
Assessment of the Extent of the Necessary Clinical Testing of New
Biotechnological Products Based on the Analysis of Scientific
Publications and Clinical Trials Reports
Roman Suvorov
1
, Ivan Smirnov
1
, Konstantin Popov
2
, Nikolay Yarygin
3
and Konstantin Yarygin
4
1
Institute of Systems Analysis of Russian Academy of Sciences, Moscow, Russia
2
Engelhardt Institute of Molecular Biology Russian Academy of Sciences, Moscow, Russia
3
State University of Medicine and Dentistry, Moscow, Russia
4
Institute of Biomedical Chemistry, Russian Academy of Medical Sciences, Moscow, Russia
Keywords:
Clinical Trials, Meta Analysis, Information Retrieval, Natural Language Processing, Machine Learning.
Abstract:
To estimate patients risks and make clinical decisions, evidence based medicine (EBM) relies upon the results
of reproducible trials and experiments supported by accurate mathematical methods. Experimental and clinical
evidence is crucial, but laboratory testing and especially clinical trials are expensive and time-consuming.
On the other hand, a new medical product to be evaluated may be similar to one or many already tested.
Results of the studies hitherto performed with similar products may be a useful tool to determine the extent
of further pre-clinical and clinical testing. This paper suggests a workflow design aimed to support such
an approach including methods for information collection, assessment of research reliability, extraction of
structured information about trials and meta-analysis. Additionally, the paper contains a discussion of the
issues emering during development of an integrated software system that implements the proposed workflow.
1 INTRODUCTION
The practice of evidence based medicine (EBM) in-
troduced in early 1990s has now become quite com-
mon. Among other things, EBM methods include ex-
amination of the outcomes of randomized clinical tri-
als, scientific literature surveys and analysis of the re-
sults of pre-clinical experiments.
Regenerative medicine is a relatively new inter-
disciplinary field of research and clinical practice. It
focuses on reparation, replacement or regeneration of
cells, tissues or even whole organs in order to recover
their functions. The general approaches employed in
regenerativemedicine include the use of small biolog-
ically activemolecules, gene therapy, stem cells trans-
plantation, tissue engineering, etc. Stem cell therapy
is now being widely tested in animal disease models
and patients with ischemic heart disease, stroke, au-
toimmune and many other medical conditions.
The evidence-based evaluation of the regenerative
medicine field demands detailed information system-
atization and analysis. The safety proof and the esti-
mation of possible harm of a method are mandatory
for this method to be allowed for use in humans. In
this case results of pre-clinical experiments with ani-
mals usually serve as the evidence basis.
Clinical and preclinical trials are expensive and
laborious. However, absolutely break through cures
based on revolutionary principles emerge rarely. Usu-
ally every novel treatment is just a development of
an already existing one or an application of a known
method to a different disease. Qualitative and quanti-
tative analysis of data already obtained by others may
help to save on pre-clinical and clinical tests.
To address the described problem we are develop-
ing a software that automates search and analysis and
aids meta-analysis of published results of pre-clinical
tests and clinical trials. This system integrates meth-
ods for metasearch, paper quality assessment, infor-
mation extraction, similarity search, classification and
other auxiliary resources like thesauri. The most dif-
ficult subtasks are selection of high-quality scientific
publications and clinical trials, information extraction
and comparison of the assessed methods on the basis
of the extracted information (classification).
The rest of paper is organized as follows: in Sec-
tion 2 we review the existing works on the subject
(including sub-subjects), in Section 3 we present the
343
Suvorov R., Smirnov I., Popov K., Yarygin N. and Yarygin K..
Assessment of the Extent of the Necessary Clinical Testing of New Biotechnological Products Based on the Analysis of Scientific Publications and
Clinical Trials Reports.
DOI: 10.5220/0005287403430348
In Proceedings of the International Conference on Pattern Recognition Applications and Methods (ICPRAM-2015), pages 343-348
ISBN: 978-989-758-077-2
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

proposed methodology for automated portfolio com-
pilation, in Section 4 we describe the proposed system
and in Section 5 we discuss the current state of work
and expected results.
2 RELATED WORK
The field of the structured information extraction
is widely exploited nowadays (Jensen et al., 2012).
Medical and clinical text mining is in the focus of the
modern research in the field (Demner-Fushman et al.,
2009). A number of shared tasks were held recently
(i2b2 and CLEF eHealth). During these shared tasks
participants were asked to develop methods to auto-
matically extract such information as medication and
diseases mentions, laboratory measurements descrip-
tions, characteristics of patients etc. Despite rather
good results were achieved, the general problem of
understanding medical texts has not been solved.
There is no doubt that these competitions will be held
in the future (Chapman et al., 2011). The most widely
used instruments for information extraction nowadays
are conditional random fields (Li et al., 2008), hand-
crafted heuristic rules (e.g. based on finite state ma-
chines), dictionary lookups (Savova et al., 2010) and
support vector machines (Kiritchenko et al., 2010).
The general problem within the mentioned ap-
proaches is the need of large annotated corpus to train
the models. There are three promising ways to over-
come this issue, i.e. crowdsourcing, bootstrapping,
(inter)active learning. Crowdsourcing has been con-
tinuously gaining its popularity during the past years,
but the confidence level of the crowdsourced corpora
is still far from ideal. (Zhai et al., 2013) reports
confidence between 0.7 to 0.9. Bootstrapping is a
very common technique used mainly for getting large
corpora from the Web and for building dictionaries
(Riloff et al., 1999). These corpora are mostly used to
learn to extract named entities and relations between
them (Etzioni et al., 2008).
The most promising approach for estimation of
possible effects of a substance on live beings or en-
vironment is the analysis of relations between struc-
ture of the substance molecule and its activity, i.e.
QSAR (Valerio Jr, 2009). Expert systems are as well
as QSAR modeling based ones are employed to solve
this task (Marchant et al., 2008). The existing models
lack prediction accuracy of such important character-
istics as carcinogenicity, genotoxicity, impact on fe-
tus, teratogenicity etc. Development fo combined ap-
proaches has been recently initiated. Such approaches
try to reconcile QSAR modeling with experimental
data in vivo and in vitro (Crump et al., 2010).
Hence, there are no production-ready systems for
safety and effectiveness estimation of the regenerative
medicine methods. However, we think that existing
models for chemical toxicity prediction may be useful
to build such a system.
3 WORKFLOW
In this section we describe the software system ex-
pected to simplify the developmentof newtreatments.
The system bases on the following principles:
• Minimization of the amount of manual labor in-
volved in search and analysis of the information.
• Unification of the methods used to solve various
sub-tasks (as much as possible).
• Intensive use the user’s feedback.
• Employment of the existing methods as effec-
tively as possible.
Figure 1 presents the flowchart of the general al-
gorithm of the system being developed.
To help user to collect information on a particu-
lar subject, we integrate a metasearch module to the
system. This module allows user to search in multi-
ple databases simultaneously. User fills in the search
query with the help of various domain-specific the-
sauri. Currently the system incorporates the UMLS
(Lindberg et al., 1993) as the thesaurus and the
Cochrane Library, PubMed and ClinicalTrials.Gov as
data sources. Most of publications in these libraries
are not freely available and thus it is impossible to add
full texts of the found documents to the local database
automatically. However, the system automates the
process of filling bibliographical information about a
publication and extracts the abstract. Finding the full
text of a paper is up to the user.
The next steps after paper retrieval are the analysis
of the document and its quality assessment.
To analyze text we employ the most up-to-date
methods. Firstly, the document is preprocessed using
the existing systems for medical and clinical text min-
ing. Currently only cTAKES (Savova et al., 2010) is
incorporated. The aim of such preprocessing is to ex-
tract as much information as possible without dupli-
cation of the efforts. Secondly, other information ex-
traction methods are applied to the information mined
as a result of the previous step. We will discuss the
way we represent and analyze the information below
in section 4. After initial analysis of a document a
number of interactive expert-controlled iterations for
information extraction follow. The system shows a
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
344
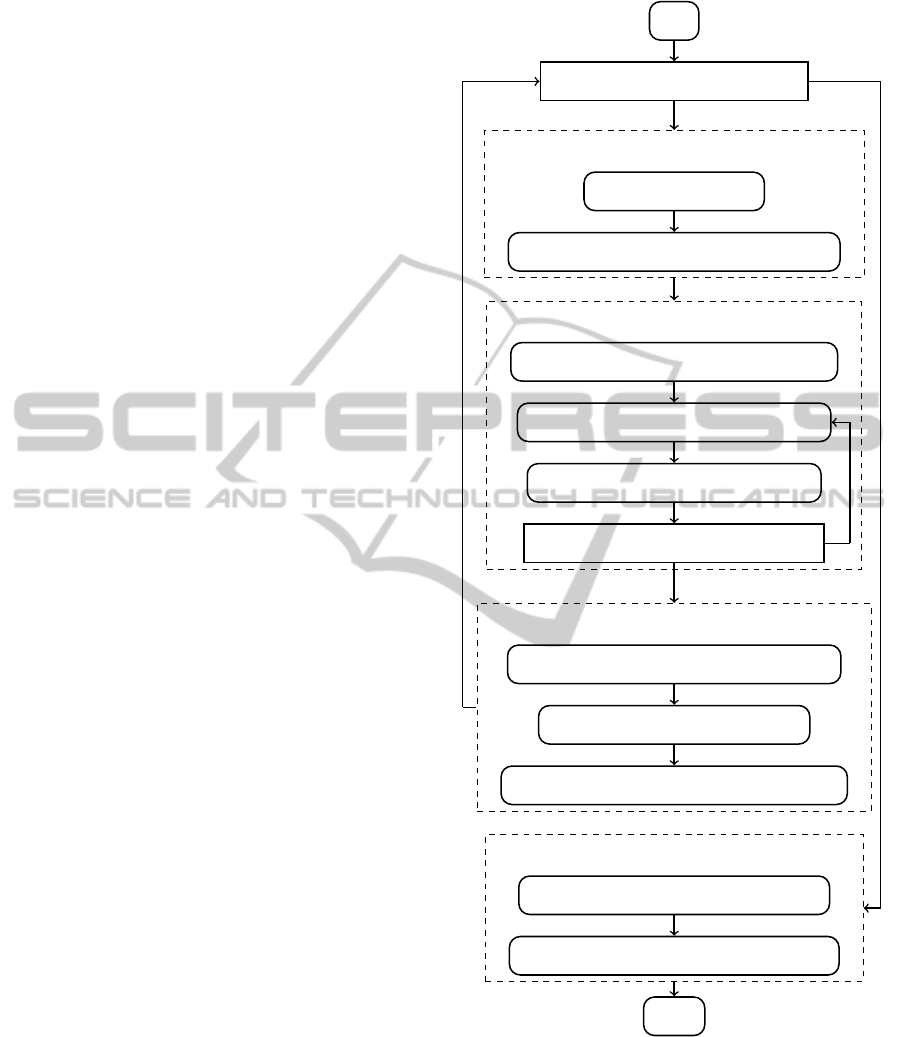
list of the extracted pieces of information, e.g. char-
acteristics and mentions of patients, methods, med-
ications, diseases etc. The expert is asked to check
each item and conclude whether it was extracted and
classified correctly or not. If the system fails to ex-
tract some data then the expert can manually annotate
these pieces of information in the text. The system
tries to update the models after each correction made
by the expert and then suggests new pieces of infor-
mation and so on.
The next step is to select reliable publications.
Quality assessment of a paper is organized through
the questionnaire filling. The expert is asked to rate
such aspects of the research presented in the analyzed
paper as adequacy of the used models, statistic rep-
resentativeness of the sample and sufficiency of the
results. The system shows to the user the content of
the paper as well as the various information extracted
from it, e.g. descriptions of methods, results of exper-
iments, various numeric characteristics. Additionally,
the paper compliance to general scientific work crite-
ria is checked (Shvets, 2014). Also, the system tries
to automatically estimate quality of the research pre-
sented on the basis of the extracted information. The
classifier automatically updates its model to predict
experts answers better.
The steps described above must be repeated until
all the relevant documents are retrieved from the data
sources.
After the information is collected and assessed,
the automatic survey is performed. The survey in-
cludes searching methods that are somehow similar
to the analyzed one, i.e. methods that target the
same nosology, use similar materials or models and
are tested in similar conditions. Thus, a method can
be represented by a vector in a N-dimensional space.
Having the meta-analysis problem defined this way,
we can employ various methods addressing the k-
nearest neighbor problem, e.g. inverted indexes, R-
trees, spatial hashing etc.
The last step is to estimate effects of the new
method. In the simplest case these effects include
probabilities to help and to harm. This problem can
be considered as a classification/regression problem.
The input features are the same as the ones used in
the survey. We propose to use a combination of both
statistical and logical methods. Statistical methods do
their best on big datasets with large number of fea-
tures. On the other hand, logical methods are capable
of providing a clear argumentation at the cost of com-
putational performance. We plan to develop an ap-
proach that is similar to the one employed by ProbLog
(De Raedt et al., 2007). It was successfully applied to
link discovery in biomedical domain. The analyzed
Metasearch
Analyze documents
Select papers
Automatic survey
Start
Is information collected enough?
User fills in the query
Automatically search Cochrane, PubMed
Preprocess the document using cTAKES
Extract information using current rules
Ask the user to assess extracted data
Does extraction works good enough?
Automatically estimate quality of a paper
Ask user to review the estimation
Update models used for quality assessment
Analyze similarity between researches
Build regression of factors and outcomes
Finish
No
No
Yes
Figure 1: The general algorithm flowchart.
graph contained 6 million vertexes and more than 15
million edges.
As a result of all these steps an user will get the
so-called clinical trials portfolio. It contains the list
of other methods in the area, information on how the
new method is related to the existing ones and estima-
tions of the outcomes of the new method. As a side
AssessmentoftheExtentoftheNecessaryClinicalTestingofNewBiotechnologicalProductsBasedontheAnalysisof
ScientificPublicationsandClinicalTrialsReports
345

effect, the proposedworkflowproduces a large dataset
that may be useful for the research in the fields of ma-
chine learning and text mining.
4 TEXT MINING ENGINE
This section covers the text mining and analytic en-
gine that we are developing to support the proposed
workflow. From the user’s point of view, this engine
must provide semi-supervised information extraction
functionality.
First, let us define the technical requirements. The
engine must:
• Integrate multiple information sources and repre-
sent data in a uniform way.
• Analyze the data fast.
• Scale seamlessly as the data size increases.
• Implement interactive machine learning paradigm
(thus response latencies must be rather small).
The mentioned information sources include vari-
ous preprocessors, ontologies and thesauri integrated
to the system. Variant is a particular piece of informa-
tion extracted from a text.
Property graph is the most suitable model for rep-
resenting highly interconnected data. Property graph
is a graph within which edges and nodes have a num-
ber of properties assigned to them, e.g. a node rep-
resenting a disease mention can have such properties
as ”umls
id”, ”normalized title” etc. This model is
implemented in a number of graph databases (Titan,
OrientDB, Neo4j etc.). Amount of time needed for a
graph database engine to execute a query does not de-
pend on the size of the database. Such model together
with efficient indexing allows representing all the data
in a uniform way and fast retrieval from very large
datasets. Currently we use a Cassandra-based setup
of Titan Database by Aurelius(TinkAurelius, 2014).
Both Cassandra and Titan support scaling inherently.
Generally the interactive text analysis is per-
formed as follows.
1. An expert uploads a document to the system.
2. The system applies preprocessors to the docu-
ment.
3. The system suggests a number of variants to the
expert.
4. The expert assesses each variant and tells whether
it was extracted right or wrong.
5. The system updates its internal models (rules,
support vectors etc.) to fit expert’s answers. Dur-
ing this update the system can ask the expert var-
ious questions regarding regularities in the data,
e.g. ”Is a hypothesis true?” or ”Was a hypothesis
a cause of a mistake?”.
6. Steps 3-6 are repeated until all the information of
interest is extracted from the document properly.
7. Steps 1-7 are repeated until sufficient coverage of
the subject is achieved.
The information extraction task includes extrac-
tion of cue, normalization and linking. To extract the
cue is to find a chunk of text that correspondsto the in-
formation of interest. Normalization consists in trans-
forming the text of cue to a single value, e.g. a canon-
ical object name or a number with unit. Linking is
the process of finding which pieces of information re-
late to which, e.g. treatments and the results of their
application. Most of these tasks can be treated as clas-
sification problems. To extract cues using classifiers
we employ the well-known BIO chunk encoding.
The most crucial part of the interactive text analy-
sis algorithm involves effective incremental update of
the classifier and suggestion of new variants.
There are a number of methods supporting incre-
mental update, including SVM (Cauwenberghs and
Poggio, 2001) and rules (Tsumoto and Tanaka, 1997).
Using vector space-based classifiers (such as SVM)
would need to convert the subgraph of interest to a
bunch of vectors. It can be done using breadth-first
traversal of the graph. Such an algorithm can con-
sider a K-neighborhood of each vertex of interest and
extract all unique simple paths beginning at it and
ending at other vertexes. K-neighborhood means that
only paths not longer than K are considered. Such
a conversion has a very subtle point - parameter K.
Large K would produce feature spaces of very high
dimensionality. Small K would lead to information
loss. Furthermore, the set of informativefeatures may
vary during the work. Thus, we propose using a rule-
based decision function.
To generalize the feature extraction process and
utilize the best of graph databases, we propose a spe-
cial algorithm for convertingthe data to a graph. Orig-
inal data is a set of interconnected objects in terms
of object-oriented programming. It originates from
the frame-system theory (Minsky, 1977). Thus, each
object has a type and a set of typed properties. The
properties can have simple types (numbers or strings)
or can refer to other objects. This algorithm bases on
the following principles.
• Type hierarchy is mapped to the graph. Each type
is mapped to a separate vertex as well as each
property is.
• Each object O is mapped to a separate vertex V
O
.
• Only a single vertex V
P,Val
is created for each dis-
tinct value Val of property P.
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
346

• Each V
O
has outcoming edges to the correspond-
ing property value verticesV
P,Val
. These edges are
labeled according to the property names.
• Each vertex V
O
and V
P,Val
have outcoming edges
to the corresponding vertices representing the
type hierarchy.
These principles lead to normalization of data dur-
ing the conversion and allow fast retrieval of objects
with the same property value.
Therefore, the rule generation can be effectively
implemented using the sequential covering technique
similar to described in (Huysmans et al., 2008). Hav-
ing data indexed like this, rules can operate very ef-
fectively. Detailed discussion of the developed text
mining engine will be a subject of a separate paper.
5 CONCLUSIONS
In this paper, we have described a problem of prepa-
ration for clinical trials, proposed a methodology and
partially described the corresponding tool that facil-
itates the safety and effectiveness estimation of the
regenerative medicine methods. Such a tool neither
existed nor proposed so far.
Our tool is in development at the moment.
Currently we have implemented components for
metasearch, linguistic processing of the downloaded
papers and a part of the quality assessment mod-
ule. The text mining engine was evaluated on CLEF
eHealth 2014 data and showed average F1-measure
when extracting the most difficult characteristic of
about 0.5 - 0.6, which is rather close to the winners of
the this year shared task. The work is still in progress,
thus the results are preliminary. The more detailed
explanation and analysis of the text mining engine is
needed and it should probably deserve a separate pa-
per.
The future work includes development of the rest
of the system; applying the system to build up a test
data set; quality assessment of the results produced by
all the implemented processing steps; improving the
methods according to the results of the quality assess-
ment. The most important problem regarding practi-
cal application of the system being developed is the
reliability of the produced estimations of the regener-
ative medicine methods. One possible solution to this
may be building a dataset that would contain papers
about the well-known and manually estimated treat-
ments. However, it is unclear how to verify that the
method extracts meaningful rules. This in turn can be
addressed either using cross-validation on large data
(hardly believable that a sufficiently large data set can
be collected) or by attracting a group of experts.
ACKNOWLEDGEMENTS
The project is supported by Russian Foundation for
Basic Research grant 13-07-12156.
REFERENCES
Cauwenberghs, G. and Poggio, T. (2001). Incremen-
tal and decremental support vector machine learning.
Advances in neural information processing systems,
pages 409–415.
Chapman, W. W., Nadkarni, P. M., Hirschman, L.,
D’Avolio, L. W., Savova, G. K., and Uzuner, O.
(2011). Overcoming barriers to nlp for clinical text:
the role of shared tasks and the need for additional
creative solutions. Journal of the American Medical
Informatics Association, 18(5):540–543.
Crump, K. S., Chen, C., and Louis, T. A. (2010). The future
use of in vitro data in risk assessment to set human ex-
posure standards: challenging problems and familiar
solutions. Environ. Health Perspect, 118:1350–1354.
De Raedt, L., Kimmig, A., and Toivonen, H. (2007).
Problog: A probabilistic prolog and its application in
link discovery. In IJCAI, volume 7, pages 2462–2467.
Demner-Fushman, D., Chapman, W. W., and McDonald,
C. J. (2009). What can natural language processing do
for clinical decision support? Journal of biomedical
informatics, 42(5):760–772.
Etzioni, O., Banko, M., Soderland, S., and Weld, D. S.
(2008). Open information extraction from the web.
Communications of the ACM, 51(12):68–74.
Huysmans, J., Setiono, R., Baesens, B., and Vanthienen, J.
(2008). Minerva: Sequential covering for rule extrac-
tion. Systems, Man, and Cybernetics, Part B: Cyber-
netics, IEEE Transactions on, 38(2):299–309.
Jensen, P. B., Jensen, L. J., and Brunak, S. (2012). Mining
electronic health records: towards better research ap-
plications and clinical care. Nature Reviews Genetics,
13(6):395–405.
Kiritchenko, S., de Bruijn, B., Carini, S., Martin, J., and
Sim, I. (2010). Exact: automatic extraction of clinical
trial characteristics from journal publications. BMC
medical informatics and decision making, 10(1):56.
Li, D., Kipper-Schuler, K., and Savova, G. (2008). Con-
ditional random fields and support vector machines
for disorder named entity recognition in clinical texts.
In Proceedings of the workshop on current trends in
biomedical natural language processing, pages 94–
95. Association for Computational Linguistics.
Lindberg, D. A., Humphreys, B. L., and McCray, A. T.
(1993). The unified medical language system. Meth-
ods of information in medicine, 32(4):281–291.
Marchant, C. A., Briggs, K. A., and Long, A. (2008). In
silico tools for sharing data and knowledge on tox-
icity and metabolism: Derek for windows, meteor,
and vitic. Toxicology mechanisms and methods, 18(2-
3):177–187.
AssessmentoftheExtentoftheNecessaryClinicalTestingofNewBiotechnologicalProductsBasedontheAnalysisof
ScientificPublicationsandClinicalTrialsReports
347

Minsky, M. (1977). Frame-system theory. Thinking: Read-
ings in cognitive science, pages 355–376.
Riloff, E., Jones, R., et al. (1999). Learning dictionaries for
information extraction by multi-level bootstrapping.
In AAAI/IAAI, pages 474–479.
Savova, G. K., Masanz, J. J., Ogren, P. V., Zheng, J., Sohn,
S., Kipper-Schuler, K. C., and Chute, C. G. (2010).
Mayo clinical text analysis and knowledge extraction
system (ctakes): architecture, component evaluation
and applications. Journal of the American Medical
Informatics Association, 17(5):507–513.
Shvets, A. (2014). A method of automatic detec-
tion of pseudoscientific publications. In Filev, D.,
Jabkowski, J., Kacprzyk, J., Krawczak, M., Popchev,
I., Rutkowski, L., Sgurev, V., Sotirova, E., Szynkar-
czyk, P., and Zadrozny, S., editors, Intelligent Sys-
tems’2014, volume 323 of Advances in Intelligent Sys-
tems and Computing, pages 533–539. Springer Inter-
national Publishing.
TinkAurelius (2014). Titan: A distributed graph database.
http://thinkaurelius.github.io/titan/.
Tsumoto, S. and Tanaka, H. (1997). Incremental learning
of probabilistic rules from clinical databases based on
rough set theory. In Proceedings of the AMIA Annual
Fall Symposium, page 198. American Medical Infor-
matics Association.
Valerio Jr, L. G. (2009). ¡ i¿ in silico¡/i¿ toxicology for
the pharmaceutical sciences. Toxicology and applied
pharmacology, 241(3):356–370.
Zhai, H., Lingren, T., Deleger, L., Li, Q., Kaiser, M.,
Stoutenborough, L., and Solti, I. (2013). Web 2.0-
based crowdsourcing for high-quality gold standard
development in clinical natural language processing.
Journal of medical Internet research, 15(4).
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
348
