
Tool Facilitating Construction of Ontologies on the KIM Platform
Roman Mou
ˇ
cek
1,2
, Jan Smitka
1
and Petr Je
ˇ
zek
1,2
1
Department of Computer Science and Engineering, Faculty of Applied Sciences, University of West Bohemia,
Univerzitn
´
ı 8, 306 14 Plze
ˇ
n, Czech Republic
2
New Technologies for the Information Society, Faculty of Applied Sciences, University of West Bohemia,
Univerzitn
´
ı 8, 306 14 Plze
ˇ
n, Czech Republic
Keywords:
Electrophysiology, Semantic Repository, Semantic Web, Ontology, KIM Platform, KIM-OWLImport,
EEG/ERP Portal, EEGbase.
Abstract:
During research based on experimental work vast amounts of data and associated metadata are usually pro-
duced. This is also the case of experimental work using the techniques of electroencephalography and event
related potentials. The collected data and associated metadata have to be stored, analyzed, and eventually
shared among research groups. Beside storing data and metadata from experiments, it is often beneficial to
collect additional information from other sources related to the kind of experiment performed. These infor-
mation sources are mostly scientific and technical publications, manuals describing the used infrastructure,
and topical discussions appearing on the web. This article deals deals with selection and use of a semantic
repository for such information sources. Development of a simple prototype ontology is shortly presented and
a tool that facilitates construction of ontologies on the KIM platform is described. Sets of test documents are
used to verify the functionality of the tool.
1 INTRODUCTION
During research based on experimental work vast
amounts of data and associated metadata are usu-
ally produced. This is also the case of experimen-
tal work using the techniques of electroencephalogra-
phy (EEG) and event related potentials (ERP). The
collected data and associated metadata have to be
stored, analyzed and eventually shared among re-
search groups.
For performing electrophysiological experiments
our research group (Neuroinformatics research group,
2014) uses the software and hardware infrastructure
described in (Moucek et al., 2014). Experimental data
and metadata are stored in EEG/ERP Portal (EEG-
base) (Jezek and Moucek, 2012) that is available as
an online tool (Neuroinformatics research group, Uni-
versity of West Bohemia, 2014) for storage, manage-
ment, and sharing of electrophysiological data. These
data and metadata, enriched by additional semantic
constructions written as a part of code annotations,
can be also published as dump files by using the lan-
guages of the Semantic Web. Through the use of
these open standards for data exchange and subse-
quent integration of the EEG/ERP Portal into the Neu-
roscience Information Framework (NIF) (Gupta et al.,
2008) the portal data are available to other researchers
via both the EEG/ERP portal and NIF interfaces.
Beside storing data and metadata from experi-
ments, it is often beneficial to collect additional in-
formation from other sources related to the kind of
experiment performed. These information sources are
scientific and technical publications, manuals describ-
ing the used infrastructure, and topical discussions ap-
pearing on the web. Search in these sources is not
easy. Leaving aside the overall number of these in-
formation sources, other troubles come with different
terminology used by individual authors. Individual
facts can be even indicated by using different key-
words. It often happens that it is necessary to refor-
mulate the search query several times and enter dif-
ferent keywords to find desired information.
The aim of this work is to find a suitable solution
for aggregating, storing and searching unstructured
electrophysiological data (mostly in the form of text
documents) that contain additional, supporting or ex-
plaining information related to the experimental work
conducted. The secondary aim is to use up and/or
extend the already existing description of the domain
and use knowledge of the Semantic Web languages
and technologies.
The article is organized in the following way. Sec-
659
Mou
ˇ
cek R., Smitka J. and Ježek P..
Tool Facilitating Construction of Ontologies on the KIM Platform.
DOI: 10.5220/0005295806590665
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2015), pages 659-665
ISBN: 978-989-758-068-0
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

tion 2 shortly introduces the basic terminology, lan-
guages, and technologies of the Semantic Web to en-
sure that even the reader unfamiliar with the Semantic
Web could follow the text. In the next section appro-
priate repositories for storing semantic data that en-
able users to use full-text search are compared. The
basic features of the selected semantic repository are
also described. Section 4 completes requirements on
the documents stored in the selected semantic reposi-
tory and introduces a part of the domain ontology that
is used to describe domain knowledge. Section 5 de-
scribes a tool named KIM-OWLImport that was cre-
ated to facilitate the ontology development. The next
section presents the verification process and results of
full-text search when using the domain ontology and
the KIM-OWLIMport tool. The last section summa-
rizes the whole work and outlines the possible further
development of the implemented solution.
2 STATE OF THE ART
This section very briefly introduces the basic termi-
nology used in the Semantic Web. At first, it is im-
portant to mention that the initial idea of the Semantic
Web has been continuously changing from the very
complex view of this phenomenon as an organized
layered system of standards, languages, and technolo-
gies to the practical (and often separate) use of spe-
cific resources.
RDF (Miller and Manola, 2004) is a language for
representation of knowledge about sources. These
sources can be identified and referenced via their Uni-
form Resource Identifier (URI) in the WWW net-
work. Knowledge is then organized as a graph struc-
ture and represented by triples (subject, predicate, ob-
ject). RDF schema (RDFs) adds a type system to
RDF; it is possible to define a hierarchy of classes.
Classes and properties defined by RDFS can be found
in (Guha and Brickley, 2004).
Web Ontology Language (OWL) is a language
based on description logic that provides means for ex-
pressing richer semantic relationships. It is used for
creating ontologies. Documents in RDF and OWL
can be stored in various syntaxes, for example RD-
f/XML, OWL/XML or Turtle. While the RDF lan-
guage was generally well adopted by a larger com-
munity, the OWL language due to its complexity is
understandable to a substantially smaller community
of experts.
Semantic repositories store Semantic Web data in
RDF graphs. These data can be queried using spe-
cific query languages. The most used language is the
SPARQL language (Harris and Seaborne, 2013) that
is standardized by W3C.
3 SELECTION OF SEMANTIC
REPOSITORY
This section describes the process of selecting seman-
tic repository that would be appropriate for our aim.
We defined the following criteria to select a semantic
repository that are ordered by their importance to our
task:
• possibility and quality of full-text search,
• performance,
• RDF and OWL support.
There are several benchmarks that deal with the
comparison of performance of semantic repositories.
We used the Berlin SPARQL Benchmark (BSBM) to
compare the performance of a selected set of semantic
repositories. There were used two data sets containing
100 000 748 triplets (100M) and 200 031 975 triplets
(200M). The results (time to import the entire data
set and the number of queries per time unit with the
different number of simultaneously connected clients)
are available in Tables 1 and 2.
Table 1: Time to import the entire data set.
Repository 100M dataset 200M dataset
4store 26min 42s 1h 12min 04s
BigData 1h 03min 47s 3h 24min 25s
BigOwlim 17min 22s 38min 36s
TDB 1h 14min 48s 2h 45min 13s
Virtuoso 1h 49min 26s 3h 59min 38s
Table 2: The number of queries per time unit.
Repository 100M dataset 200M dataset
4store 5589 4593
BigData 2428 1795
BigOwlim 3534 1795
TDB 2274 1443
Virtuoso 7352 4669
Finally we chose the semantic repository OWLIM
with the KIM platform (named BigOwlim in Tables 1
and 2) that provided extended search and automated
annotation of documents. This repository also pro-
vided good performance results as it can be seen in
Tables 1 and 2.
The KIM platform is a product that enables users
to upload documents in various formats (e.g. HTML,
XML, RTF or PDF documents are supported) into
a semantic repository. It also provides resources
for automated annotation of repository documents
according to prepared ontologies and resources for
HEALTHINF2015-InternationalConferenceonHealthInformatics
660

subsequent search in them. The principle of auto-
mated annotation and its implementation is available
in (Kiryakov et al., 2003). The KIM platform works
over the OWLIM repository. Processing and anno-
tation of input documents is implemented using the
tools of the GATE project. The functionality of the
KIM platform is provided using the SOAP web ser-
vices and JAVA RMI interface. The OWLIM reposi-
tory is used for example for BBC Sport web (Rayfield,
2012) or the National Archives in Great Britain (On-
totext AD, 2012a).
The knowledge base used in the OWLIM repos-
itory is based on the PROTON ontology (Ontotext
AD, 2012b). This ontology is divided into three mod-
ules and create a suitable cornerstone for the ontolo-
gies specific to elaborated domains. The KIM World
Knowledge Base as a part of the KIM platform is also
based on this ontology. Documents can be stored in
three supported repositories: Apache Lucene, Seman-
tic Annotation Repository, and Mimir.
To use the semantic repository for a specific do-
main thus means to define a domain ontology. All
classes of this ontology have to be subclasses of the
class Entity from the Proton ontology.
4 REQUIREMENTS AND
DOMAIN ONTOLOGY
Before we started to create a domain ontology we
had to decide what documents and in which for-
mats would be stored in the selected semantic repos-
itory. Finally we decided to index two types of doc-
uments: scientific and other technical publications in
pdf format and discussions in expert electrophysiol-
ogy groups published in the social network LinkedIn.
The typical domain information searched in these
sources could be, for example, as the following one:
”I want to find a discussion about the matching pur-
suit method that is used to investigate the existence
of the P3 component”. The aim of the proposed so-
lution is to find the relevant information for this kind
of query. It is also necessary to easily find the query
results in original documents.
Beside necessary installation and configuration of
the KIM platform the domain ontology that enables
an advanced search has to be created. Since the on-
tology serves for the evaluation of functionality of the
semantic repository, it was not proposed to be con-
structed using the best principles for creating ontolo-
gies like looking for terms in the ontologies cover-
ing similar domains. As a result a simple prototype
of the domain ontology using the data model of the
EEG/ERP Portal enriched by defining terms and rela-
tionships from specific parts of the domain was devel-
oped.
The base of this ontology is a collection of evoked
potentials components (Figure 1) and methods used
for EEG/ERP signal processing. The graph descrip-
tion of the components includes individual compo-
nents, their polarity and their group membership.
Some components also have their aliases which can
be found in the literature. When creating these aliases
we took into account some terminological customs,
for example the component P3 is usually used as an
alias to the component P3b and not as a superior term
for all P3 components. That is why the component P3
is considered as an alias to the P3b component in the
graph structure and not as a super class denoting the
whole family of components. This graph as well as
the graph representing the signal processing methods
was expressed as an ontology in the OWL language.
The KIM platform imposes additional require-
ments on the form of ontologies, for example, all
created terms have to be marked as trusted. How-
ever, when creating an ontology is better to focus
on description of knowledge and extend the ontol-
ogy by additional information later. A tool named
KIM-OWLImport was proposed and developed to fa-
cilitate development of ontologies for the KIM plat-
form and OWLIM repository. It enables users to focus
on the ontology development itself while it automat-
ically transforms it into/from the OWLIM repository
in structures relevant to the Proton ontology.
5 KIM-OWLImport
The KIM-OWLImport is a tool that allows extend-
ing an existing ontology in a way that it can be used
within the KIM platform (trusted resources have to be
defined, all classes have a supeclass Entity or eventu-
ally a more specific class from a limited set, visibility
in the web interface is ensured). Moreover, the tool is
designed to be easily extensible by the possible future
conditions defined within the KIM platform. The tool
has a graphical user interface in which users can add,
create and/or edit their ontologies. For each ontology
it is possible to define a set of rules that are applied
back to the ontology. The tool does not work with
the basic RDF/XML syntax but works directly with
triples using the Sesame library (Sesame developers,
2012). Sesame provides API that is used (in this case)
to access a semantic repository OWLIM-Lite. Sesame
also supports its query language SeRQL. The follow-
ing query (Figure 2) shows the case when individual
entities are extended with the property generatedby;
the property value is a trusted resource.
ToolFacilitatingConstructionofOntologiesontheKIMPlatform
661
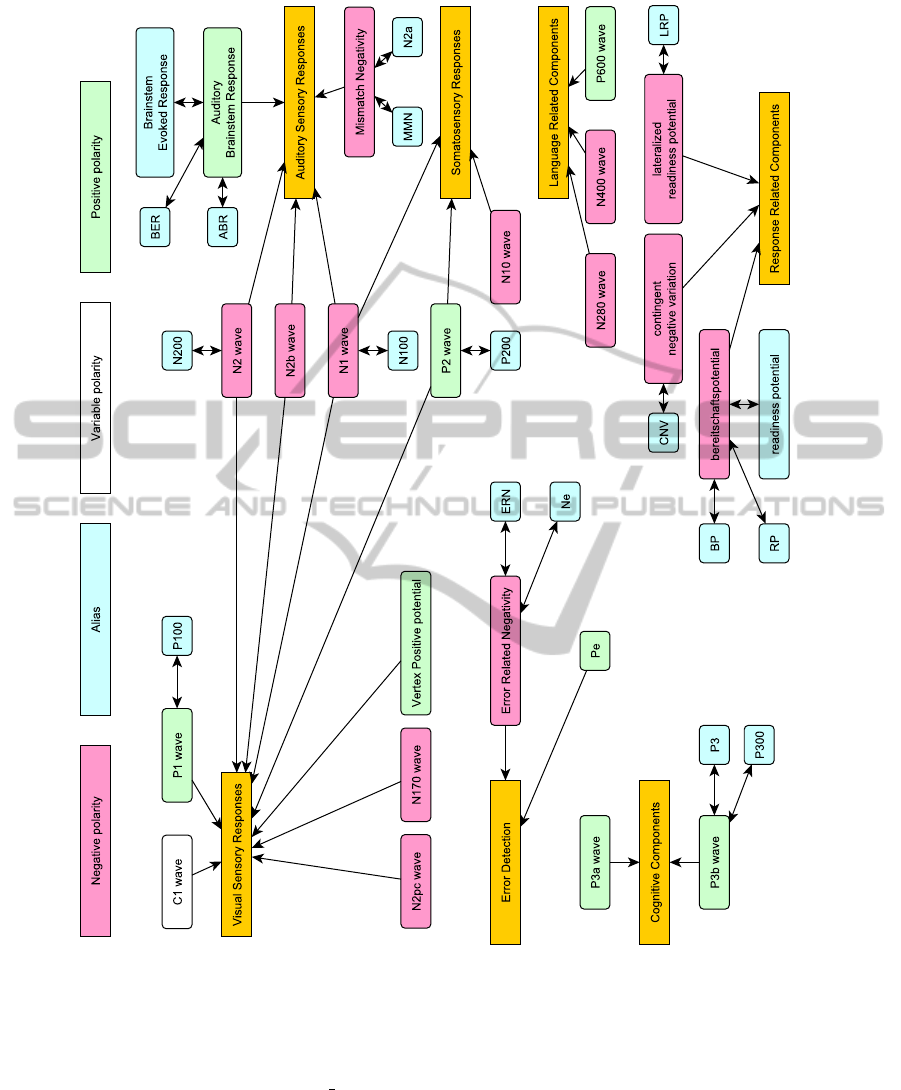
Figure 1: Ontology of ERP Components.
The query finds all the entities which type is a di-
rect instance of the owl:Class type. For the entities
which are found the triplet appearing in the CON-
STRUCT part is generated. The parameter sourceUri
has to be completed with the URI of a trusted re-
source. The outputs from all queries are stored us-
ing the Sesame API that provides classes for storing
triplets in various formats.
The UML diagram of the most important classes
and interfaces of the KIM-OWLImport is shown in
Figure 3.
The files containing ontologies are uploaded into
semantic repositories. The class RepositoryManager
ensures the management of these repositories. Each
created repository has its own configuration and it
is accessed using the RepositoryWrapper class. An
ontology is represented by the class AbstractSource.
Ontology construction is ensured by the implementa-
tion of the ISourceFactory interface. The parameters
necessary for creating a specific resource (URL, file
path) are passed by the class implementing the ISour-
ceParams interface. All resources and their factories
HEALTHINF2015-InternationalConferenceonHealthInformatics
662

CONSTRUCT DISTINCT { E n t i t y } p r o t o n s : g e n e rate d B y { s o u r c e U r i }
FROM { E n t i t y } r d f : t y p e {Type } , { Type } sesame : d i r e c t T y p e { C l a s s T y p e }
WHERE C l a s s T y p e = owl : C l a s s
USING NAMESPACE
p r o t o n s = < h t t p : / / p r o t o n . se m a n t ic w e b . or g / 2 0 0 6 / 0 5 / p r o t o n s #>
Figure 2: Entities extended with the property generatedby.
GUI
SourceManager
«interface»
ISourceFactory
AbstractSource
RuleManager
«interface»
IRuleFactory
RepositoryWrapper
RepositoryManager
AbstractRule
creates
1
1..*
creates
imported to
creates and shutdowns
1
1..*
queries
exported by
11..*
creates
11
1
1
Figure 3: KIW-OWLImport - UML diagram of the most
important classes and interfaces.
are managed by the SourceManager class.
A number of rules, which ensure construction of
triplets, can be assigned to each resource. The archi-
tecture is very similar to the management of ontolo-
gies. Specific rules that implement the AbstractRule
class are created by the factories implementing the
IRuleFactory interface. Rules parameters are repre-
sented by the classes implementing the IRuleParams
interface. Individual factories are managed by the
class RuleManager, the rules are aggregated in a col-
lection belonging to the resource. Rules parameters
are configured after the construction of the rule, while
ontology resources are configured at the construction
time.
The rules method getStatements() performs
queries on the semantic repository and returns a re-
sult. Individual triplets are then stored in the collec-
tion implementing the interface Iteration that is pro-
vided by Sesame.
6 RESULTS
The functionality of the KIM-OWLImport tool was
verified using two following approaches. The first ap-
proach involved the semantic annotation according to
created ontology, while in the second approach a spe-
cific query was tested. Both tests were performed
using the test documents in which the results had
been known in advance and using the real documents
stored in the semantic repository.
For the verification of the functionality of the se-
mantic annotation the test document containing all the
terms (including aliases) from the ontology was cre-
ated. Each term was placed in a simple sentence that
simulates the neighbourhood of the term. The whole
text was uploaded to the KIM platform. The anno-
tated document is available in Figure 4.
Figure 4: A part of the annotated document shown in the
KIM user interface. Annotated terms are highlighted in
bold.
Except for one single term all keywords were cor-
rectly recognized. The difficulty was with the term
ART2 , which contains letters and numerals. Only the
number identified as a numeral was recognized in this
term, the rest of the term was not recognized. The
terms containing one letter followed by one or more
digits were recognized correctly.
Then 76 documents from public sources were up-
loaded to the semantic repository and annotated. An
example of annotated document is available in Fig-
ure 5.
The search in the semantic repository was verified
using the search scenario: ”I want to find a discus-
sion about the matching pursuit method that is used to
investigate the existence of the P3 component.” The
query contains the following keywords: ”matching
pursuit”, ”P3”, and ”existence”. For testing a set of
ToolFacilitatingConstructionofOntologiesontheKIMPlatform
663
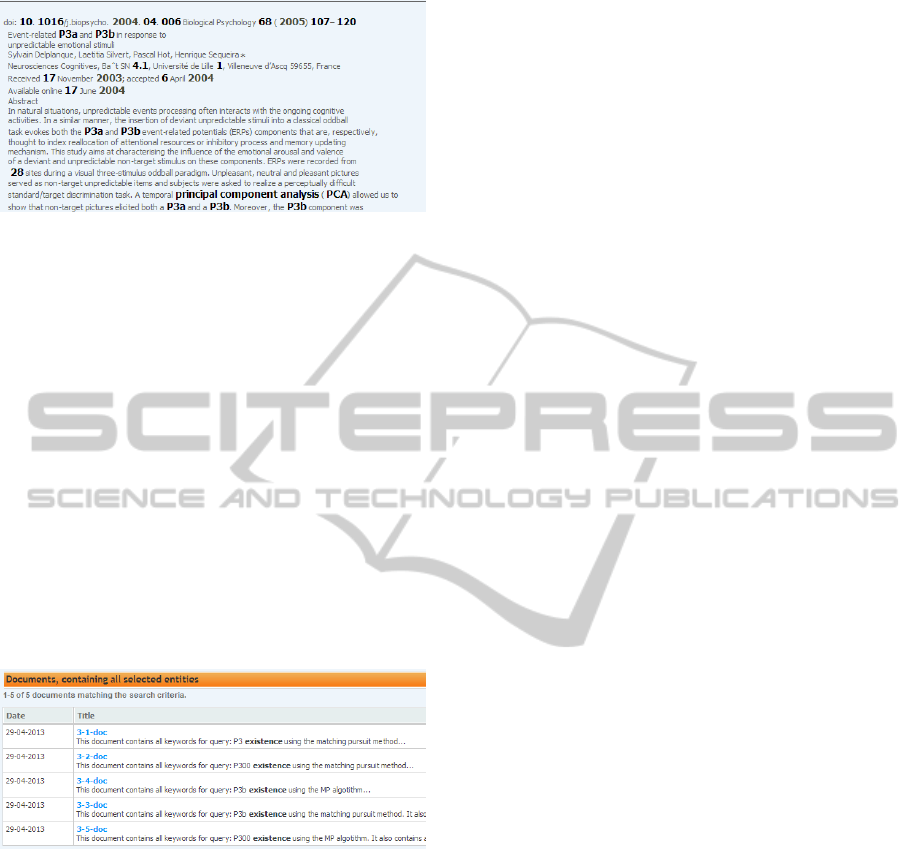
Figure 5: Annotated document dealing with P3a and P3b
components shown in the KIM Platform user interface.
15 documents was created. These documents were
divided into three groups:
• Documents containing none of the keywords in
the query.
• Documents containing any subset of the keywords
in the query, eventually the aliases of the entities
”P3” and ”matching pursuit”.
• Documents containing all the keywords from the
query and aliases of the entities ”P3” and ”match-
ing pursuit”.
The set of documents was uploaded to the KIM plat-
form and the query described above was applied. The
results are shown in Figure 6. As it was expected the
search results included only documents that contained
all the keywords.
Figure 6: Results of searching entities ”P3b”, ”matching
pursuit”, and the keyword ”existence”.
When forming a query any alias for the selected
entity can be entered. Apart from ontology entities it
is also possible to enter any keyword, which will be
subsequently used for full-text search.
7 CONCLUSIONS
This article at first briefly deals with the principles and
technologies of the Semantic Web. It is followed by
a short overview of the widespread semantic reposito-
ries, which are compared in performance tests. Based
on the features and the use in real projects the seman-
tic repository OWLIM and the KIM Platform were se-
lected for storing documents in the electrophysiology
domain.
The KIM Platform allows semantic annotation of
documents based on ontology, which is stored in the
semantic repository. The annotated documents can be
searched and by using ontology terms it is possible to
get more relevant results than in the case of common
full-text search. The used ontology must be based
on the PROTON ontology and has to meet additional
conditions for the full functionality of the platform.
Semantic annotation thus requires an ontology,
which contains definitions and classification of do-
main terms. Since the ontology fully covering the
electrophysiology domain does not exist yet, a proto-
type ontology containing a part of domain knowledge
was developed.
To facilitate the development of the ontology,
which meets the requirements of the KIM Platform,
a tool named KIM-OWLImport was designed and im-
plemented. This tool is able to load the selected ontol-
ogy in the semantic repository in memory and extend
it according to defined rules in a way so that it can
be used for semantic annotation of documents (in our
case scientific and technical documents and discus-
sions from the social network LinkedIn).
Downloaded documents are annotated and in-
dexed within the KIM Platform. Subsequent search is
made possible through the web interface, which is the
part of the KIM platform. Search functionality was
verified on a set of test documents and on scientific
publications dealing with research in event related po-
tentials domain.
The tool KIM-OWLImport thus can be used for
automated transfer to any ontology structure, which
corresponds to the PROTON ontology required by the
KIM Platform. The tool is easily extensible by addi-
tional rules and may become a full-fledged transfor-
mation tool.
Within the further development it is necessary to
replace the prototype ontology with the complex on-
tology, which will include a larger number of the key
terms from the electrophysiological domain. This on-
tology is currently being developed within Ontology
for Experimental Neurophysiology (OEN) group.
ACKNOWLEDGEMENTS
This work was supported by the UWB grant SGS-
2013-039 Methods and Applications of Bio- and
Medical Informatics and by the European Regional
Development Fund (ERDF), Project ”NTIS - New
Technologies for Information Society”, European
Centre of Excellence, CZ.1.05/1.1.00/02.0090.
HEALTHINF2015-InternationalConferenceonHealthInformatics
664

REFERENCES
Guha, R. V. and Brickley, D. (2004). RDF vocabulary de-
scription language 1.0: RDF schema. Recommenda-
tion, W3C.
Gupta, A., Bug, W., Marenco, L., Qian, X., Condit, C., Ran-
garajan, A., Mller, H., Miller, P., Sanders, B., Grethe,
J., Astakhov, V., Shepherd, G., Sternberg, P., and Mar-
tone, M. (2008). Federated access to heterogeneous
information resources in the neuroscience informa-
tion framework (nif). Neuroinformatics, 6:205–217.
10.1007/s12021-008-9033-y.
Harris, S. and Seaborne, A. (2013). SPARQL 1.1 Query
Language. Recommendation, W3C.
Jezek, P. and Moucek, R. (2012). System for EEG/ERP
Data and Metadata Storage and Management. Neural
Network World, 22(3):277–290.
Kiryakov, A., Popov, B., Ognyanoff, D., Manov, D., Kir-
ilov, A., and Goranov, M. (2003). Semantic annota-
tion, indexing, and retrieval. In 2nd International Se-
mantic Web Conference. Springer-Verlag Berlin Hei-
delberg.
Miller, E. and Manola, F. (2004). RDF primer. Recommen-
dation, W3C.
Moucek, R., Bruha, P., Jezek, P., Mautner, P., Novotny,
J., Papez, V., Prokop, T., Rondk, T., tebetk, J., and
Vareka, L. (2014). Software and hardware infrastruc-
ture for research in electrophysiology. Frontiers in
Neuroinformatics, 8(20).
Neuroinformatics research group (2014 [cited
10. 10. 2014]). Neuroinformatics research group web
portal.
Neuroinformatics research group, University of West Bo-
hemia (2014). EEG/ERP Portal (EEGBase) eeg-
database.kiv.zcu.cz.
Ontotext AD (2012a). The national archives: Semantic
knowledge base.
Ontotext AD (2012. [cit. 21. 10. 2012]b). Proton ontology.
Rayfield, J. (2012). Sports refresh: Dynamic semantic pub-
lishing.
Sesame developers (2012. [cited 21. 10. 2012]). Sesame
User Guide: rdf:about Sesame 2. Sesame developers.
ToolFacilitatingConstructionofOntologiesontheKIMPlatform
665
