
Ontology Selection for Semantic Similarity Assessment
Montserrat Batet and David Sánchez
Department of Computer Engineering and Mathematics, Universitat Rovira i Virgili,
Av. Països Catalans, 26, 43007, Tarragona, Catalonia, Spain
Keywords: Knowledge Representation, Ontologies, Semantic Similarity.
Abstract: The assessment of the semantic similarity between concepts is a key tool to improve the understanding of
text. The structured knowledge that ontologies provide has been extensively used to estimate similarities
with encouraging results. However, in many domains, several ontologies modelling the same concepts in
different ways are available. In such scenarios, the most suitable ontology for similarity calculation should
be selected. In this paper we tackle this task by proposing an unsupervised method to select the ontology
that seems to enable the most accurate similarity assessments. By studying the ontology features that most
influence the similarity accuracy, we propose a score that captures them in a mathematically coherent way.
Then, the most suitable ontology can be selected as that with the highest score. We also report the results of
the proposed method for several well-known ontologies and a widely-used semantic similarity benchmark.
1 INTRODUCTION
A key element to text understanding is the
assessment of the semantic similarity between the
concepts referred in the text (Resnik, 1995).
Semantic similarity is understood as the level of
taxonomic proximity between concepts (Batet and
Sánchez, 2014). To enable this assessment in an
automatic way, ontologies provide a formal and
machine-readable representation of the knowledge
related to a domain, from which similarities can be
estimated (Batet and Sánchez, 2014).
Traditionally, ontology-based similarity
measures relied on the knowledge modeled in a
single ontology (Wu and Palmer, 1994; Jiang and
Conrath, 1997; Resnik, 1995; Batet et al., 2011b).
Thus, the similarity results strongly depended on the
accuracy of the knowledge modeled in the ontology.
To overcome this limitation and, given the
availability of several complementary and
overlapping knowledge bases in many domains,
some authors have recently proposed methods to
exploit multiple ontologies for semantic similarity
assessment (Rodríguez and Egenhofer, 2003;
Petrakis et al., 2006; Al-Mubaid and Nguyen, 2009).
The motivation is that the additional knowledge and
the complementary views that several knowledge
sources provide of a certain domain could lead to
more accurate similarity estimations.
Semantic similarity computation from multiple
ontologies faces two main challenges. First, in many
situations, a single ontology does not model the
concepts to be compared, so that, their similarity
should be computed across different ontologies.
Second, in cases in which the pair of terms to be
compared belong to several ontologies at the same
time (and, thus, different similarity results can be
obtained for the several ontologies), it is necessary
to select the best knowledge source.
In this work we focus on the latter problem. This
situation is especially relevant in domains in which
several ontologies are available (e.g., in
biomedicine, we can find overlapping knowledge
bases such as MeSH (Nelson et al., 2001) or
SNOMED-CT (Spackman, 2004), which model the
same medical concepts). However, ontologies are
usually independently created from a wide variety of
sources and with different goals and quality criteria.
Thus, different ontologies can model the same
domain of knowledge in significantly different ways
because the scope of the ontology, and the point of
view and design principles followed by knowledge
engineers may differ. Consequently, overlapping
ontologies usually present different levels of detail,
completeness and semantic structure, thus enabling
more or less accurate similarity assessments.
Because of the many factors that are involved in the
knowledge modelling, it is difficult to select a priori
569
Batet M. and Sanchez D..
Ontology Selection for Semantic Similarity Assessment.
DOI: 10.5220/0005284205690576
In Proceedings of the International Conference on Agents and Artificial Intelligence (ICAART-2015), pages 569-576
ISBN: 978-989-758-074-1
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

the most suitable ontology (Al-Mubaid and Nguyen,
2009; Sánchez et al., 2012b).
In this paper, we tackle this issue by proposing a
method to assess the suitability of an ontology as a
source for similarity assessments. This is done by
analysing the taxonomic structure of the ontology
and results in a numerical score that enables the
selection of the ontology that seem to enable the best
similarity assessments. The theoretical premises
have been empirically evaluated by applying our
method to a set of well-known ontologies. Results
suggest that those ontologies with the highest score
also enable the most accurate similarity assessments.
The rest of the paper is organised as follows.
Section 2 discusses related works on semantic
similarity. Section 3 details the proposed
mechanism. Section 4 presents and discusses the
empirical results. The final section contains the
conclusions of the work.
2 RELATED WORK
In the literature, ontology-based semantic similarity
measures are usually classified in different
paradigms according to the knowledge sources they
exploit and the theoretical principles in which they
rely. In this work, we focus on methods that only
rely in the knowledge modelled in an ontology.
Earliest approaches measure the distance (i.e.,
the opposite to similarity) of a pair of concepts as a
function of the length of the shortest path
connecting those concepts by means of taxonomic
relationships (Wu and Palmer, 1994; Li et al., 2003).
One limitation of these approaches is that they omit
much of the knowledge represented in the ontology,
because only the shortest path is considered.
To overcome this limitation, some authors have
proposed measures in which different ontological
features are considered (Sánchez et al., 2012a; Batet
et al., 2011b; Pirró, 2009; Rodríguez and Egenhofer,
2003; Petrakis et al., 2006). They usually measure
similarity as a ratio between the number of features
that the concepts to be compared have or do not
have in common. Because these measures exploit
more ontological knowledge, feature-based methods
provide, in general, more accurate results than
measures based on taxonomic paths (Sánchez et al.,
2012a).
Other similarity paradigms rely, not only on the
taxonomic knowledge provided by an ontology, but
also on the Information Content of concepts, which
is computed as the inverse of the probability of
appearance of such concepts in a corpus (Jiang and
Conrath, 1997; Resnik, 1995). The main limitation
of these methods is that they require representative
textual corpora in which concepts have been tagged,
so that the probabilities required to compute their
information content and estimate similarities
assessed (Batet and Sánchez, 2014).
In any case, the results provided all these
methods depend on the coverage, completeness and
level detail of the ontology in which they rely (Al-
Mubaid and Nguyen, 2009; Sánchez et al., 2012b).
In recent years, researchers have tackled this
limitation by considering multiple ontologies.
In (Rodríguez and Egenhofer, 2003; Petrakis et
al., 2006) two ontologies are connected by means of
an imaginary root node that subsumes the root nodes
of each ontology. In (Rodríguez and Egenhofer,
2003) similarity is computed according to the
overlapping between a set of non-taxonomic features
(e.g. synonyms, meronyms). In (Petrakis et al.,
2006) the Jaccard index is used to calculate the
degree of overlapping between concept glosses and
synonym sets. Overlapping features are found by
means of the terminological matching of concept
labels. The simplistic solution used to join
ontologies is a main drawback of these approaches.
Moreover, they do not consider the case in which
concept pairs appear in different ontologies. Finally,
their dependency on the availability of non-
taxonomic features, which are rarely found in
ontologies (Ding et al., 2004), limit the practical
applicability of these methods, which are focused on
the more general notion of semantic relatedness
rather than strict taxonomic similarity.
In (Saruladha et al., 2010), the similarity is
assessed as a function of the concreteness of the
most specific concept in the taxonomy that
subsumes the pair of concepts to be compared.
When each concept belongs to a different ontology,
the common subsuming concept is obtained by
means of a terminological matching of the labels of
the subsumers of each concept. Similarly to the
previous approach, in (Al-Mubaid and Nguyen,
2009) authors retrieve concepts that act as bridges
between ontologies. First, the user selects a primary
ontology, which she believes it the most accurate
one. Then, if the pair of concepts are found in the
primary ontology or in an unique secondary
ontology, the similarity is computed using the
ontology to which the concepts belong; if one of the
concepts is found in the primary ontology and the
other one in a secondary ontology, the two
ontologies are connected using bridge nodes and the
resulting structure is used as if it was a unique
ontology; finally, when concepts appear in several
ICAART2015-InternationalConferenceonAgentsandArtificialIntelligence
570

secondary ontologies, the similarity is computed
using the ontology with the highest alikeness to the
primary one. Because the method relies on a path-
based measure, the authors face the problem that
different ontologies may have different granularity
degrees. To solve this problem, they propose to
normalize similarity values by scaling the part of the
path corresponding to a secondary ontology taking
as reference the primary ontology. In all cases they
assume that the ontology that has been –manually-
selected as primary will lead to better similarity
estimations than any secondary ontology. However,
this requires the user to deeply understand the
knowledge structure of the ontologies.
3 ONTOLOGY SELECTION
In this section, we propose an unsupervised method
to quantify the suitability of an ontology to measure
semantic similarity. First, we analyse the semantic
and structural features of ontologies that seem to
most influence the accuracy of semantic similarity
assessments. Then, we propose a numerical score
that quantify the suitability of an ontology to guide
similarity assessments.
3.1 Problem Analysis
From the analysis of related works on semantic
similarity, one can realize that all of them rely on the
number of differences (which are inversely
proportional to similarity) and commonalities (which
are proportionally to similarity) that can be
identified in the semantic structures associated to the
concepts in their respective ontology/ies. Since we
are focusing on semantic similarity, which is a
function of taxonomic features, in the following we
will study the effect that the modelling of such
taxonomic structure in an ontology has over the
evaluation of similarities/distances.
In the simplest case, in which the taxonomy is
perfectly balanced, the root node is the geometric
centre and all concepts are direct specializations of
the root node, the distance between all pairs of
concepts is exactly the same. In figure 1 we show an
example of this scenario for a set of medical
concepts. Because all concepts are modelled in the
same way, in this taxonomy all concepts pairs will
appear to be equally distant/similar for any semantic
similarity measure, a result that would be unlikely
realistic. In fact a knowledge representation as
simple as this is not much different to a flat list of
concepts with any semantic structure at all.
Figure 1: Sample ontology O
1
.
In order to better represent the differences in
semantics inherent to the concepts and, coherently
with the principles of cognitive saliency (i.e.
concepts are specialised when they must be
differentiated from other ones), in figure 2 we have
added a new inner taxonomic level (mental disorder)
that separates the set paranoia, schizophrenia and
affective psychosis from the set melanosis, albinism
and vitiligo, which are subsumed by disorder of
pigmentation. Even though more taxonomic levels
have been included the taxonomy is still balanced
and, thus, the root node is still the geometric.
However, now the semantic distance between
paranoia and albinism will be larger than the
distance between paranoia and schizophrenia
because we are able to distinguish concepts that are
mental disorder from those that are disorder of
pigmentation. Thanks to the better differentiation
between concepts, distance/similarity results
obtained from this structure will be more diverse
than in the previous case and, assuming that the
representation is semantically coherent, results will
offer a better understanding concept semantics.
Figure 2: Sample ontology O
2
.
Finally, in figure 3 we show how these concepts are
represented in SNOMED-CT (Spackman, 2004).
Notice that in this case the concepts schizophrenia
and affective psychosis has been better differentiated
from paranoia by adding a new inner node
(psychotic disorder). As result, the root node is not
the geometric centre of the taxonomy anymore,
because the branch on the left goes deeper than the
OntologySelectionforSemanticSimilarityAssessment
571
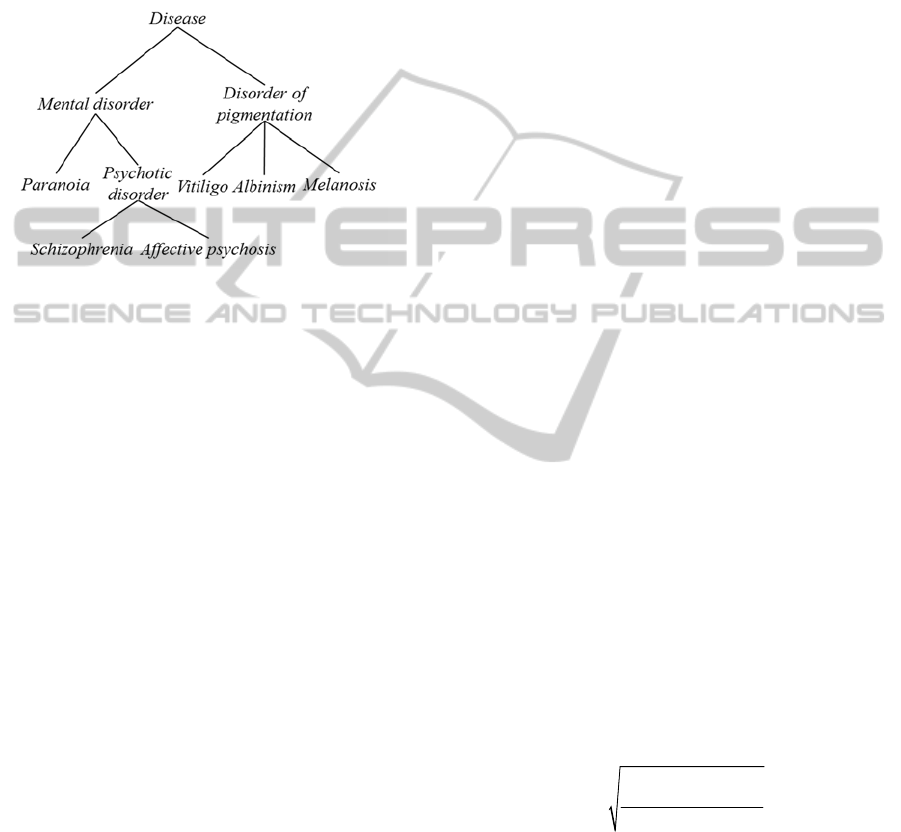
one on the right. This taxonomy better represents the
fact that, for example, albinism and schizophrenia
are more different than albinism and paranoia, a
dimension that can be captured by similarity
measures by evaluating the number of common and
disjoint subsumers (Batet et al., 2011b) and that
results in even more diverse similarity/distance
results than in the previous cases.
Figure 3: Sample ontology O
3
.
3.2 A Score for Ontology Selection
From the above discussion, we can conclude that a
taxonomy with an accurate knowledge modelling is
likely to better differentiate concepts from each
other. Inversely, an accurately modelled ontology
will unlikely have a homogenous taxonomy because,
in such structure, concepts are distributed uniformly
and, hence, they are hardly distinguishable. This
assumption is coherent with the principles of
knowledge modelling, whose main aim is to make
concepts well-differentiated in order to minimize the
ambiguity of the semantic inferences (Pirró, 2009).
Likewise, human judgements on semantic similarity
(which computerized measures try to mimic) are
rarely homogenous and tend to be highly diverse
because of the informal nature of semantics
(Pedersen et al., 2007).
Given these arguments, in the following we
propose a score that aims to measure the degree of
concept differentiation that the taxonomic structure
of an ontology provides. Since this level of
differentiation positively influences the diversity of
the similarity assessments, which we hypothesise as
an indication of accuracy in such assessments, our
score can be used to compare and select the most
suitable ontology for semantic similarity calculation.
To measure this level of differentiation, we
evaluate the dispersion of the taxonomic structure.
From a semantic point of view, the centre of an
ontology can be seen as the root of the taxonomic
tree. As shown in the previous section, in a perfectly
balanced ontology (figures 1 and 2), the root node
corresponds to the geometric centre of the
taxonomy. As stated in (Martínez et al., 2012), this
central node is the one that minimizes the distances
with respect to all the concepts in the ontology. In
such balanced structure, the level of differentiation
between concepts will tend to be low, because
“sibling” or “cousin” concepts will be all equally
distant/similar. On the contrary, in a taxonomy in
which the different branches have different depths
and branching factors, the root node will not match
with the geometrical centre of the structure (as in
figure 3). Here, the degree of differentiation between
concepts will tend to be higher than in the previous
case, thus producing more diverse similarities.
From a mathematical perspective the dispersion
of a sample quantifies the variability of the values of
that sample with regard to the central value. A high
dispersion indicates that values are very different
from each other. By considering the set of concepts
in an ontology as a sample of values and the root
node as their centre, we can adapt the mathematical
notion of dispersion to quantify to what extent the
concepts modelled in taxonomy are dispersed or
differentiated. Consequently, we propose a score
that quantifies that degree of differentiation by
measuring the dispersion of the taxonomic structure
of the ontology. For numerical values, the dispersion
of a sample is the normalized aggregation of their
distances towards the central value. When dealing
with ontologies, such distance should be a measure
of the semantic distance of each concept towards the
root node of the taxonomy. In particular, our score is
based on the standard numerical deviation, which
has the advantage that the results are expressed in
the same units as the distance.
Formally, we quantify to what extent the whole
set of concepts C of an ontology O
i
are
differentiated, as the square root of the average
squared semantic distance between each concept c
i
in C and the root node of O
i
.
2
()
(, ( ))
||
i
ii
cC
i
Score O
dc RootO
C
(1)
In the above expression, |C| is the number of
concepts in the ontology O
i
without considering the
root node, which does not contributes to the
numerator, and function d(.,.) is any semantic
distance measure to be applied between each
concept c
i
in C and the root node (Root(O
i
)). Notice
that the contribution of the most scattered concepts,
which are those that contribute most to the
unbalancing of the taxonomy, is greater because of
ICAART2015-InternationalConferenceonAgentsandArtificialIntelligence
572
the squared semantic distances. Numerically,
equation (1) is zero when all the values are identical.
In contrast, a high Score suggests that concepts are
far apart from the root node and form each other,
and thus, that they are well differentiated.
By means of the proposed Score, we can select
the most suitable ontology O
s
to be used to compute
the similarity of a set of concepts (modelled in a set
of ontologies O) as the one with the max value.
arg max ),(()
sii
ScoreOOOO
(2)
The proposed method can also be applied only to
taxonomic branches of different ontologies. This is
relevant because the different scopes and goals by
which ontologies are designed can produce more or
less accurate or detailed taxonomic branches, even
in ontologies modelling the same domain. With our
method, this partial comparison can be done by
using the common generalization of that branch as
the root, and by computing the distances towards all
of its taxonomic specializations.
3.3 Measuring Distances
In order to apply the proposed Score, we should be
able to estimate the semantic distance (function
d(.,.)) between each concept c
i
and the root node.
The semantic distance should meet some
requirements in order to be suitable to compare
ontologies. First, it should not be affected by the size
of the ontology in order to fairly compare the degree
of concept differentiation of ontologies with
different sizes. On the other hand, the semantic
distance should only rely on taxonomic knowledge
because similarity assessment only relies on this
feature, and also because many ontologies does not
model other knowledge than taxonomic
relationships (Ding et al., 2004).
As stated in section 2, many different measures
have been proposed (Batet and Sánchez, 2014).
Since path-based approaches provide absolute
distance values between concepts, semantic
distances tend to be larger as the ontology size
increases. Thus, they cannot be used to compare
ontologies with different sizes. Moreover, since only
the shortest path is considered, they omit a lot of
knowledge explicitly modelled in the ontology
(Batet et al., 2011b).
Feature-based measures are able to overcome
these limitations. They compute similarity or
distance according to a normalized ratio of semantic
commonalities and differences between concepts
and, thus, they evaluate a larger number of semantic
evidences. In this work, we instantiate d(.,.) with the
feature-based measure defined in (Sánchez et al.,
2012a) because it solely relies on taxonomic
features. This measure computes the distance
d(c
1
,c
2
) between two concepts
as the logarithm of the
number of non-common subsumers of c
1
and c
2
divided by their total number of subsumers:
12 12
12 2
12
() () () ()
(, ) log 1
() ()
Tc Tc Tc Tc
dc c
Tc Tc
(3)
where T(c
i
) is the set of taxonomic subsumers of
concept c
i
in the ontology, including itself.
This measure captures more knowledge than the
methods based on shortest paths, since it implicitly
considers all the paths connecting the two concepts,
which are represented by all their subsumers. As a
result, and according to a set of empirical
experiments, it approximates human judgments of
similarity better than other ontology-based measures
(Sánchez et al., 2012a; Batet et al., 2011b).
Numerically, thanks to the normalizing
denominator, this distance results in positive
normalized values in the [0,1] range, thus making it
suitable to compare the degree of concept
differentiation of ontologies with different sizes.
3.4 Example
Let us illustrate how the Score proposed in section
3.2 behaves with regard to the taxonomic structure
of an ontology. To do so, we will use the different
knowledge representations shown in figures 1, 2 and
3 for the same set of concepts. By applying the
Score to the taxonomy in figure 1 (the one in which
concepts are the least differentiated), we obtain:
22
22
1
22
22
222
()
(,)( ,)
( ,)( ,)/6
(,)(,)
111
log 1 log 1 log 1
222
Score O
d paranoia dis d schizophrenia dis
d affective psychosis dis d melanosis dis
d albinism dis d vitiligo dis
2
222
222
/6 0.585
111
log 1 log 1 log 1
222
By applying the same calculation to the taxonomy
shown in figure 2, which offers a better
differentiation between mental and pigmentation
disorders, the Score increases accordingly:
OntologySelectionforSemanticSimilarityAssessment
573

22
22
2
22
22
()
(,)( ,)
( ,)( ,)
/8
(,)(,)
( ,)( ,)
6lo
Score O
d paranoia dis d schizophrenia dis
d affective psychosis dis d melanosis dis
d albinism dis d vitiligo dis
d mental dis dis d dis of pigmentation dis
22
22
21
g1 2 log1
32
0.702
8
Finally, for the taxonomy shown in figure 3, which
offers the best differentiation between concepts, we
also obtain the highest Score.
22
22
22
3
22
()
(,)( ,)
( ,)( ,)
(,)(,)
( ,)( ,)
( ,
Score O
d paranoia dis d schizophrenia dis
d affective psychosis dis d melanosis dis
d albinism dis d vitiligo dis
d mental dis dis d dispigmentation dis
d psychotic disorder di
2
222
222
/9
)
321
2log1 5log1 2log1
432
0.723
9
s
Thus, according to the method presented in section
3.2, the ontology O
3
shown in figure 3 will be
selected as the base to compute semantic similarities
between the modelled concepts.
4 EXPERIMENTS
The goal of the experiments is to show that, in
practice, the ontology (from a set of overlapping
ones) with the highest Score is also the one that
enables the most accurate similarity assessments.
We focused on the biomedical domain because, as
mentioned in the introduction, several standard
ontologies modelling the same concepts exist.
To measure the accuracy of similarity
assessments, that is, up to which level the similarity
results mimic human judgements, related works
measure the Pearson correlation between human
similarity ratings and computerized results for a
given set of concept pairs. In the literature, several
benchmarks providing human ratings for a set of
concepts of different domains have been proposed.
For the biomedical domain, the Pedersen et al.’s
benchmark (Pedersen et al., 2007) has become the
de facto standard for similarity evaluation. It
consists of 30 pairs of medical terms, whose
similarity has been assessed, in the range [1..4], by a
group of experts of the Mayo Clinic.
Table 1: Medical term pairs that can be found both in
SNOMED-CT and MeSH, with averaged experts’
similarity scores from the Pedersen et al. benchmark
(Pedersen et al., 2007). In boldface we represent those
pairs that specifically correspond to diseases.
Term 1 Term 2 Sim.
Renal failure Kidney failure 4.0
Heart Myocardium 3.3
Stroke Infarct 3.0
Abortion Miscarriage 3.0
Delusion Schizophrenia 3.0
Congestive heart
failure
Pulmonary edema 3.0
Metastasis Adenocarcinoma 2.7
Calcification Stenosis 2.7
Mitral stenosis Atrial fibrillation 2.3
Rheumatoid
arthritis
Lupus 2.0
Brain tumor Intracranial
hemorrhage
2.0
Carpal tunnel
syndrome
Osteoarthritis 2.0
Diabetes mellitus Hypertension 2.0
Acne Syringe 2.0
Antibiotic Allergy 1.7
Cortisone Total knee
replacement
1.7
Pulmonary
embolus
Myocardial
infarction
1.7
Pulmonary fibrosis Lung cancer 1.7
Cholangiocarcinoma Colonoscopy 1.3
Lymphoid
hyperplasia
Laryngeal cancer 1.3
Multiple sclerosis Psychosis 1.0
Appendicitis Osteoporosis 1.0
Xerostomia Alcoholic cirrhosis 1.0
Peptic ulcer disease Myopia 1.0
Depression Cellulitis 1.0
Varicose vein Entire knee meniscus 1.0
Hyperlipidemia Metastasis 1.0
As ontologies to be compared, we use SNOMED-
CT (Spackman, 2004) and MeSH (Nelson et al.,
2001), which semantically model biomedical
concepts with a large degree of overlapping. Since
we focus in the scenario in which concepts appear in
several ontologies at the same time, in table 1 we
show the pairs of terms of the Pedersen et al.’s
benchmark that can be found both in SNOMED-CT
and MeSH. The last column provides the averaged
similarity ratings provided by the experts. Moreover,
those pairs that are diseases in both ontologies are
shown in boldface. According to this set of terms,
we configured two scenarios:
(a) Scenario 1: ontology selection evaluation. All
the term pairs in table 1 are evaluated. Since
those terms are spread through the different
branches of SNOMED-CT and MeSH, in this
ICAART2015-InternationalConferenceonAgentsandArtificialIntelligence
574

scenario we aim to select the ontology that is
best suited to compute semantic similarities.
(b) Scenario 2: branch selection evaluation. This
scenario is designed to show how our method
can also be applied to a particular branch of an
ontology. In this case, only the set of term pairs
that are diseases are considered. Likewise, only
the taxonomic branches of SNOMED-CT and
MeSH that model diseases have been
evaluated. The reference root nodes are now
the Disease (disorder) concept in SNOMED-
CT and C-Disease in MeSH.
Table 2 shows the Scores resulting from the
evaluation of each ontology/branch and also the
accuracy (Correlation) of the similarity assessments
obtained for the same ontology/branch in the two
scenarios detailed above. In all cases, distances and
similarities have been computed using equation (3).
From table 2, we can see that our Score and the
accuracy of the semantic similarity assessments are
positively correlated. This supports our hypothesis
and suggests that the most appropriate ontology to
compute semantic similarities would be the one that
better differentiates concepts, a dimension that our
Score quantifies. In fact, this better differentiation
provides semantic similarity measures with more
degrees of freedom to evaluate concepts and
produces more diverse similarity results that, as
shown in the experiments, better correlate with
human ratings of similarity.
Table 2: Pearson correlation coefficients for each scenario
and ontology/branch between the experts’ similarity
ratings in (Pedersen et al., 2007) and the measure from Eq.
(3). The last column shows the Score (Eq. (1)) for each
ontology/branch.
Ontology/branch Correlation
Scenario 1
Correlation
Scenario 2
Score
SNOMED-CT 0.69 0.938
MeSH
0.65 0.903
SNOMED-CT
disease
0.83 0.951
MeSH disease 0.77
0.886
Looking at the numeric scales, we can also see that,
thanks to the normalized values provided by
equation (3), Score values are not dependant on the
ontology size. Specifically, even though SNOMED-
CT models 300,000 concepts and MeSH just around
22,000, Score values do not differ proportionally to
these sizes (0.938 vs. 0.903). These Score values are
however quite proportional to the differences
observed in semantic similarity accuracies for both
ontologies (0.69 vs. 0.66). The same behaviour is
also observed for taxonomic branches.
5 CONCLUSIONS
In this paper we presented an unsupervised method
to assess the suitability of ontologies as sources to
measure semantic similarity in the scenario in which
the concepts to be compared appear in several
ontologies. Given that similarity measures benefit
from knowledge structures that better
(taxonomically) differentiate concepts, we propose a
quantitative Score that measures the degree of
taxonomic differentiation of concepts in an
ontology. To do so, the Score adapts to the semantic
domain the mathematical notion of numerical
dispersion of a sample. By means of this Score, we
can select the ontology that likely provides the most
accurate similarities from a set of overlapping ones.
The results of the empirical experiments carried
out using biomedical ontologies and a widely-used
semantic similarity benchmark, supported our
hypotheses: in all cases, those ontologies (or
taxonomic branches) with the highest Score also
enabled the most accurate similarity assessments.
As future work, we plan to evaluate the proposed
method in other domains in which multiple
overlapping ontologies are available. Moreover, we
will also evaluate the behaviour of other distance
functions in the Score calculus, such as those based
on word vectors (Mikolov et al., 2013). Finally, we
plan to study its suitability as a predictor of results
accuracy in specific tasks that require from semantic
similarity assessments and in which different
knowledge-bases are available, such as semantic
clustering (Batet et al., 2011a) or textual data
anonymisation (Batet et al., 2013).
ACKNOWLEDGEMENTS
This work was partly supported by the European
Commission under FP7 project Inter-Trust and
H2020 project CLARUS, by the Spanish Ministry of
Science and Innovation through project ICWT
TIN2012-32757 and by the Government of
Catalonia under grant 2014 SGR 537. This work was
also made possible through the support of a grant
from Templeton World Charity Foundation. The
opinions expressed in this paper are those of the
authors and do not necessarily reflect the views of
Templeton World Charity Foundation.
OntologySelectionforSemanticSimilarityAssessment
575

REFERENCES
Al-Mubaid, H. & Nguyen, H. A. 2009. Measuring
Semantic Similarity between Biomedical Concepts
within Multiple Ontologies. IEEE Transactions on
Systems, Man, and Cybernetics, Part C: Applications
and Reviews, 39(4), pp 389-398.
Batet, M., Erola, A., Sánchez, D. & Castellà-Roca, J.
2013. Utility preserving query log anonymization via
semantic microaggregation. Information Sciences,
242(1), pp 49-63.
Batet, M., Gibert, K. & Valls, A. Semantic clustering
based on ontologies: an application to the study of
visitors in a natural reserve. In: Filipe, J. & Fred, A. L.
N., eds. 3th International Conference on Agents and
Artificial Intelligence (ICAART'11), 2011a Rome,
Italy. SciTePress, 283-289.
Batet, M. & Sánchez, D. 2014. A review on semantic
similarity. Encyclopedia of Information Science and
Technology, Third Edition. IGI Global.
Batet, M., Sánchez, D. & Valls, A. 2011b. An ontology-
based measure to compute semantic similarity in
biomedicine. Journal of Biomedical Informatics,
44(1), pp 118-125.
Ding, L., Finin, T., Joshi, A., Pan, R., Cost, R. S., Peng,
Y., Reddivari, P., Doshi, V. & Sachs, J. Swoogle: A
Search and Metadata Engine for the Semantic Web.
In: Grossman, D. A., Gravano, L., Zhai, C., Herzog,
O. & Evans, D. A., eds. thirteenth ACM international
conference on Information and knowledge
management, CIKM 2004, 2004 Washington, D.C.,
USA. ACM Press, 652-659.
Jiang, J. J. & Conrath, D. W. Semantic Similarity Based
on Corpus Statistics and Lexical Taxonomy.
International Conference on Research in
Computational Linguistics, ROCLING X, Sep 1997
Taipei, Taiwan. 19-33.
Li, Y., Bandar, Z. & McLean, D. 2003. An Approach for
Measuring Semantic Similarity between Words Using
Multiple Information Sources. IEEE Transactions on
Knowledge and Data Engineering, 15(4), pp 871-882.
Martínez, S., Valls, A. & Sánchez, D. 2012. Semantically-
grounded construction of centroids for datasets with
textual attributes. Knowledge-Based Systems, 35(1),
pp 160-172.
Mikolov, T., Chen, K., Corrado, G. & Dean, J. Efficient
Estimation of Word Representations in Vector Space.
International Conference on Learning Representations,
2013. 1-12.
Nelson, S. J., Johnston, D. & Humphreys, B. L. 2001.
Relationships in Medical Subject Headings.
Relationships in the Organization of Knowledge. K.A.
Publishers.
Pedersen, T., Pakhomov, S., Patwardhan, S. & Chute, C.
2007. Measures of semantic similarity and relatedness
in the biomedical domain. Journal of Biomedical
Informatics, 40(3), pp 288-299.
Petrakis, E. G. M., Varelas, G., Hliaoutakis, A. &
Raftopoulou, P. 2006. X-Similarity:Computing
Semantic Similarity between Concepts from Different
Ontologies. Journal of Digital Information
Management, 4(1), pp 233-237.
Pirró, G. 2009. A semantic similarity metric combining
features and intrinsic information content. Data &
Knowledge Engineering, 68(11), pp 1289-1308.
Resnik, P. Using Information Content to Evalutate
Semantic Similarity in a Taxonomy. In: Mellish, C. S.,
ed. 14th International Joint Conference on Artificial
Intelligence, IJCAI 1995, 1995 Montreal, Quebec,
Canada. Morgan Kaufmann Publishers Inc., 448-453.
Rodríguez, M. A. & Egenhofer, M. J. 2003. Determining
semantic similarity among entity classes from different
ontologies. IEEE Transactions on Knowledge and
Data Engineering, 15(2), pp 442–456.
Sánchez, D., Batet, M., Isern, D. & Valls, A. 2012a.
Ontology-based semantic similarity: A new feature-
based approach. Expert Systems with Applications,
39(9), pp 7718-7728.
Sánchez, D., Solé-Ribalta, A., Batet, M. & Serratosa, F.
2012b. Enabling semantic similarity estimation across
multiple ontologies: An evaluation in the biomedical
domain. Journal of Biomedical Informatics, 45(1), pp
141-155.
Saruladha, K., Aghila, G. & Bhuvaneswary, A. 2010.
Computation of Semantic Similarity among Cross
Ontological Concepts for Biomedical Domain.
Journal of Computing, 2(8), pp 111-118.
Spackman, K. A. 2004. SNOMED CT milestones:
endorsements are added to already-impressive
standards credentials. Healthcare Informatics, 21(9),
pp 54-56.
Wu, Z. & Palmer, M. Verb semantics and lexical
selection. In: Pustejovsky, J., ed. 32nd annual Meeting
of the Association for Computational Linguistics,
1994 Las Cruces, New Mexico. Association for
Computational Linguistics, 133 -138.
ICAART2015-InternationalConferenceonAgentsandArtificialIntelligence
576
