
Compressed Domain ECG Biometric Identification using JPEG2000
Yi-Ting Wu, Hung-Tsai Wu and Wen-Whei Chang
Institute of Communications Engineering, National Chiao-Tung University, Hsinchu, Taiwan
Keywords:
ECG Biometric, Person Identification, JPEG2000.
Abstract:
In wireless telecardiology applications, electrocardiogram (ECG) signals are often represented in compressed
format for efficient transmission and storage purposes. Incorporation of compressed ECG based biometric
enables faster person identification as it by-passes the full decompression. This study presents a new method
to combine ECG biometrics with data compression within a common JPEG2000 framework. To this end, ECG
signal is considered as an image and the JPEG2000 standard is applied for data compression. Features relat-
ing to ECG morphology and heartbeat intervals are computed directly from the compressed ECG. Different
classification approaches are used for person identification. Experiments on standard ECG databases demon-
strate the validity of the proposed system for biometric identification with high accuracies on both healthy and
diseased subjects.
1 INTRODUCTION
The demand for improved security for person identi-
fication has been growing rapidly and among the po-
tential alternatives is employing innovative biomet-
ric traits. Biometric identification is reliant on pat-
tern recognition approaches by extracting physiolog-
ical or behavioral traits of human body and match-
ing them with enrollment database. Various biomet-
rics have been proposed for use in person identifica-
tion, such as voice, face, and fingerprint. However,
these biometrics either cannot provide reliable perfor-
mance or not robust enough against falsification. Re-
cent studies have suggested the possibility of using
electrocardiogram (ECG) as a new biometric modal-
ity for person identification (Biel et al., 2001; Israel
et al., 2005; Odinaka et al., 2012; Plataniotis et al.,
2006; Chiu et al., 2009). ECG signal is a record-
ing of the electrical activity of human heart, which
is individual-specific in the sense of amplitude and
time durations of the fiducial points. Furthermore,
ECG signal can deliver the proof of subject’s being
alive as extra information which other biometrics can-
not deliver as easily. It is believed that ECG bio-
metric would be particularly effective in health care
applications, as the signal can be used for diagno-
sis of cardiac diseases and also be used to identify
subjects before granting them medical services. Re-
cently, ECG biometric recognition has been success-
fully commercialized as products in mobile applica-
tions such as health care and online payment. For ex-
ample, Nymi wristband (Nymi Inc., 2013) is a wear-
able biometric authentication device that recognizes
unique ECG patterns and interfaces directly with mo-
bile devices as a replacement for passwords. In 2015,
Linear Dimensions also announced a family of bio-
metric authentication devices including ECG Biolock
and ECG optical wireless mouse (Linear Dimensions
Inc., 2015). Both devices offer proven security and
will authenticate users by learning their unique bio-
metric signature in ECG waveform pattern. Based on
the features that are extracted from ECG signals, we
can classify ECG biometrics as either fiducial points
dependent (Biel et al., 2001; Israel et al., 2005; Odi-
naka et al., 2012) or independent (Plataniotis et al.,
2006; Chiu et al., 2009). Fiducial-based approaches
rely on local features linked to the peak and time dura-
tions of the P-QRS-T waves. On the other hand, non-
fiducial approaches extract statistical features based
on the overall morphology of ECG waveform.
In wireless telecardiology scenarios, compressed
ECG packets are often preferred for efficient trans-
mission and storage purposes. Most of ECG com-
pression methods adopt one-dimensional (1-D) repre-
sentation for ECG signals and focus on the utilization
of the intra-beat correlation between adjacent samples
(Jalaleddine et al., 1990). To better exploit both intra-
beat and inter-beat correlations, 2-D compression al-
gorithms have been proposed by converting ECG sig-
nals into data arrays and then applying vector quan-
tization (Sun and Tai, 2005) or the JPEG2000 im-
age coding standard (Bilgin et al., 2003). Irrespective
5
Wu Y., Wu H. and Chang W..
Compressed Domain ECG Biometric Identification using JPEG2000.
DOI: 10.5220/0005499500050013
In Proceedings of the 12th International Conference on Signal Processing and Multimedia Applications (SIGMAP-2015), pages 5-13
ISBN: 978-989-758-118-2
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

of the underlying method used for data compression,
compressed ECG imposes a new challenge for per-
son identification as most of existing algorithms have
implicitly considered that biometric features are ex-
tracted from raw ECG signals (Biel et al., 2001; Israel
et al., 2005; Odinaka et al., 2012; Plataniotis et al.,
2006; Chiu et al., 2009). Full decompression is then
required to convert compressed data into ECG signals
prior to feature extraction. This step is undesirable in
health care systems, as the hospital may have thou-
sands of enrolled patients and decompression of all
their ECG packets is an enormous work. Thus, there
has been a new focus on biometric techniques which
directly read the compressed ECG to obtain unique
features with good discrimination power. Apart from
its advantage of by-passing the full decompression,
reduced template size also enables faster biometric
matching compared to the non-compressed domain
approaches. In (Sufi and Khalil, 2011), the authors
proposed a clustering method for compressed-domain
ECG biometric using specially designed compression
algorithms. The method starts with the detection of
cardiac abnormality and only the normal compressed
ECG data are used for person identification. It is ex-
pected that considering the ECG as images and then
applying the JPEG2000 will lead to further better and
generalized results. As the discrete wavelet transform
(DWT) is an embedded part of the JPEG2000, and
DWT itself is one of the best features for ECG bio-
metrics (Chiu et al., 2009), working in DWT domain
remains to be the most promising area for compressed
ECG based biometric.
Another problem which requires further investiga-
tion is to test the feasibility of ECG biometrics with
diseased patients in cardiac irregular conditions. Pre-
vious works have shown that ECG biometric prob-
lem for healthy persons can be satisfyingly solved
with high recognition accuracies, but a much lower
accuracy may be achieved for cardiovascular disease
(CVD) patients. This is mainly because that CVD
may cause irrecoverable damage to the heart and in-
curs different forms of distorted ECG morphologies.
Recently, there have been initial studies of ECG bio-
metrics for diseased patients with ECG irregularities.
In (Chiu et al., 2009), the authors proposed a DWT-
based algorithm and reported overall accuracies of
100% and 81% on 35 normal subjects and 10 arrhyth-
mia patients, respectively. In (Agrafioti and Hatzi-
nakos, 2009), the authors obtained 96.42% recogni-
tion rate using autocorrelation method when tested on
26 healthy subjects from two databases and 30 pa-
tients with atrial premature contraction and premature
ventricular contraction. Another recent study (Yang
et al., 2013) indicated that a normalization and inter-
Figure 1: Block diagram of the 2-D ECG compression
scheme (Bilgin et al., 2003).
polation algorithm can achieve 100% and 90.11% in
accuracies on 52 healthy subjects and 91 CVD pa-
tients, respectively. The main purpose of this study is
to extract features in a way that the intra-subject vari-
ability is minimized and the inter-subject variability
is maximized.
The rest of this paper is organized as follows. Sec-
tion 2 describes the ECG fundamentals and presents
a preprocessor which converts ECG signals to 2-D
images. Also included is a short overview of the
JPEG2000 encoding algorithm. Details of the al-
gorithms for the proposed compressed-domain ECG
biometric system are provided in Section 3. Section
4 presents the experimental results on standard ECG
databases for both healthy and diseased subjects. Fi-
nally, Section 5 gives our conclusions.
2 2-D ECG DATA COMPRESSION
2-D ECG data compression algorithms are designed
to exploit both intra-beat and inter-beat correlations
of ECG signals. To begin, we apply a preprocessor
which can be viewed as a cascade of two stages. In
the first stage, the QRS complex in each heartbeat
is firstly detected for segmenting and aligning 1-D
ECG signals to 2-D images and in the second stage,
the constructed ECG images are compressed by the
JPEG2000 (Taubman and Marcellin, 2001). Figure 1
shows the block diagram of the 2-D ECG compres-
sion scheme described in (Bilgin et al., 2003).
2.1 Signal Preprocessing
ECG itself is 1-D in the time-domain, but can be
viewed as a 2-D signal in terms of its implicit period-
icity. ECG signals tend to exhibit considerable simi-
larity between adjacent heartbeats, along with short-
term correlation between adjacent samples. Thus,
by dividing ECG signals into segments with lengths
equal to the heartbeats, there should be a large cor-
relation between individual segments. Typical ECG
waveform of a heartbeat consists of a P wave, a QRS
complex, and a T wave (Thaler, 2012). The QRS
complex is the most characteristic wave in an ECG
SIGMAP2015-InternationalConferenceonSignalProcessingandMultimediaApplications
6

waveform and hence, its peak can be used to iden-
tify each heartbeat. To begin, ECG signals are band-
pass filtered to remove various noises. Afterwards
we apply the Biomedical Signal Processing Toolbox
(Aboy et al., 2002) to detect the R peak of each QRS
complex. Accordingly, ECG signals are divided into
heartbeat segments and each segment is stored as one
row of a 2-D data array. Having constructed the data
array as such, the intra-beat correlation is in the hor-
izontal direction of the array and the inter-beat cor-
relation is in the vertical direction. Since the heart-
beat segments may have different lengths, each row
of the data array is period normalized to a fixed length
of N
p
= 200 samples via cubic spline interpolation.
This choice was based on the observation that the av-
erage heartbeat length is about 0.8 second, which cor-
responds to 200 samples for a sampling frequency of
250 Hz. Note that the original heartbeat lengths were
represented with 9 bits and transmitted as side infor-
mation. Finally, we proceed to construct ECG im-
ages of dimension N
c
× 200 by gathering together N
c
rows of the data array and normalizing the amplitude
of each component to an integer ranging from 0 to
255. The constructed grayscale ECG images are then
ready to be further compressed by the JPEG2000 cod-
ing standard.
2.2 JPEG2000 Encoding Algorithm
The JPEG2000 coding standard supports lossy and
lossless compression of grayscale and color imagery.
Although the standard was originally developed for
still image compression (Taubman and Marcellin,
2001), its applicability for ECG compression has been
proposed in (Bilgin et al., 2003). As shown in Figure
2, the JPEG2000 encoding process consists of several
operations: preprocessing, 2-D DWT, quantization,
entropy coding and bit-stream organization. It begins
with a preprocessor which divides the source image
into disjoint rectangular regions called tiles. For each
tile, the DC level of image samples is shifted to zero
and color space transform is performed to de-correlate
the color information. The 2-D DWT can be viewed
as applying a 1-D DWT decomposition along the lines
then the columns to generate an approximation sub-
band and three detail subbands oriented horizontally,
vertically and diagonally. With respect to the lifting
realization of 1-D DWT (Daubechies and Sweldens,
1998), prediction and update steps are performed on
the input signal to obtain the detail and the approx-
imation signals. A multiresolution representation of
the input image over J decomposition levels is ob-
tained by recursively repeating these steps to the re-
sulting approximation coefficients.
Figure 2: Fundamental building blocks of JPEG2000 en-
coder (Taubman and Marcellin, 2001).
With J-level wavelet decomposition, the image
will have a total of 3J + 1 subbands. For notational
convenience, the subbands S
j
are numbered from 1
to 3J + 1, with 1 and 3J + 1 corresponding to the
top-left and bottom-right subbands, respectively. Let
S
j
= {s
j
(m,n),1 ≤ m ≤ M
j
,1 ≤ n ≤ N
j
} represent the
j-th subband whose row and column dimensions are
denoted by M
j
and N
j
with j ∈ {1,2,...,3J +1}. For
each coefficient s
j
(m,n) located at position (m,n), a
mid-tread uniform quantizer is applied to obtain an
index v
j
(m,n) as follows:
v
j
(m,n) = sign[s
j
(m,n)] ·
|s
j
(m,n)|
∆
j
, (1)
where ∆
j
denotes the quantizer step size for the j-th
subband. A different quantizer is employed for the
coefficients of each subband and therefore, a bit al-
location among the subbands is carried out in order
to meet a targeted coding rate ρ. The last step in
JPEG2000 encoding consists in entropy coding of the
quantizer indexes with two tier encoders. The tier-1
encoder employs a context modeling to cluster the bits
of quantizer indexes into groups with similar statistics
in order to improve the efficiency of the arithmetic
coder. In the tier-2 encoder, the output of the arith-
metic coder is collected into packets and a bit-stream
is generated according to a predefined syntax.
3 COMPRESSED DOMAIN ECG
BIOMETRIC
Person identification is essentially a pattern recogni-
tion problem consisted of two stages: feature extrac-
tion and classification. Under the JPEG2000 frame-
work, the person identification problem is analo-
gous to a content-based image retrieval (CBIR) prob-
lem. Concerning compressed-domain biometric tech-
niques, the JPEG2000 code-stream is subject to par-
tial decoding and then features relating to ECG mor-
phology are computed directly from the dequantized
wavelet coefficients. In the classification stage, the
query ECG of an unknown subject will be compared
with the enrollment database to find a match. The
CompressedDomainECGBiometricIdentificationusingJPEG2000
7

Figure 3: The proposed ECG biometric system.
block diagram of the proposed ECG biometric system
is shown in Figure 3.
3.1 Feature Extraction in DWT Domain
Feature extraction is the first step in applying ECG
biometrics to person identification and one that con-
ditions all the subsequent steps of system implemen-
tation. For large image databases, color, shape and
texture features are considered the most important
content descriptors in CBIR problems. Due to the
grayscale nature of ECG images, we only focus on
the texture features that characterize smooth, coarse-
ness and regularity of the specific image. One ef-
fective tool for texture analysis is the DWT as it
provides good time and frequency localization abil-
ity. Its multi-resolution nature also allows the de-
composition of an ECG image into different scales,
each of which represents particular coarseness of the
signal. Furthermore, DWT coefficients can be ob-
tained without involving a full decompression of the
JPEG2000 code-stream. This is a favorable property
as the inverse DWT and subsequent decoding pro-
cesses could impose intensive computational burden.
Different texture features such as energy, significance
map, and modelling of wavelet coefficients at the out-
put of wavelet filter-banks have been successfully ap-
plied to CBIR (Smith and Chang, 1994; Do and Vet-
terli, 2002). In general, any measures that provide
some degree of class separation should be included in
the feature set. However, as more features are added,
there is a trade-off between classification performance
and computational complexity. In this work, three dif-
ferent feature sets derived from the compressed ECG,
denoted by FS1, FS2, and FS3, are presented and in-
vestigated.
We began by using the subband energies as a first
step towards an efficient characterization of texture
in ECG images. It has been suggested (Smith and
Chang, 1994) that the texture content of images can
be represented by the distribution of energy along
the frequency axis over scale and orientation. For
each subband, the dequantized wavelet coefficients
ˆs
j
(m,n) are computed as
ˆs
j
(m,n) = {v
j
(m,n) + δ · sign[v
j
(m,n)]} · ∆
j
, (2)
where δ ∈ [0,1) is a user defined bias parameter. Al-
though the value of δ is not normatively specified in
the standard, it is likely that many decoders will use
the value of one half. Then, the energy of subband j
is defined as
E
j
=
1
M
j
N
j
M
j
∑
m=1
N
j
∑
n=1
ˆs
2
j
(m,n). (3)
The resulting subband energy-based features are used
as a morphological descriptor of ECG signals. An-
other feature of interest is the average time elapse
between two successive R peaks, referred to as the
RR
av
. Certain ectopic heartbeats, such as premature
ventricular contraction and atrial premature beats, are
related with premature heartbeats that provide shorter
RR-intervals than other types of ECG signals. Thus,
changes in the RR-interval plays an important role
in characterizing the dynamics information around
the heartbeats. Notice that the RR
av
can be calcu-
lated from the heartbeat lengths which are transmitted
as side information along with the JPEG2000 code-
stream. With J-level wavelet decomposition, a to-
tal of (3J + 2) features are used to form a biomet-
ric identification vector (BIV) of the subject. The
BIV used for the FS1 will be denoted as b
(1)
=
{E
1
,E
2
,...,E
3J+1
,RR
av
}.
The second feature set FS2 is obtained by ap-
plying principal component analysis (PCA) (Bishop,
2006) on wavelet coefficients from the lowpass sub-
band S
1
. The validity of using S
1
is supported by
the fact that the lowpass subband represents the ba-
sic figure of an image, which features a high sim-
ilarity among the ECGs of the same person. In
order to achieve dimension reduction, PCA finds
projection vectors in the directions of highest vari-
ability such that the projected samples retain the
most information about the original data samples.
To begin, consider a training dataset consisting of
ECG images of dimension 200 × 200. Let M
1
and
N
1
denote the row and column dimensions of the
lowpass subband S
1
after the wavelet decomposi-
tion, respectively. Before applying the PCA, M
1
rows of the subband S
1
are concatenated to form a
wavelet coefficient vector of length M
1
× N
1
, i.e.,
u = {ˆs
1
(1,1), ˆs
1
(1,2),..., ˆs
1
(M
1
,N
1
)}. Then, the
mean vector
¯
u and the covariance matrix Σ
u
are com-
puted using the training dataset. Following the eigen-
decomposition, we obtain an eigenvalue matrix D and
its corresponding eigenvector matrix V with its col-
umn vectors sorted in the descending order of eigen-
values. Finally, the wavelet vector u is projected
on the eigenvectors to obtain the principal compo-
nent vector p = V
T
(u −
¯
u). In this work, only the
first seven principal components are selected based
on containment of 99% of the total variability. To-
gether with the RR-interval, the BIV for the FS2 is
SIGMAP2015-InternationalConferenceonSignalProcessingandMultimediaApplications
8

composed of eight features and is denoted as b
(2)
=
{p(1),..., p(7),RR
av
}. As for the feature set FS3, it
is a combined version of FS1 and FS2 and the corre-
sponding BIV is denoted as b
(3)
= {b
(1)
,b
(2)
}.
3.2 Enrollment and Recognition
Enrollment and recognition are two important stages
of the person identification system. In the enrollment
stage, BIVs of each subject are taken as representa-
tions of the subject and enrolled into a database. In
the recognition stage, the query BIV of an unknown
subject is compared with the enrollment database to
find a match. The recognition performance depends
on the underlying classifier used for person identi-
fication. First, nearest-neighbor (NN) classifiers are
exploited for testing various feature sets in discrimi-
nating different subjects. The NN classifier tends to
search for the most similar class to a given query BIV
with similarity defined by the normalized Euclidean
distance (Smith and Chang, 1994).
Another classification approach considered here
is the support vector machine (SVM) which has
shown effective in many pattern recognition problems
(Bishop, 2006). This is partly due to their good gen-
eralization capability derived from the structural risk
minimization principle. Since person identification
involves the simultaneous discrimination of several
subjects, we considered the one-against-one method
for solving multiclass SVM problems (Bishop, 2006).
For a K-class problem, the method constructs K(K −
1)/2 binary SVM classifiers where each one is trained
on the training data from two classes. Training the
binary SVM consists of finding a separating hyper-
plane with maximum margin and can be posed as the
quadratic optimization problem. For the t-th ECG
image, suppose that the pair (x
t
,y
t
) contains the fea-
ture vector x
t
∈ {b
(1)
,b
(2)
,b
(3)
} and its corresponding
class label y
t
∈ {1,2,...,K}. Given a set of T training
data pairs {(x
t
,y
t
),t = 1, 2, . . . , T } from classes i and
j, SVM algorithm can be formulated as the following
primal quadratic optimization problem
min
w
i j
,b
i j
,ξ
i j
t
1
2
||w
i j
||
2
+C
T
∑
t=1
ξ
i j
t
,
subject to: (w
i j
)
T
x
t
+ b
i j
≥ 1 − ξ
i j
t
, if y
t
= i,
(w
i j
)
T
x
t
+ b
i j
≤ −1 + ξ
i j
t
, if y
t
= j,
ξ
i j
t
≥ 0, (4)
where C is a regularization parameter, w
i j
, b
i j
and ξ
i j
t
are the weight vector, bias and slack variable, respec-
tively. Due to various complexities, a direct solution
to minimize the objective function (4) with respect to
w
i j
is usually avoided. A better approach is to apply
the method of Lagrange multipliers to solve the dual
problem as follows:
max
a
i j
T
∑
t=1
a
i j
t
−
1
2
T
∑
t=1
T
∑
t
0
=1
a
i j
t
a
i j
t
0
y
t
y
t
0
x
T
t
x
t
0
,
subject to:
T
∑
t=1
a
i j
t
y
t
= 0,
0 ≤ a
i j
t
≤ C, (5)
where a
i j
= (a
i j
1
,a
i j
2
,...,a
i j
T
) is the vector of Lagrange
multipliers. In this work, the dual problem to SVM
learning is solved using the sequential minimal opti-
mization method (Bishop, 2006). After obtaining the
optimum values of Lagrange multipliers a
i j
, it is then
possible to determine the corresponding weight vec-
tor w
i j
and the decision function f
i j
(x
t
) as follows:
w
i j
=
T
∑
t=1
a
i j
t
y
t
x
t
, (6)
f
i j
(x
t
) = (w
i j
)
T
x
t
+ b
i j
. (7)
Finally, the class label y for a query BIV x of a new
subject is determined based on the max-wins voting
strategy (Bishop, 2006). More precisely, each binary
SVM casts one vote for its preferred class and the final
result is the class with the highest vote, i.e.,
y = arg max
i
K
∑
j6=i, j=1
sign[ f
i j
(x)]. (8)
4 EXPERIMENTAL RESULTS
Computer simulations were conducted to evaluate the
performances of the proposed ECG biometric sys-
tem for both healthy and diseased subjects. ECG
records from the PhysioNet QT Database (Laguna
et al., 1997) were chosen to represent a wide vari-
ety of QRS and ST-T morphologies. First, 10 healthy
subjects that are originally from the MIT-BIH Normal
Sinus Rhythm Database are used in the experiments
and denoted as dataset D1. Subjects that are added to
the database to examine the effects of diseased ECG
consist of 10 records from the MIT-BIH Arrhythmia
Database, 10 records from the MIT-BIH Supraven-
tricular Arrhythmia Database, and 10 records from
the Sudden Cardiac Death Holter Database. These
three groups of diseased subjects are denoted as D2,
D3, and D4, respectively. Each of these records
is 15 minutes in duration and are sampled at 250
Hz with a resolution of 11 or 12 bits/sample. The
JPEG2000 simulation was run on the open-source
CompressedDomainECGBiometricIdentificationusingJPEG2000
9

Table 1: Average CR and PRD (%) performances for JPEG2000 with coding rate ρ = 0.15 and ρ = 0.08.
Rate ρ = 0.15 Rate ρ = 0.08
Dataset N
c
50 100 150 200 50 100 150 200
D1
CR 12.19 13.45 14.04 14.28 21.62 21.90 21.84 21.84
PRD 3.59 3.19 3.19 3.08 8.72 5.91 5.55 5.55
D2
CR 9.03 9.37 9.49 9.70 16.81 16.92 16.84 16.84
PRD 4.19 3.48 3.36 3.26 11.36 7.95 7.18 7.18
D3
CR 10.07 10.06 10.06 10.61 18.90 19.19 18.87 18.87
PRD 4.53 3.56 3.45 3.18 11.86 8.75 7.99 7.94
D4
CR 10.11 10.23 10.42 10.05 19.11 19.13 19.13 19.13
PRD 4.21 3.32 3.19 3.29 12.35 8.76 7.92 7.92
software JasPer version 1.900.0 (Adams, 2002). Each
ECG image is regarded as a single tile and the dimen-
sion of the code-block is 64 × 64. ECG images were
compressed in lossy mode using Daubechies 9/7 fil-
ter with 5-level wavelet decomposition. Besides, the
targeted coding rate ρ was empirically determined to
be 0.15 and 0.08 in order to achieve the compression
ratio in the region of 10 and 20, respectively.
A preliminary experiment was first conducted to
examine the performance dependence of 2-D com-
pression on the number N
c
of heartbeats employed in
constructing an ECG image. The system performance
is evaluated in terms of the compression ratio (CR)
and the percent root mean square difference (PRD).
The CR is defined as CR = N
ori
/N
com
, where N
ori
and
N
com
represent the total number of bits used to code
the original and compressed ECG data, respectively.
The PRD is used to evaluate the reconstruction distor-
tion and is defined by
PRD(%) =
s
∑
L
l=1
[x
ori
(l) − x
rec
(l)]
2
∑
L
l=1
x
ori
(l)
2
× 100, (9)
where L is the total number of original samples in
the record and x
ori
and x
rec
represent the original
and reconstructed ECG signals, respectively. Table
1 presents the average results of CR and PRD for
JPEG2000 simulation with coding rate ρ = 0.15 and
ρ = 0.08. As expected, the system yielded better
performance with an increase in the heartbeat num-
ber N
c
. However, the increasing delay may limit its
practical applicability as the JPEG2000 must buffer a
total of N
c
× 200 samples before it can start encod-
ing. Thus, we empirically choose N
c
= 200 as the
best compromise in the sequel. Note that while the 2-
D compression method works well for normal ECGs,
it may suffer from ECG irregularities due to the false
QRS detection in the preprocessor stage. For the case
of ρ = 0.08 and N
c
= 200, the PRD performance has
dropped from 5.55% in the D1 to 7.92% in the D4.
In order to justify the efficiency of the proposed
method, we also analyze the run-time complexity
of JPEG2000 decoder for ECG data. According to
the JPEG2000 coding standard, its full decompres-
sion process can be highlighted as: entropy decod-
ing, dequantization to obtain the DWT coefficients,
and inverse DWT to reconstruct blocks of pixels. By
studying the code execution profiles, we can see that
the decoder spends most of its time on the inverse
DWT (typically 61.5% or more). By contrast, the
amount of time consumed by entropy decoding and
de-quantization is about 30.8%. This observation is
in accord with the results for natural and synthetic im-
agery produced by the Jasper software implementa-
tion, reported earlier (Adams and Kossentini, 2000).
It was found that the execution time breakdown for
the JPEG2000 decoder depends heavily on the par-
ticular image and coding scenarios employed. In
the case of lossless image compression, tier-1 decod-
ing usually requires the most time (typically more
than 50%), followed by the inverse DWT (typically
30 − 35%), and then tier-2 decoding. For lossy im-
age compression, as in our case, the inverse DWT ac-
counts for the vast majority of the decoder’s execution
time (typically 60 − 80%). The complexity analysis
results demonstrate that the proposed method has the
advantage of by-passing the inverse DWT operation.
The next step is to evaluate the recognition per-
formances of NN classifiers on the feature sets FS1
and FS2. Both feature sets have in common the RR
av
to provide dynamics feature of ECG signals. Mor-
phological features to be computed for the FS1 are
subband energies {E
j
,1 ≤ j ≤ 16}, and PCA-based
features {p(i),1 ≤ i ≤ 7} for the FS2. The system per-
formance is evaluated in terms of the recognition rate,
which is normally defined as the ratio of the num-
ber of correctly identified subjects to the total num-
ber of testing subjects. First of all, the proposed sys-
tems were individually tested on datasets from D1
to D4. Since ECG records from the QT database
may vary in the number of heartbeats, a total of 4
to 8 ECG images of dimension 200 × 200 would be
generated for different individuals. For a fair assess-
ment, 1000 trials of repeated random sub-sampling
were implemented to eliminate possible classification
SIGMAP2015-InternationalConferenceonSignalProcessingandMultimediaApplications
10
biases. In each trial, four compressed ECG images
per subject were randomly selected for feature extrac-
tion. Among them, the first two ECG images are used
for training in the enrollment stage, and the other two
are used for testing in the recognition stage. Table 2
presents the average recognition rates associated with
various datasets for JPEG2000 coding with ρ = 0.15
and ρ = 0.08. Compared with the subband energy-
based FS1, the improved performances of FS2 indi-
cate that morphological features of ECG signals are
better to be exploited in the DWT domain. The re-
sults also show that the recognition performances are
affected by ECG variations caused by cardiovascu-
lar diseases. For the case of FS2 and ρ = 0.15, the
recognition rate has dropped from 100% for D1 to
91.77% for D4. The possible reasons resulting in a
lower recognition rate for diseased subjects could be
unstable QRS-complex that appear aperiodically in
ECGs. It is important to note that the lower value
of ρ = 0.08 did not result in a significant performance
degradation. This can be attributed to the fact that
the JPEG2000 supports a lower coding rate by en-
larging the quantizer step sizes within high frequency
subbands. This has little effect on the recognition
performance, as the frequency content of the QRS
complex is most concentrated in low frequency sub-
bands. To elaborate further, we also include in Ta-
ble 2 the recognition performances for the case where
normal and diseased subjects are jointly enrolled and
tested. As the table shows, the additional inclusion
of 30 diseased subjects has dropped the recognition
rate by 4.45%. The results indicate that the com-
bined use of FS2 and NN classifier is still not robust
enough to handle the inclusion of diseased patients
in the database. In addition to the above-mentioned
schemes, we also examine how the recognition per-
formance changes as a function of the number of
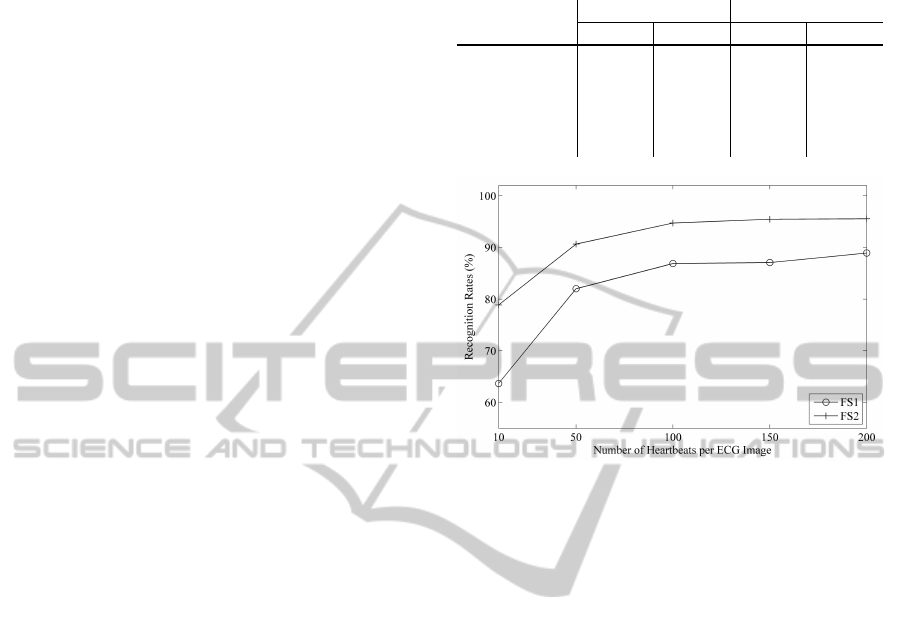
heartbeats used in constructing an ECG image. The
results are illustrated in Figure 4 for various image
sizes using an NN classifier and a combined dataset
from D1 to D4. Our general conclusion is that better
performances can be achieved with an increase in the
heartbeat number N
c
, but the performance has a ten-
dency to become flattened for N
c
≥ 100. This would
be beneficial for future works on real-time implemen-
tation of the proposed ECG biometric systems.
Another problem which requires further investi-
gation is to test the proposed system for the situa-
tion where subjects were identified solely by means
of DWT-based morphological features. The perfor-
mances of NN classifiers for FS1 and FS2 without in-
corporating the RR-interval are summarized in Table
3. A comparison between Table 2 and 3 indicates that
the inclusion of RR-interval is likely to help in dis-
Table 2: Recognition rates (%) of the feature sets FS1 and
FS2 using an NN classifier.
Dataset
ρ = 0.15 ρ = 0.08
FS1 FS2 FS1 FS2
D1
96.16 100 95.29 99.98
D2
86.82 91.82 86.73 91.75
D3
91.61 95.62 91.75 95.50
D4
84.50 91.77 83.52 91.91
D1, D2, D3, D4
88.92 95.55 88.85 95.57
Figure 4: Recognition rates for different ECG image sizes
using JPEG2000 with ρ = 0.15.
criminating between subjects with greater accuracy.
However, we should state that the issue of whether
it is better to use RR-interval in the ECG biomet-
ric does not appear to be settled and may be prob-
lem dependent (Odinaka et al., 2012). Prior works on
ECG biometric (Biel et al., 2001; Israel et al., 2005),
(Sufi and Khalil, 2011) have suggested the use of the
RR-interval as a second source of biometric data to
supplement morphological features of ECG signals.
However, almost all these works exploited a database
containing ECG signals in rest conditions, which rep-
resents also a limitation for practical issues of bio-
metric systems. Recent studies (Odinaka et al., 2012;
Poree et al., 2014) have shown that ECG signals ac-
quired in different physiological conditions allow for
substantial variability in heart rates, which could sig-
nificantly impact the performance of the biometric
system. Continuing this research, we will address
ourselves to the study of robust ECG biometric al-
gorithms utilizing biometric features without the RR-
interval.
We next compare the performances of NN and
SVM classifiers trained on the combined feature set
FS3. With 5-level wavelet decomposition, a total of
24 features are used to form the FS3, including RR
av
,
{p(i),1 ≤ i ≤ 7}, and {E
j
,1 ≤ j ≤ 16}. Table 4 gives
the recognition performances of various datasets us-
ing NN and SVM for classification. With respect to
the implementation of SVM classifiers, the simula-
CompressedDomainECGBiometricIdentificationusingJPEG2000
11

Table 3: Recognition rates (%) of NN classifiers using feature sets FS1 and FS2 without RR-interval.
Dataset
JPEG2000 (ρ = 0.15) JPEG2000 (ρ = 0.08)
FS1 FS2 FS1 FS2
(no RR
av
) (no RR
av
) (no RR
av
) (no RR
av
)
D1
95.77 99.73 94.93 99.65
D2
85.61 88.18 85.93 88.40
D3
91.27 95.34 90.84 95.57
D4
80.98 90.20 77.73 89.69
D1, D2, D3, D4
86.32 94.58 86.46 94.56
Table 4: Recognition rates (%) of the feature set FS3 using
NN and SVM classifiers.
Dataset
ρ = 0.15 ρ = 0.08
NN SVM NN SVM
D1
100 100 99.99 100
D2
98.99 100 98.93 100
D3
99.85 100 99.99 100
D4
97.37 100 97.44 100
D1, D2, D3, D4
99.09 100 99.15 100
QT database
96.06 99.50 96.15 99.48
tion was run on the open-source software LIBSVM
(Chang and Lin, 2011), which supports SVM formu-
lations for various classification problems. The re-
sults clearly demonstrate the improved performances
achievable using the FS3 in comparison to those of
FS1 and FS2, even with a simple NN classifier. The
main reason for this is that the FS3 features a high
similarity among the BIVs of the same person, ei-
ther healthy or with CVD, but a much lower simi-
larity between two BIVs of different persons. The
results also indicate that the SVM classifiers trained
on FS3 are very effective with 100% recognition rate
in all test datasets. Compared with NN classifier,
the SVM shows better generalization ability on the
limited training data, which is indeed the case per-
formed in this study. Also included in the table are
the performances when 86 ECG records from the QT
database are jointly enrolled and tested. The proposed
system can achieve 99.50% and 99.48% in accura-
cies for JPEG2000 compression with ρ = 0.15 and
ρ = 0.08, respectively. The results indicate that the
combined use of FS3 and SVM classifier is more in-
variant to ECG irregularities induced by cardiovascu-
lar diseases.
5 CONCLUSIONS
This paper proposed a robust method for biometric
identification using wavelet-based features extracted
from the JPEG2000 compressed ECG. Under the
JPEG2000 framework, the person identification prob-
lem is analogous to a content-based image retrieval
problem. Morphological features of ECG signals
are derived directly from the DWT coefficients with-
out involving full decompression of JPEG2000 bit-
stream. Also, dynamic feature such as RR-interval
proved to be beneficial for ECG biometric. Combined
performance of both healthy persons and diseased pa-
tients indicate that the proposed system enables faster
biometric identification in compressed domain and in
the same time it is more invariant to ECG irregulari-
ties induced by cardiovascular diseases.
It should be noted that the results presented in this
work, while promising, were obtained from a mod-
erate size of datasets. Also, the experiments were
conducted off-line, using ECG signals collected under
controlled conditions from public databases. In prac-
tical applications, however, a range of issues would
require further investigations. First, the ECG biomet-
ric system needs to be tested in more realistic environ-
ments, varying with respect to the type and quantity of
data collected. Second, the effects of varying mental
and physiological conditions on the recognition accu-
racy, as well as the delay induced by JPEG2000 cod-
ing also need to be resolved.
ACKNOWLEDGEMENTS
This research was supported by the National Science
Council, Taiwan, ROC, under Grant NSC 102-2221-
E-009-030-MY3.
REFERENCES
Aboy, M., Crespo, C., McNames, J., Bassale, J., Jenkins,
L., and Goldsteins, B. (2002). A biomedical signal
processing toolbox. In Biosignal’02. (pp. 49-52).
Adams, M. D. (2002). The JasPer Project Home
Page. Retrieved may 22, 2015, from http://www.ece.
uvic.ca/∼frodo/jasper/.
Adams, M. D. and Kossentini, F. (2000). JasPer: A
software-based JPEG-2000 codec implementation.
In International Conference on Image Processing.
(pp. 53-56). IEEE.
SIGMAP2015-InternationalConferenceonSignalProcessingandMultimediaApplications
12

Agrafioti, F. and Hatzinakos, D. (2009). ECG biometric
analysis in cardiac irregularity conditions. Signal, Im-
age, Video Process., 3(4):329–343.
Biel, L., Pettersson, O., Philipson, L., and Wide, P. (2001).
ECG analysis: A new approach in human identifica-
tion. IEEE Trans. Instrum. Meas., 50(3):808–812.
Bilgin, A., Marcellin, M. W., and Altbach, M. I.
(2003). Compression of electrocardiogram signals
using JPEG2000. IEEE Trans. Consum. Electron.,
49(4):833–840.
Bishop, C. M. (2006). Pattern Recognition and Machine
Learning. Springer, New York.
Chang, C. C. and Lin, C. J. (2011). LIBSVM: A li-
brary for support vector machines. ACM Trans. In-
tell. Syst. Techn., 2(3):27.
Chiu, C. C., Chuang, C. M., and Hsu, C. Y. (2009). Dis-
crete wavelet transform applied on personal identity
verification with ECG signal. Int. J. Wavelets Multi.,
7(3):341–355.
Daubechies, I. and Sweldens, W. (1998). Factoring wavelet
transforms into lifting steps. J. Fourier Anal. Appl.,
4(3):245–267.
Do, M. N. and Vetterli, M. (2002). Wavelet-based tex-
ture retrieval using generalized gaussian density and
kullback-leibler distance. IEEE Trans. Image Pro-
cess., 11(2):146–158.
Israel, S. A., Irvine, J. M., Cheng, A., Wiederhold, M. D.,
and Wiederhold, B. K. (2005). ECG to identify indi-
viduals. Pattern Recognit., 38(1):133–142.
Jalaleddine, S. M., Hutchens, C. G., Strattan, R. D.,
and Coberly, W. (1990). ECG data compres-
sion techniques - a unified approach. IEEE
Trans. Biomed. Eng., 37(4):329–343.
Laguna, P., Mark, R. G., Goldberger, A. L., and Moody,
G. B. (1997). A database for evaluation of algorithms
for measurement of QT and other waveform inter-
vals in the ECG. In Computers in Cardiology 1997.
(pp. 673-676). IEEE.
Linear Dimensions Inc. (2015). Biometric ECG secu-
rity devices. Retrieved may 22, 2015, from http://
www.lineardimensions.com/securitycardfinal.pdf.
Nymi Inc. (2013). Nymi white paper. Retrieved may
22, 2015, from https://www.nymi.com/wp-content/
uploads/2013/11/nymiwhitepaper-1.pdf.
Odinaka, I., Lai, P. H., Kaplan, A. D., OSullivan, J. A.,
Sirevaag, E. J., and Rohrbaugh, J. W. (2012). ECG
biometric recognition: A comparative analysis. IEEE
Trans. Inform. Forensic Secur., 7(6):1812–1824.
Plataniotis, K. N., Hatzinakos, D., and Lee, J. K. (2006).
ECG biometric recognition without fiducial detection.
In BSYM’06, Biometrics Symposium: Special Session
on Research at the Biometric Consortium Conference.
(pp. 1-6). IEEE.
Poree, F., Kervio, G., and Carrault, G. (2014). ECG biomet-
ric analysis in different physiological recording condi-
tions. J. Signal, Image and Video Process.
Smith, J. R. and Chang, S. F. (1994). Transform features for
texture classification and discrimination in large im-
age databases. In Int. Conf. Image Process. (pp. 407-
411). IEEE.
Sufi, F. and Khalil, I. (2011). Faster person identifica-
tion using compressed ECG in time critical wireless
telecardiology applications. J. Netw. Comput. Appl.,
34(1):282–293.
Sun, C. C. and Tai, S. C. (2005). Beat-based ECG com-
pression using gain-shape vector quantization. IEEE
Trans. Biomed. Eng., 52(11):1882–1888.
Taubman, D. S. and Marcellin, M. W. (2001). JPEG
2000: Image Compression Fundamentals, Standards,
and Practice. Kluwer Academic Publishers, Norwell,
MA.
Thaler, M. S. (2012). The Only EKG Book You’ll Ever Need.
Lippincott Williams & Wilkins, Philadelphia, 7th edi-
tion.
Yang, M., Liu, B., Zhao, M., Li, F., Wang, G., and Zhou,
F. (2013). Normalizing electrocardiograms of both
healthy persons and cardiovascular disease patients
for biometric authentication. PloS one, 8(8):e71523.
CompressedDomainECGBiometricIdentificationusingJPEG2000
13
