
1
exp.
exp.
),,,,,,
(4)
where the coefficients d
attractant
, ω
attractant
, h
repellant
,
ω
repellant
are control parameters.
The objective function
l,k,jP,f
cc
is added
to the original objective function to represent a
time varying objective function in that if many
cells come close together there will be a high
amount of attractant and hence an increasing
likelihood that other cells will move towards the
group. This produces the swarming effect
(Passino, 2002).
Reproduction: Through this process the least
healthy bacteria die out and the healthier ones
will replicate themselves. This guarantees that the
size of the bacterial swam will remain constant.
Elimination and dispersal: There might be a
gradual or sudden change in the environment
where the bacteria live. As a result, a small
percentage of the bacteria in a certain region will
be liquidated or a group might be dispersed into
another location. This has two effects on
chemotaxis: the first is destroying the
chemotactic progress, the second is that the new
bacteria might be placed at locations with a better
food source, thus assisting chemotaxis.
3.3 CGA
GA has the advantage of quickly locating high
performance regions of vast and complex search
spaces, but they are not well suited for fine-tuning
solutions (Gendreau and Potvin, 2005), (Kazarlis et
al., 2001).
There have been several attempts to hybridize
GA with other optimization algorithms. In (Mahfoud
and Goldberg, 1995) the authors present the Parallel
Recombinative Simulated Annealing (PRSA) which
combines elements from the simulated annealing
algorithm with others from GA. Another hybrid was
presented in (Lee and Lee, 2005). This method
hybridizes GA with Ant Colony Optimization
(ACO).
On the other hand, BF possesses a poor
convergence behavior over multi-modal and rough
fitness landscapes. Its performance is also heavily
affected with the growth of problem dimensionality
(Biswas et al., 2007).
To take advantage of the two optimizers, (Kim et
al., 2007) proposed a hybrid of GA and BF (called
GA-BF). They validated their method on several test
functions.
In another paper (Das and Mishra, 2013)
proposed another hybrid of GA and BF which they
called the Chemo-inspired Genetic Algorithm
(CGA). Their motivation is that chemotaxis actually
contributes the most in the search process, so instead
of taking the whole BF to hybridize with GA, they
only integrate the chemotaxis step in the hybrid with
GA. CGA has five major steps: selection, crossover,
mutation, elitism and chemotaxis. In addition, CGA
employs three mechanisms: a) adaptive step size b)
squeezed search space c) fitness function criterion.
4 EXPERIMENTS
We conducted intensive experiments to compare
CGA with DEWPAA. The experiments were the
same as those conducted in (Muhammad Fuad,
M.M., 2012) and (Muhammad Fuad, M.M., 2013);
i.e. classification task experiments of time series.
The aim of the experiments is to show that using
CGA in the optimization process of determining the
weights of the segments in equation (2) will result in
a lower classification error than that of DEWPAA.
As in (Muhammad Fuad, M.M., 2012), we
computed the classification error using WPAAD for
different compression ratios (1:8,1:12,1:16). We
conducted our experiments using the UCR archive
(Keogh et al., 2011), which is the same archive used
to test DEWPAA.
The two methods were tested on a classification
task based on the first nearest-neighbor (1-NN) rule
using leaving-one-out cross validation.
Table 2 shows some of the results of our
experiments. As we can see from the table, CGA
outperforms DEWPAA on almost all the datasets
tested and for the different compression ratios.
5 CONCLUSIONS
In this paper we used a hybrid of genetic algorithm
and bacterial foraging (CGA) to calculate the
weights given to different segments of the time
ICORES2015-InternationalConferenceonOperationsResearchandEnterpriseSystems
208
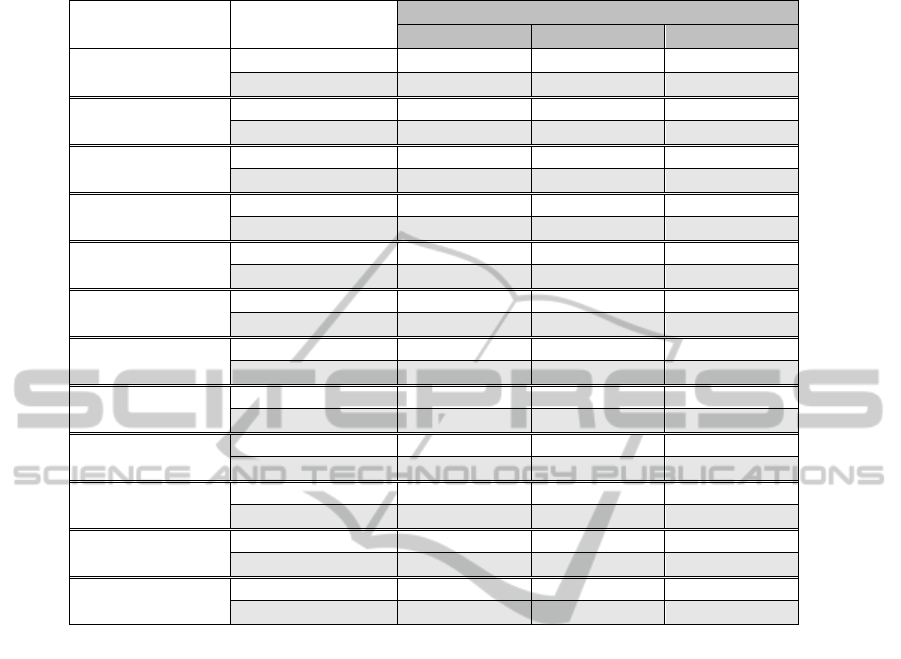
Table 2: Comparison of the classification error between CGA and DEWPAA for different compression ratios.
Dataset Method
Compression Ratio
1:8 1:12 1:16
Lighting7
CGA 0.329 0.342 0.356
DEWPAA 0.370 0.397 0.427
MedicalImages
CGA 0.308 0.308 0.321
DEWPAA 0.337 0.337 0.378
Gun_Point
CGA 0.06 0.06 0.06
DEWPAA 0.053 0.067 0.087
Coffee
CGA 0.179 0.179 0.179
DEWPAA 0.179 0.179 0.25
Lighting2
CGA 0.164 0.180 0.197
DEWPAA 0.213 0.230 0.246
MALLAT
CGA 0.077 0.082 0.094
DEWPAA 0.094 0.094 0.095
FacesUCR
CGA 0.238 0.238 0.238
DEWPAA 0.238 0.316 0.366
FISH
CGA 0.194 0.194 0.194
DEWPAA 0.194 0.229 0.240
synthetic_control
CGA 0.067 0.067 0.100
DEWPAA 0.110 0.113 0.113
CBF
CGA 0.017 0.021 0.021
DEWPAA 0.017 0.021 0.037
ECG
CGA 0.090 0.100 0.100
DEWPAA 0.110 0.120 0.130
Trace
CGA 0.100 0.120 0.120
DEWPAA 0.140 0.180 0.190
series represented by the piecewise aggregate
approximation representation method. The weights
were obtained through an optimization process using
chemo-inspired genetic algorithm (CGA) as
optimizer and the fitness function is the
classification error. Compared with differential
evolution, another optimizer that we used in a
previous work to solve the same optimization
problem, CGA gives better results.
REFERENCES
Biswas, A., Dasgupta, S., Das, S. and Abraham, A., 2007.
Synergy of PSO and bacterial foraging optimization: a
comparative study on numerical benchmarks. HAIS
2007.
Cai, Y., and Ng, R., 2004. Indexing spatio-temporal
trajectories with chebyshev polynomials. In SIGMOD.
Chan, K., and Wai-chee Fu, A., 1999. Efficient time series
matching by wavelets. In Proc. 15th. Int. Conf. on
Data Engineering.
Das, K. N., and Mishra, R., 2013. Chemo-inspired genetic
algorithm for function optimization. Applied
Mathematics and Computation, 220, 394–404.
Faloutsos, C., Ranganathan, M., and Manolopoulos, Y.,
1994. Fast subsequence matching in time-series
databases. In Proc. ACM SIGMOD Conf.,
Minneapolis.
Gendreau M., Potvin., J. Y,. 2005. Annals of operations
research 140(1)pp189–213.
Kazarlis, S.A., Papadakis. S. E., Theocharis, J.B., Petridis,
V., 2001. IEEE transactions on evolutionary
computation 5(3)pp 204–217.
Keogh, E., Chakrabarti, K., Pazzani, M., and Mehrotra,S.,
2000. Dimensionality reduction for fast similarity
search in large time series databases. J. of Know. and
Inform. Sys.
Kim, D. H., Abraham, A., Cho, J. H., 2007. A hybrid
genetic algorithm and bacterial foraging approach for
global optimization. Information Sciences, Vol. 177
(18), 3918-3937.
Lee, Z .J., and Lee, C. Y., 2005. A hybrid search algorithm
with heuristics for resource allocation problem,
Information Sciences 173.
Keogh, E., Zhu, Q., Hu, B., Hao. Y., Xi, X., Wei, L. &
Ratanamahatana, C.A., 2011. The UCR Time Series
Classification/Clustering Homepage: www.cs.ucr.edu/
~eamonn/time_series_data/
Lin, J., Keogh, E., Lonardi, S., Chiu, B. Y., 2003. A
symbolic representation of time series, with
implications for streaming algorithms. DMKD 2003.
Chemo-inspiredGeneticAlgorithmforOptimizingthePiecewiseAggregateApproximation
209

Mahfoud, S., and Goldberg, D., 1995. Parallel
recombinative simulated annealing: a genetic
algorithm. Parallel Computing, vol. 21, pp. 11-28.
Morinaka, Y., Yoshikawa, M., Amagasa, T., and Uemura,
S., 2001: The L-index: An indexing structure for
efficient subsequence matching in time sequence
databases. In Proc. 5th Pacific Asia Conf. on
Knowledge Discovery and Data Mining.
Muhammad Fuad, M. M., 2014a. A hybrid of bacterial
foraging and differential evolution -based distance of
sequences. In Procedia Computer Science 2014.
Volume 35. pp 101-110.
Muhammad Fuad, M. M., 2013. A pre-initialization stage
of population-based bio-inspired metaheuristics for
handling expensive optimization problems. ADMA
2013, December 14-16, 2013, Zhejiang, China.
Lecture Notes in Computer Science Volume 8347,
2014, pp 396-40.
Muhammad Fuad, M. M., 2014b. A synergy of artificial
bee colony and genetic algorithms to determine the
parameters of the Σ-Gram distance. In DEXA 2014,
Munich, Germany. Lecture Notes in Computer Science
Volume 8645, 2014, pp 147-154.
Muhammad Fuad, M. M., 2014c. A weighted minimum
distance using hybridization of particle swarm
optimization and bacterial foraging. In PRICAI 2014,
Gold Coast, QLD, Australia. Lecture Notes in
Computer Science Volume 8862, 2014, pp 309-319.
Muhammad Fuad, M. M., 2012. Using differential
evolution to set weights to segments with different
information content in the piecewise aggregate
approximation. In KES 2012, San Sebastian, Spain,
(FAIA). IOS Press.
Passino, K. M., 2002. Biomimicry of bacterial foraging for
distributed optimization and control, IEEE Control
Syst. Mag., vol. 22, no. 3, pp. 52–67.
Yi, B. K., and Faloutsos, C., 2000. Fast time sequence
indexing for arbitrary Lp norms. Proceedings of the
26th International Conference on Very Large
Databases, Cairo, Egypt.
ICORES2015-InternationalConferenceonOperationsResearchandEnterpriseSystems
210
