
BigTexts
A Framework for the Analysis of Electronic Health Record Narrative Texts based
on Big Data Technologies
Wilson Alzate Calder
´
on
1
, Alexandra Pomares Quimbaya
1
, Rafael A. Gonzalez
1
and Oscar Mauricio Mu
˜
noz
2
1
Departamento de Ingenier
´
ıa de Sistemas, Pontificia Universidad Javeriana, Bogot
´
a, Colombia
2
Departamento de Medicina, Pontificia Universidad Javeriana, Hospital Universitario San Ignacio, Bogot
´
a, Colombia
Keywords:
Electronic Medical Record, Big Data, Natural Language Processing, Text Mining, Framework.
Abstract:
In the healthcare domain the analysis of Electronic Medical Records (EMR) may be classified as a Big Data
problem since it has the three fundamental characteristics: Volume, Variety and Speed. A major drawback is
that most of the information contained in medical records is narrative text, where natural language processing
and text mining are key technologies to enhance the utility of medical records for research, analysis and
decision support. Among the tasks performed for natural language processing, the most critical, in terms of
time consumption, are the pre-processing tasks that give some structure to the original non-structured text.
Studying existing research on the use of Big Data techniques in the healthcare domain reveals few practical
contributions, especially for EMR analysis. To fill this gap, this paper presents BigTexts, a framework that
provides pre-built functionalities for the execution of pre-processing tasks over narrative texts contained in
EMR using Big Data techniques. BigTexts enables faster results on EMR narrative text analysis improving
decision making in healthcare.
1 INTRODUCTION
Nowadays, the amount of information stored in the
world is estimated at around of 1,200 exabytes (10
18
bytes), which come from multiple sources; for exam-
ple, Google processes over 24 petabytes (10
15
bytes)
of data per-day and Facebook gets more than 10
million photos every hour (Mayer-Schonberger and
Cukier, 2013). Given these large amounts of data
that are currently being generated, the paradigm of
Big Data has been proposed to describe the phe-
nomenon and suggest the need for organizations to
adopt innovative tools that can help to accurately de-
termine opportunities for improvement and creating
value (Moore et al., 2013). A Big Data problem is
one that complies with the three Vs: Volume (scale
of data), Variety (different forms of data) and Veloc-
ity (on data flow analysis) (McAfee and Brynjolfsson,
2012). As in other domains, the healthcare domain
is generating increasingly large amounts of data, sig-
nificantly within Electronic Medical Records (EMR).
Such data becomes an invaluable source for analy-
sis and research to address public health issues, im-
prove service quality and increase the coverage and
efficiency of health services (Sengupta, 2013).
Considering the large volume of data contained
in EMR, the variety going from structured to un-
structured data, and the speed which certain stud-
ies require, the analysis of EMR using computational
techniques may be classified as a Big Data problem
(Moore et al., 2013). In particular, the existence of
narrative text in fields such as nursing notes, treatment
plans, and lab tests, among others, generates a major
challenge that needs to be addressed natural language
processing technologies, text mining and Big Data.
To face this requirement from the healthcare do-
main, the aim of this paper is to present Big Texts,
a Big Data framework for the pre-processing of nar-
rative texts contained in EMRs. Big Texts provides
pre-built functions for the execution of pre-processing
tasks required to apply text mining techniques like
tokenization and Part-of-Speech recognition over the
text. In order to improve the performance of pre-
processing tasks, Big Texts uses big data technologies
like Map Reduce, Yarn and HDFS, and can be used
by running a standalone application or using its Java
Application Programming Interface (API).
In order to present Big Texts, this paper is or-
ganized as follows: Section 2 presents an overview
129
Alzate Calderón W., Pomares Quimbaya A., A. Gonzalez R. and Muñoz O..
BigTexts - A Framework for the Analysis of Electronic Health Record Narrative Texts based on Big Data Technologies.
DOI: 10.5220/0005434101290136
In Proceedings of the 1st International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AgeingWell-
2015), pages 129-136
ISBN: 978-989-758-102-1
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

of Big Data technologies and some related previous
work. Section 3 specifies the characteristics and ar-
chitecture of Big Texts. Then Section 4 describes the
evaluation of the framework and its use for the analy-
sis of patient with a chronic disease. Finally, Section 5
presents conclusions and future work.
2 RELATED WORKS
Big data techniques have already been used by dif-
ferent projects. This section describes the most rele-
vant techniques related to Big Data; then, it presents
a set of related proposals for using them to improve
the analysis of data in the healthcare domain as well
as those created to enhance the execution of pre-
processing tasks and text mining techniques in any
context.
2.1 Big Data Technologies
Big Data is not a technology by itself, it is rather a
paradigm that has promoted the creation of several
products and frameworks; Table 1 presents a subset
of them including their classification according to the
V property they are intended for.
Table 1: Big Data Technologies Classification.
Technology Sub-technology Velocity Variety Volume
NoSQL X X
Analytics
Exploratory Data Analysis (EDA) X X
Confirmatory Data Analysis (CDA) X X
Qualitative Data Analysis (QDA) X X X
MapReduce X X
Apache Hive X X
Apache Pig X X
MapReduce: is a computing style that can be
used to manage large-scale calculations in a way that
is hardware fault tolerant (Rajaraman and Ullman,
2012).
NoSQL: or Not Only SQL, is a data manage-
ment approach and database design that is useful for
very large datasets that are distributed (Sadalage and
Fowler, 2012).
Analytics: is the science of examining raw data
with the purpose of obtaining conclusions and convert
them to information (Maheshwari, 2015).
Hadoop: is a framework that allows distributed
processing of large sets of data across computer
clusters using simple programming models (White,
2012).
Apache Hive: provides a SQL-based dialect
called Hive Query Language (HiveQL) to query data
stored in a Hadoop cluster (Capriolo et al., 2012).
Apache Pig: provides an engine for a parallel data
flow execution in Hadoop and a language called Pig
Latin for the expression of such data flows (Gates,
2011).
2.2 Big Data Applied to the Health Care
Domain
Table 2 summarizes relevant projects related to the ap-
plication of Big Data technologies in the healthcare
domain. One of the main conclusion of this anal-
ysis is that they are mostly theoretical proposals of
desirable uses of Big Data technologies in that do-
main. Similarly, there are few tangible practices and
approaches of using this paradigm in products or pro-
totypes that make the expected benefits a reality. Also
it was evident that one of the recurring factors in pa-
pers is the high amount of data required to analyze by
medical professionals and administrative personal to
make better decisions. Having in mind that one of the
major sources of information is the narrative text in
the EMRs, it is worth reviewing previous work that
use the Big Data paradigm to enhance the manage-
ment of textual data (Section 2.3).
2.3 Big Data Solutions to Improve Text
Analysis
The literature review of solutions that provide natu-
ral language processing and/or text mining techniques
indicates that there is an important platform called
GATECloud.net that provides several strategies to im-
prove response time in natural language processing,
among which has two options: the use of MapReduce
and the Cloud Computing technologies management.
From these two options the authors of these prod-
ucts recommend the second one, under a model PaaS,
since it is not necessary to rewrite the processing al-
gorithms to be executed. GATECloud.net is a Web
platform where scientists can conduct experiments on
text mining, and whose services are collected through
payment for use. It is important to note that this prod-
uct does not provide libraries (APIs) that can be used
from other systems (Tablan et al., 2012).
In addition to this proposal, there are other
projects that create customized solutions that use Big
Data technologies to analyze a specific set of nar-
rative texts. It is the case of the project presented
in (Das and Mohan Kumar, 2013) that proposes the
use of Big Data technologies and text mining tech-
niques to analyze public tweets. It uses technologies
such as HBase, Java, REST and Hadoop to enhance
the performance of the analysis. Even though these
kind of projects are important to mature the conjunc-
tion of Big Data and Text Mining, they do not pro-
vide open products or frameworks that can be used by
ICT4AgeingWell2015-InternationalConferenceonInformationandCommunicationTechnologiesforAgeingWelland
e-Health
130
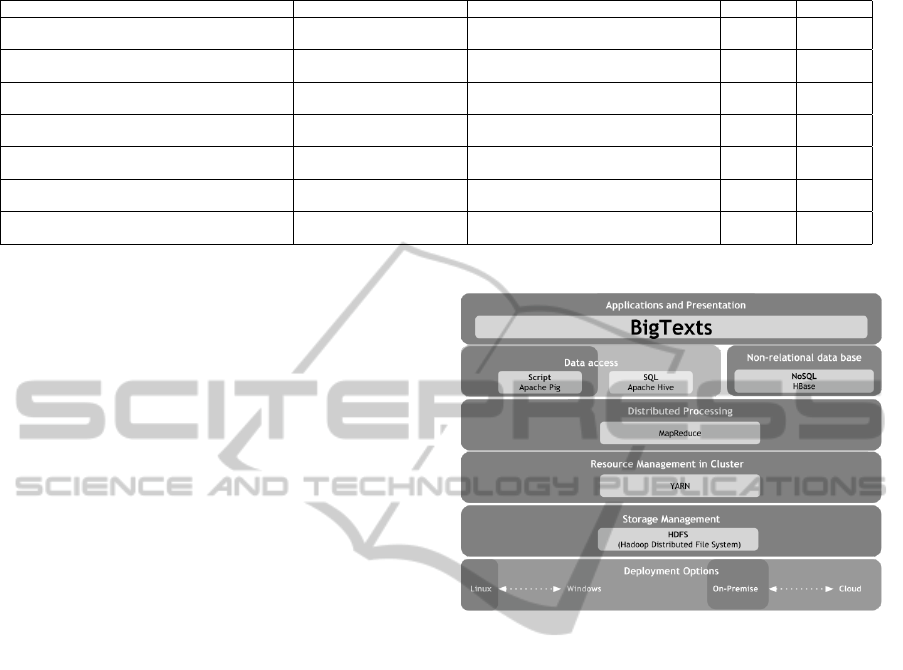
Table 2: Comparison of Big Data Projects in the HealthCare Domain.
Project Technology Problem Solved Type Content
Use of Big Data and Cloud Computing for opera-
tional Health BI (Purkayastha and Braa, 2013)
Cloud Computing Bridging the digital divide for Medical In-
formation Systems in developing countries
Practical Text
Big Data in personalized health care (Chawla and
Davis, 2013)
Data mining Risk Profile Custom diseases Theoretical
- Practical
Text
Big Data in Functional Echocardiography (Sen-
gupta, 2013)
Cloud computing, Robots,
Artificial Intelligence
To have more variables in the functional
echocardiography
Theoretical Images
Acquisition, compression, encryption and storage
of Big Data in Health (Brinkmann et al., 2009)
File processing Compressing data from electrophysiologi-
cal recordings
Practical Images
Big Data in medical devices (Meeker and Hong,
2014)
Sensors Reliability and availability of medical de-
vices
Theoretical Text
Development of an ecosystem of healthcare using
IPHIs (Liyanage et al., 2012)
Ontologies Ecosystem health interoperability for het-
erogeneous systems
Theoretical Text
Big Data to assess the impact of medical pro-
grams (Fox, 2011)
Analytics Allocate and keep real patients Theoretical Text
other projects, which is our need to analyze medical
records.
As is evident, although there are approaches that
try to unify the paradigm of Big Data to text process-
ing, they are not still mature enough or do not provide
APIs through which existing systems can run tasks
(such as pre-processing) in large amounts of text, pro-
viding a great opportunity to attacked by BigTexts
(Section 3), the proposal of this paper.
3 BIGTEXTS
BigTexts is a framework that allows the execution of
pre-processing tasks over narrative texts required to
apply text mining techniques. It was developed using
Big Data technologies, which allows it to improve the
performance when it is necessary to analyze large vol-
umes of medical records and texts contained in them.
BigTexts allows the pre-processing of hundreds, thou-
sands or millions of narrative texts when a research
required the analysis at institutional, regional or na-
tional level. BigTexts provides pre-built functional-
ities that can be used as an independent application
(with a graphical user interface - GUI) or as a library
(API) from an existing application.
Figure 1 shows highlighted the BigData technolo-
gies used in BigTexts. BigTexts uses Apache Pig
at the data access level, MapReduce for distributed
processing, YARN as a resource manager, HDFS for
distributed storage and Linux Ubuntu as the operat-
ing system. BigTexts, takes the EMR texts and par-
tition them in the Hadoop cluster using HDFS. Us-
ing MapReduce to execute the pre-processing tasks
in parallel, in order to finally obtain the set of pre-
processed documents.
As it is shown in Figure 2, typically the analysis
of EMRs are executed sequentially (As-Is). This way
of running generates a bottle neck when the number
of EMR is large. With BigTexts the execution is par-
allelized across the set of machines available in the
Figure 1: BigTexts in the Reference Architecture.
cluster where BigTexts is running. BigTexts takes the
EMR texts and partitions them in the Hadoop cluster
using HDFS. Using MapReduce it executes the pre-
processing tasks in parallel. Finally, it obtains the set
of pre-processed texts.
The following sections explain the functionalities
provided by BigTexts and its architecture.
3.1 BigTexts Tasks
As it was mentioned, BigTexts provides pre-
processing tasks required to give some structure to
the originally unstructured texts. These tasks allow
for example the division of the text in tokens, to iden-
tify sentences, to recognize the role of each word in
a sentence, to recognize patterns in the text, etc. The
following list explains the current tasks provided by
BigTexts, which are based on the Stanford CoreNLP
library (The Stanford NLP Group, 2014b) and the
Weka library (The University of Waikato, 2014). It
is important to notice that these tasks can be extended
according to the requirements of the analysis.
• Identification of co-reference: It identifies when
two or more expressions in a text refer to the
BigTexts-AFrameworkfortheAnalysisofElectronicHealthRecordNarrativeTextsbasedonBigDataTechnologies
131
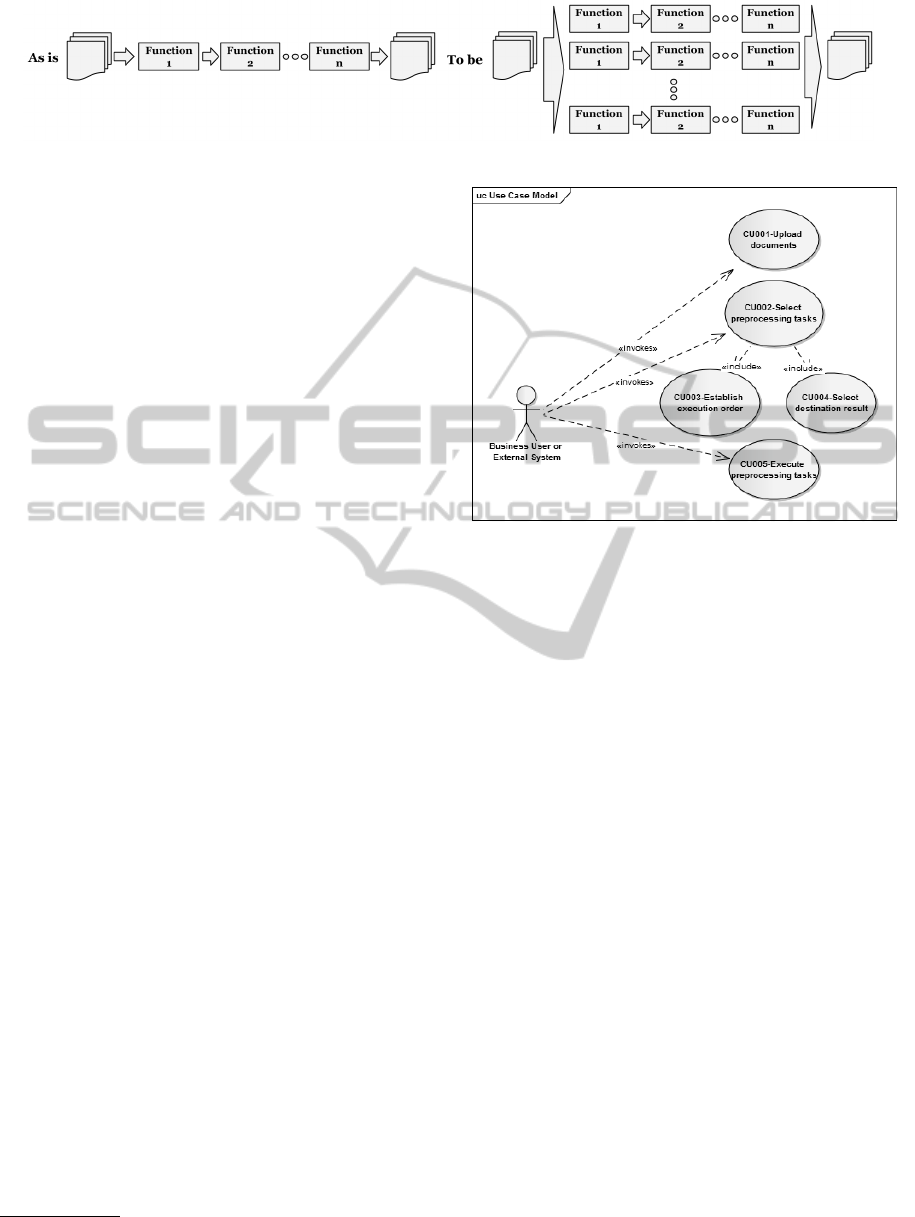
Figure 2: Current Situation vs. BigTexts Situation.
same person or thing (The Stanford NLP Group,
2014a).
• Lemmatisation Lovins: It reduces a word to its
lemma, root or canonical form using the Lovins
Stemmer algorithm (Lancaster University, 2014).
• Named Entity Recognition: It locates and gives
a label to some elements in text taking into ac-
count pre-defined categories such as the names of
persons, organizations, locations, dates, monetary
values, etc (The Stanford NLP Group, 2014d).
• Part of Speech: It recognizes and gives a label in-
dicating the grammar role a word plays in a sen-
tence (The Stanford NLP Group, 2014c).
• Named Entity Recognition based on regular ex-
pressions (NE Transducer): It locates and gives a
label to some elements in text taking into account
rules described using regular expressions.
• Lemmatisation Snowball: It reduces a word to its
lemma, root or canonical form using the snowball
stemmer algorithm.
• Partition text by phrases: It splits a text into its
sentences using a set of configurable terminators
like ., ?.
• Tokenization: It breaks down a text into tokens
using a set of configurable separators like space,
punctuation tokens, among others.
• Switch a text to upper case.
The implementation of the pre-processing tasks
was made using Apache Pig, particularly the option
for creating User Defined Functions (UDFs). Each
pre processing tasks is represented by a Java class
that inherits from org.apache.pig.EvalFunc and pack-
aged using Apache Maven. After that the resulting
jar is added to the Pig pipeline and the UDF is called
inside of Pig instructions. As mentioned before, the
pre-processing tasks use two NLP libraries, Stanford
CoreNLP and Weka.
The addition of a new pre-processing task is de-
signed to be a simple and flexible process. To do this,
a new class is created in which the functionality is im-
plemented and added to the preprocessing tasks cata-
log
1
.
1
An XML file with all the pre-processing tasks and their
parameters.
Figure 3: BigTexts Use Cases.
Each one of the previous tasks can be executed
using an API provided by BigTexts or directly using
its graphical user interface. Figure 3 describe the use
cases of BigTexts:
1. CU001-Upload documents: The actor can load a
set of documents for further processing.
2. CU002-Select preprocessing tasks: The actor can
select the pre-processing tasks he/she wants to ex-
ecute in BigTexts.
3. CU003-Establish execution order: The actor can
give the order in which the selected tasks must be
executed and configured on the CU002.
4. CU004-Select destination result: The actor
chooses the way the results are going to be deliv-
ered, with two options: FTP or HDFS directory.
5. CU005-Execute preprocessing tasks: The actor
can launch the execution of the pre-processing
task list over the uploaded documents.
3.2 BigText Process and Architecture
Figure 4 shows an overview of how BigTexts interacts
with the BigData technology. As it can be seen it is
composed by a Client component (BigTexts Client)
and a Server component(BigTexts).
The Client offers a graphical user interface
through which the user can select the files to be pro-
cessed, the pre-processing tasks to run (and their set-
ICT4AgeingWell2015-InternationalConferenceonInformationandCommunicationTechnologiesforAgeingWelland
e-Health
132
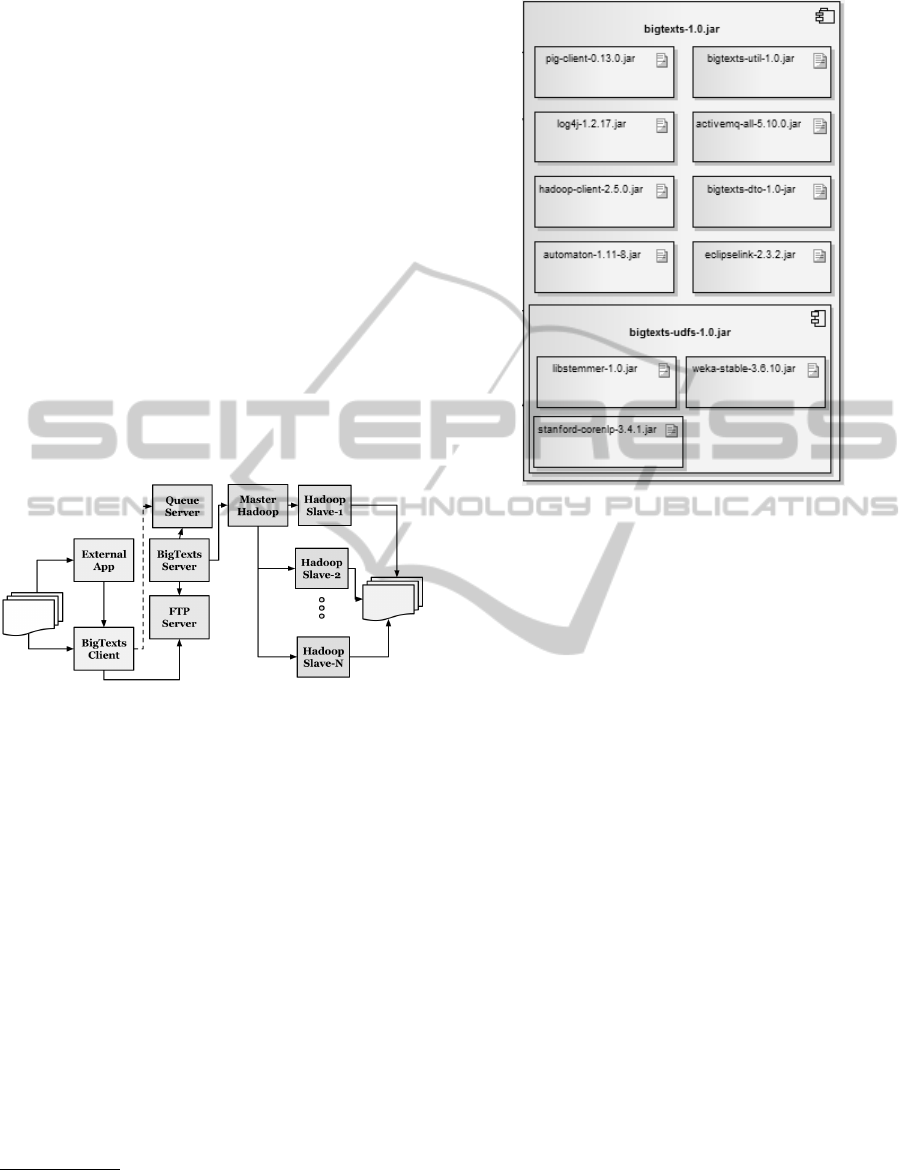
tings) and the delivery method. This client applica-
tion can also be used by an external application as a
library. Once a user sends the files to the server using
FTP and indicates the tasks he/she wanted to execute,
the client component sends an XML format message
to the server queue.
The BigTexts server component connects to the
server queue (ActiveMQ) and gets the queued mes-
sage, transforming it from XML (using the library
JAXB
2
) to objects. Subsequently, the converted mes-
sage to object is executed in the Hadoop cluster
by an Apache Pig script that uses UDFs (User De-
fined Functions) with the pre-processing tasks. The
Hadoop cluster is composed of a main machine (the
master), which manages both storage and parallel pro-
cessing, and a number of secondary machines (slaves)
that stores data blocks and process them in parallel.
Finally, a set of pre-processed documents are deliv-
ered in the HDFS or in the FTP directory, according
to what the user has selected.
Figure 4: BigTexts.
The internal architecture of BigTexts Server
3
is
presented in Figure 5.
1. pig-client-0.13.0.jar: It is the library that allows
the connection with the installation of Pig.
2. bigtexts-util-1.0.jar: BigTexts utility classes.
3. log4j-1.2.17.jar: The application log manager.
4. hadoop-client-2.5.0.jar: Library that allows inter-
action with HDFS and Hadoop.
5. bigtexts-dto-1.0-jar: Package with the classes that
allow the communication between the BigTexts
client and server.
6. automaton-1.11-8.jar: Finite state automata li-
brary, used by the Pig client.
7. eclipselink-2.3.2.jar: Java Persistence API imple-
mentation created by The Eclipse Foundation
2
Java Architecture for XML Binding
3
bigtexts-1.0.jar: Server application, is responsible for ex-
ecuting the pre-processing tasks into the BigData infras-
tructure.
Figure 5: BigTexts Physical View.
8. bigtexts-udfs-1.0.jar: Component with the imple-
mentations of the pre-processing tasks.
(a) libstemmer-1.0.jar: Library with the Snowball
Stemmer implementation.
(b) stanford-corenlp-3.4.1.jar: Natural Language
Processing library.
(c) weka-stable-3.6.10.jar: Library for the imple-
mentation of Lovins Stemmer.
4 CASE STUDY AND
VALIDATION
In order to validate the functionality and the perfor-
mance of BigTexts, the framework was used for the
analysis of EMRs contained in a Hospital EMR Sys-
tem. The goal of the analysis was to identify a set of
patients with specific characteristics required by med-
ical researchers for a retrospective cohort study. This
section presents first the scenario in which the vali-
dation of the system was performed (Section 4.1) and
then its results (Section 4.2).
4.1 Validation Scenario
The validation scenario requires searching, from a set
of EMRs, those records that match different charac-
teristics required to include them in a study on Heart
Failure. The input for the analysis of each record in-
cludes terms related to the diagnosis, personal and
BigTexts-AFrameworkfortheAnalysisofElectronicHealthRecordNarrativeTextsbasedonBigDataTechnologies
133
family antecedents, lab exams, medications, symp-
toms, treatment plans complications and outcomes.
The intention of the case study was to find the set of
EMRs that include most of the characteristics defined
by the group of researchers to include them in a ret-
rospective cohort study around this chronic disease.
BigTexts was used for this purpose. It was config-
ured for the pre-processing of 10.000 narrative texts
provided by the hospital. In order to assure the confi-
dentiality of the patients the records were previously
anonymized and provided in 4 files. In addition, the
Named Entity Recognition task of BigTexts was con-
figured for the identification of terms related to the
searched diagnosis like Cardiac Failure, Cardiopathy,
chronic heart failure, CHF, among others;related to
lab exams like bnp, echocardiogram; related to symp-
toms like Paroxysmal Nocturnal Dyspnea, and related
to complications as death.
From the technical point of view, the machines
used for the configuration of the Hadoop cluster (Ta-
ble 3) were not high specification computers (server
type), but rather common computers in order to simu-
late real hospital environments, where was not pos-
sible to have dedicated servers. A main machine
(master) was used and a set of secondary machines
(slaves). The client application was installed on the
master machine as well as the BigTexts server. All
computers were installed with the Ubuntu operating
system 14.0.LTS. The machines were connected in a
local area network (LAN) establishing static IP ad-
dresses. All machines had Intel processors.
The size of the files provided was the following:
1.945.886 bytes (1.945 Mb), 1.423.803 bytes (1.423
Mb), 437.643 bytes (437.643 Kb) and 761 bytes. The
validation consisted of the execution of five iterations
per file in each of the following pre-processing tasks:
• RegexNamedRecognition: Given the list of terms
related to the diagnosis and the lab exams with a
label for each of them, the system identified using
regular expressions if a word was in the token list
and set the corresponding label.
• Tokenizer: Partition files into tokens.
• Tokenizer + POS-Tagger: Two preprocessing
tasks were performed. The first one broke up the
file into tokens and then applied Part of Speech
task identifying whether each word was a verb, an
adjective, an adverb, etc.
• Tokenizer + SnowballStemmer: It breaks up the
file into tokens and then identified the root of each
one of them.
4.2 Perfomance Results
The analysis of the data, using figures of lines to iden-
tify trends is shown below. For a clear display on the
graph the file size was divided for 10.000, having all
of them in the same scale. There was used a combi-
nation of N, M and L machines, where N < M < L.
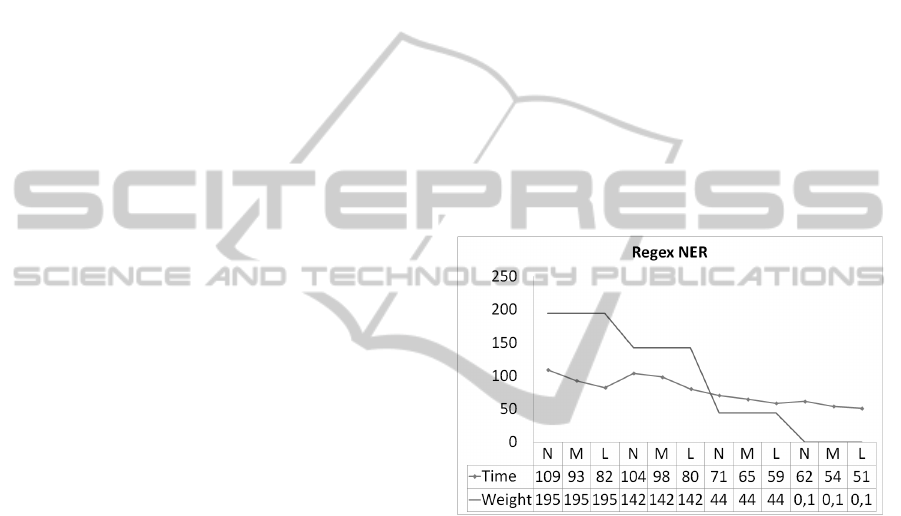
Finding that the Regex Entity Recognition task
(Figure 6) execution with the 1,945,886 bytes file and
N machines obtained an average time of 109.13 sec-
onds, for M machines of 92.78 seconds and for L
machines of 82.25 seconds. For the 1.423.803 bytes
file, the times were 104.07, 98.33 and 80.45 sec-
onds. Likewise, the file of 437,643 bytes had times
of 70.54,65.13 and 58.71 and the file of 761 bytes had
times of 61.57, 54 and 51 seconds for N, M and L ac-
tive machines in the cluster. As it can be seen, there
is a downward trend on time when the number of ma-
chines is increased.
Figure 6: Validation - RegexNER.
For the tokenization task (Figure 7) for the file
of 1.945.886 bytes, times obtained were 50.22, 46.63
and 36 seconds. For the file of 1.423.803, time av-
erage was 56.25, 41.33 and 33.6 seconds. Similarly,
for the 437.643 files and 761 bytes, execution times
improved when the number of machines in the cluster
was increased.
Regarding the tokenization task added to the Part
of Speech (Figure 8) task, for the 1.945.886 bytes
files, times were 181, 113.75 and 85.67 seconds; for
the 1.423.803 bytes the times were 118.33, 96.67 and
76.33 seconds; and for the 761 bytes the times were
69.67 , 65 and 52.5 seconds. These results show an
improvement when the number of available machines
in the cluster were increased. However, in the case of
the 437.643 bytes file there was a not an improvement
in time when the number of machines was increased
slightly, but a substantial improvement was presented
when the number of machines was changed radically.
For the tokenization task added to Stemming using
ICT4AgeingWell2015-InternationalConferenceonInformationandCommunicationTechnologiesforAgeingWelland
e-Health
134
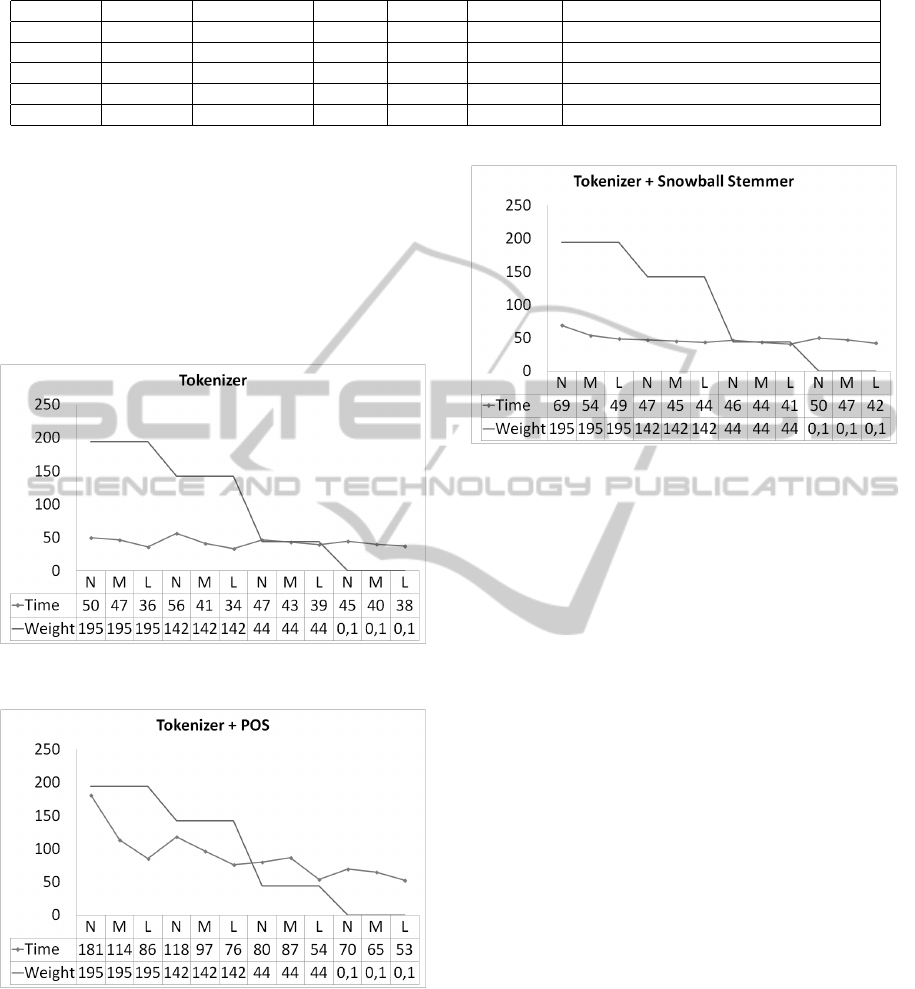
Table 3: Cluster Machines.
Name Hostname IP Interna Brand Memory Hard drive Processor
bigtexts-1 master 192.168.0.101 HP 3,8 GiB 155,3 GB Intel Core 2 Duo CPU E7200 2.53GHz x 2
bigtexts-2 slave-2 192.168.0.102 HP 1.9 GiB 155.3 GB Intel Core 2 Duo CPU E7200 2.53GHz x 2
bigtexts-3 slave-3 192.168.0.103 Lenovo 1.9 GiB 76.5 GB Intel Core 2 CPU 4400 2.00GHz x 2
bigtexts-4 slave-4 192.168.0.104 HP 1.9 GiB 155.3 GB Intel Core 2 Duo CPU E4600 2.40GHz x 2
bigtexts-5 slave-5 192.168.0.105 HP 1.9 GiB 155.3 GB Intel Core 2 Duo CPU E7200 2.53GHz x 2
Snowball algorithm (Figure 10) the following times
for N, M and L machines were obtained, respectively:
• 1.945.886 bytes: 69.15, 53.86 and 49 seconds
• 1.423.803 bytes: 47.05, 45.4 and 43.71 seconds
• 437.643 bytes: 46.46, 43.6 and 40.67 seconds
• 761 bytes: 50.21, 47.42 and 42.25 seconds
Figure 7: Validation - Tokenizer.
Figure 8: Validation - Tokenizer + POS.
Allowing identifying a downward trend as they
were adding available machines to the cluster as a
possible option.
As it can be seen the execution time for the same
file in a same task, differs depending on the number
of available slaves. It improves as new slaves were
added to the cluster. Besides, it can be seen that the
improvement is much more noticeable when complex
tasks need to be executed like RegexNER and Part Of
Figure 9: Validation - Tokenizer + Snowball.
Speech. Additionally, the larger is the file, the greater
the processing speed, for example, for the 761 bytes
file the times for different scenarios (N, M and L ma-
chines) are very similar.
From the point of view of the requirements of the
medical researchers BigTexts allowed the identifica-
tion of the set of EMRs that include all or part of the
terms required for the analysis. The result allowed re-
ducing the time previously required to select records
and analyze which of them actually met the charac-
teristics desired for the study.
5 CONCLUSIONS AND FUTURE
WORK
Shortening large EMR data analysis helps clinical and
administrative data management in the health domain
with both decision making and quality improvement
as benefits.
BigTexts demonstrated its utility to improve effi-
ciency in narrative text analysis of electronic medi-
cal records, this will not only allow performing more
analysis projects in the health sector but also al-
low generating new products that can be constructed
based on BigTexts.
Additionally, BigTexts showed that the addition of
an abstraction layer to the entire ecosystem of tech-
nology of Big Data and text mining avoids installa-
tion, configuration and integration efforts of the Big
Data components that, although powerful, are limited
BigTexts-AFrameworkfortheAnalysisofElectronicHealthRecordNarrativeTextsbasedonBigDataTechnologies
135

in terms of accurate and reliable documentation.
The validation results let us to conclude that it
is worth to use the entire infrastructure of Big Data
when the analysis include large files or complex tasks,
because it was found that only in those situations the
addition of nodes into the cluster markedly improves
performance.
As future work it is proposed to use cloud comput-
ing to address the problems encountered in the present
project, to evaluate the changes in development ef-
fort, and to promote scalability of BigTexts. Addi-
tionally, another field of work is the development of
more UDFs to increase the catalogue of BigTexts pre-
processing tasks.
REFERENCES
Brinkmann, B. H., Bower, M. R., Stengel, K. A., Wor-
rell, G. A., and Stead, M. (2009). Large-scale elec-
trophysiology: Acquisition, compression, encryption,
and storage of big data. Journal of Neuroscience
Methods, 180(1):185–192.
Capriolo, E., Wampler, D., and Rutherglen, J. (2012). Pro-
gramming Hive. O’Reilly Media, 1 edition edition.
Chawla, N. V. and Davis, D. A. (2013). Bringing big
data to personalized healthcare: A patient-centered
framework. Journal of General Internal Medicine,
28(S3):660–665.
Das, T. and Mohan Kumar, P. (2013). Big data analytics:
A framework for unstructured data analysis. School
of Information Technology and Engineering, VIT Uni-
versity.
Fox, B. (2011). Using big data for big impact. leveraging
data and analytics provides the foundation for rethink-
ing how to impact patient behavior. Health manage-
ment technology, 32(11):16.
Gates, A. (2011). Programming Pig. O’Reilly Media, 1
edition edition.
Lancaster University (2014). What
is stemming? Retrieved from
http://www.comp.lancs.ac.uk/computing/research
stemming/general/.
Liyanage, H., Liaw, S.-T., and de Lusignan, S. (2012). Ac-
celerating the development of an information ecosys-
tem in health care, by stimulating the growth of safe
intermediate processing of health information (IPHI).
Informatics in primary care, 20(2):81–86.
Maheshwari, A. (2015). Data Analytics Made Accessible.
Mayer-Schonberger, V. and Cukier, K. (2013). Big Data: A
Revolution That Will Transform How We Live, Work,
and Think. Houghton Mifflin Harcourt, Boston.
McAfee, A. and Brynjolfsson, E. (2012). Big data: The
management revolution. Harvard business review,
90(10):p60–68.
Meeker, W. Q. and Hong, Y. (2014). Reliability meets big
data: Opportunities and challenges. Quality Engineer-
ing, 26(1):102–116.
Moore, K. D., Eyestone, K., and Coddington, D. C. (2013).
The big deal about big data. Healthcare financial
management: journal of the Healthcare Financial
Management Association, 67(8):60–66, 68. PMID:
23957187.
Purkayastha, S. and Braa, J. (2013). Big data analytics for
developing countries – using the cloud for operational
BI in health. The Electronic Journal of Information
Systems in Developing Countries, 59(0).
Rajaraman, A. and Ullman, J. D. (2012). Mining of mas-
sive datasets. Cambridge University Press, New York,
N.Y.; Cambridge.
Sadalage, P. J. and Fowler, M. (2012). NoSQL Distilled:
A Brief Guide to the Emerging World of Polyglot Per-
sistence. Addison-Wesley Professional, Upper Saddle
River, NJ, 1 edition edition.
Sengupta, P. P. (2013). Intelligent platforms for dis-
ease assessment. JACC: Cardiovascular Imaging,
6(11):1206–1211.
Tablan, V., Roberts, I., Cunningham, H., and Bontcheva,
K. (2012). GATECloud.net: a platform for large-
scale, open-source text processing on the cloud.
Philosophical Transactions of the Royal Society A:
Mathematical, Physical and Engineering Sciences,
371(1983):20120071–20120071.
The Stanford NLP Group (2014a). Coref-
erence resolution. Retrieved from
http://nlp.stanford.edu/projects/coref.shtml.
The Stanford NLP Group (2014b). Stan-
ford CoreNLP. Retrieved from
http://nlp.stanford.edu/software/corenlp.shtml.
The Stanford NLP Group (2014c). Stanford log-
linear part-of-speech tagger. Retrieved from
http://nlp.stanford.edu/software/tagger.shtml.
The Stanford NLP Group (2014d). Stanford named
entity recognizer (NER). Retrieved from
http://nlp.stanford.edu/software/CRF-NER.shtml.
The University of Waikato (2014). Weka 3
- data mining with open source machine
learning software in java. Retrieved from
http://www.cs.waikato.ac.nz/ml/weka/.
White, T. (2012). Hadoop: The Definitive Guide. Yahoo
Press, Beijing, third edition edition edition.
ICT4AgeingWell2015-InternationalConferenceonInformationandCommunicationTechnologiesforAgeingWelland
e-Health
136
