Biomedical Question Types Classification using Syntactic and Rule
based Approach
Mourad Sarrouti
1
, Abdelmonaime Lachkar
2
and Said El Alaoui Ouatik
1
1
Department of Computer Science, FSDM, Sidi Mohamed Ben Abdellah University, Fez, Morocco
2
Department of Electrical and Computer Engineering, ENSA, Sidi Mohamed Ben Abdellah University, Fez, Morocco
Keywords: Biomedical Question Answering System, Biomedical Question Types Classification, Answer Types
Classification, Natural Language Processing, Syntactic Patterns, Unified Medical Language System.
Abstract: Biomedical Question Types (QTs) Classification is an important component of Biomedical Question
Answering Systems and it attracted a notable amount of research since the past decade. Biomedical QTs
Classification is the task for determining the QTs to a given Biomedical Question. It classifies Biomedical
Questions into several Questions Types. Moreover, the Question Types aim to determine the appropriate
Answer Extraction Algorithms. In this paper, we have proposed an effective and efficient method for
Biomedical QTs Classification. We have classified the Biomedical Questions into three broad categories.
We have also defined the Syntactic Patterns for particular category of Biomedical Questions. Therefore,
using these Biomedical Question Patterns, we have proposed an algorithm for classifying the question into
particular category. The proposed method was evaluated on the Benchmark datasets of Biomedical
Questions. The experimental results show that the proposed method can be used to effectively classify
Biomedical Questions with higher accuracy.
1 INTRODUCTION
Unlike Information Retrieval (IR) System that
retrieve a large number of documents that are
potentially relevant for the questions posed by the
inquirers, Question Answering (QA) System aims to
provide inquirers with direct, precise answers to
their questions, by employing Information
Extraction (IE) and Natural Language Processing
(NLP) methods (Athenikos and Han, 2009).
Typically an automated QA System consists of
three main phases (Athenikos and Han, 2009;Gupta
and Gupta, 2012): Questions Processing, Documents
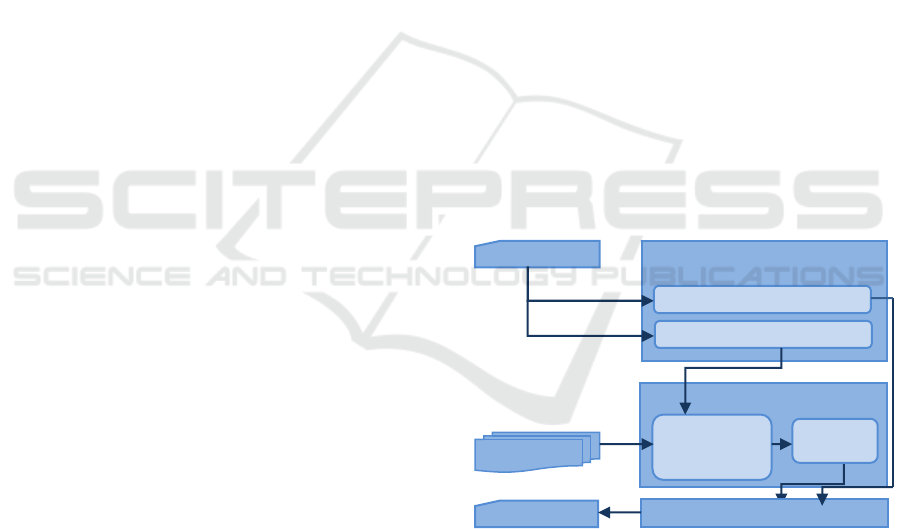
Processing and Answers Processing phases. Figure 1
illustrates the generic architecture of Biomedical QA
System.
Biomedical QTs Classification is a crucial task
of any Biomedical QA System. It classifies
Biomedical Questions into several Biomedical QTs.
In addition, the main goal of Biomedical QTs
Classification is to determine the Excepted Answer
Type to a given Biomedical Question, such as
whether the answer should be a Biomedical Entity
Names, short text summarization, paragraph or just
“Yes” or “No”.
Documents Processing
Questions
Questions Processing
Query Reformulation
Question Classification
Documents
And Passa
g
es
Candidate
Passages
Answers Processing
Answer
(
s
)
Documents
Figure 1: Generic Architecture of Question Answering
System.
Indeed, in order to extract the answer for a given
Biomedical Question, the system should know in
advance the Excepted Answer Type that allows a
Biomedical QA System to use type-specific answer
retrieval algorithms and to reject possible answers of
the wrong type. Therefore, Biomedical QTs
Classification task plays a vital role in Biomedical
QA System that can strongly affect positively or
negatively the Answers Processing phase and hence
Sarrouti, M., Lachkar, A. and Ouatik, S..
Biomedical Question Types Classification using Syntactic and Rule based Approach.
In Proceedings of the 7th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2015) - Volume 1: KDIR, pages 265-272
ISBN: 978-989-758-158-8
Copyright
c
2015 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
265

determines the quality and overall performance of
the Biomedical QA System.
In recent years, several works have been done in
this field for open domain QA System (Zhang et al.,
2007; Tomuro, 2004), as well as for Biomedical QA
System. Ely et al. (1999,2000,2002) have proposed a
generic taxonomy of common Medical QTs and
another taxonomy which classifies questions into
Clinical vs Non-Clinical, General vs Specific,
Evidence vs No Evidence, and Intervention vs No
Intervention. For instance, question taxonomies have
some expressiveness limits. For example, Ely et al.,
(2000)’s question taxonomy provides only some
forms of expression for each question category,
when in the real world we may often retrieve several
other expressions for the same categories.
In fact, there are important factors that
distinguish Biomedical QA System from open-
domain QA System. Those factors include: (1) size
of data, (2) domain context, and (3) resources.
Indeed, in biomedical domain QA, the domain of
application provides a context for the QA process.
This involves domain-specific terminologies and
domain-specific types of questions, which also differ
between domain experts and non-expert users.
Athenikos and Han (2009) report the following
characteristics for restricted-domain QA in the
biomedical domain: (1) large-sized textual corpora,
(e.g., MEDLINE) , (2) highly complex domain
specific terminology, that is covered by domain-
specific lexical, terminological, and ontological
resources, (e.g., Unified Medical Language System
(UMLS)), (3) tools and methods for exploiting the
semantic information (e.g., MetaMap), and (4)
Domain-specific format and typology of questions.
Therefore from the above remarks, the
Biomedical QTs Classification block needs its own
methods that are different from others used for open
domain QA System that have been proposed for the
following Question Types: Location, Date, Person,
Organization, etc (Khoury, 2011).
In light of this, there are two main approaches of
Questions Classification: Syntactic Patterns-based
approach and Machine Learning. Due to the limited
number of Question Types and due to the lack of
quantity of labeled data, the Syntactic Patterns-based
methods have become the most popular methods in
QTs Classification System (Sung et al., 2008).
As far as we know
, there are no studies that have
discussed QTs Classification problem in the biomedical
domain and that is why several Biomedical QA System
that have been presented deal only with one Question
Type
(Weissenborn et al., 2013; Yang et al., 2015).In
this paper, we have proposed an efficient and
effective method for Biomedical QTs Classification.
Indeed, we have taken into account all Biomedical
QTs. We have defined the Syntactic Patterns of
each Biomedical QTs. In particular, we have
classified the Biomedical Questions into three broad
categories: Yes/No, Factoid and Summary Questions.
We denote that all the Excepted Answer Types are
summarized by these categories (Tsatsaronis et al.,
2012).
Yes/No Questions: These are questions that
require “Yes” or “No” as an answer. For example,
“Is COL5A2 gene associated to ischemic heart
disease?” is a Yes/No Question.
Factoid Questions: These are questions that
require a particular one or more of Biomedical
Entity Names (e.g., of a disease, drug, gene, a list of
gene names, etc.), a number(s), or a similar short
expression as an answer. For example, “Which genes
have been found mutated in Gray platelet syndrome
patients?” is a Factoid Question.
Summary Questions: These are questions that
can only be answered by phrase extracted from
relevant document or by producing a short text
summarizing the most prominent relevant
information. For example, “What is the role of
anhedonia in coronary disease patients?” is a
Summary Question (Tsatsaronis et al., 2012).
The rest of this paper is organized as follows: In
Section 2, we review a representative sample of
work done on the task of QTs Classification. Our
proposed method for Biomedical QTs Classification
is presented in Section 3. The experimental results
are presented and discussed in Section 4. Finally, we
conclude the paper and describe future work in
Section 5.
2 RELATED WORK
Although research on QA System has boomed in
recent years, Question Classification has been a
large part in the research community of text mining
after the introduction of QA Track in the Text
REtrieval Conference (TREC) in 1999 as well as the
presentation of Biomedical QA in the BioASK
(Tsatsaronis et al., 2012). Several works that have
been presented for open domain QTs Classification
are usually based on Syntactic Patterns or rule-based
approach such as (Prager et al., 1999; Khoury, 2011;
Haris and Omar, 2012; Biswas et al., 2014).
However, laborious researcher’s effort is
required to create these rules. Some researchers have
used machine-learning approach. Li and Roth (2002)
have presented a hierarchical classifier for open
KDIR 2015 - 7th International Conference on Knowledge Discovery and Information Retrieval
266

domain QTs Classification based on the Sparse
Network of Winnows .Two classifiers were involved
in this work: the first one classified questions into
the coarse categories; and the other classified
questions into fine categories. Several syntactic and
semantic features were extracted and compared in
their experiments. Their result showed that the
hierarchical classifier achieved an accuracy of 90%.
Yu et al., (2005) have improved the Bayesian
model by applying the tf-idf measure to deal with
the weight of words for Chinese Question
Classification. They have achieved an accuracy of
72.4%. Xu et al., (2006) have employed affiliated
ingredients as the features of the model and used the
results obtained by the syntactic analysis for
extracting the question word. They have also
achieved an accuracy of 86.62% for the coarse
grained categories. Sun et al., (2007) have got
features for classification using HowNet as the
Semantic resource, whereas Yu et al., (2005) have
used Support Vector Machine model, and choose
word, part of speech, chunk, Named Entity, word
meaning, synonyms and Categories coherence word.
Li et al., (2008) have classified open domain
what-type questions into proper semantic categories
using Conditional Random Fields (CRFs). They
have used several syntactic and semantic features
were extracted and compared in their experiments.
They have used the CRFs model to label all the
words in a question, and then choose the label of
head noun as the question category and achieved an
accuracy of 85.60%.
Yu et al., (2007) have presented their
implemented medical QA system, MedQA, which
generates paragraph-level answers from both the
MEDLINE collection and the Web. The system in
its current implementation deals with just
definitional questions (e.g., “What is X?”).
Jacquemart and Zweigenbaum (2003) have
described semantic based approach toward the
development of a French Language Medical QA
System. They have proposed a semantic model of
Medical Questions Classification. In fact, they have
modeled the forms of the 100 Medical Questions as
Syntactic-Semantic Patterns and with one hundred
questions that have been used in their study cannot
cover all QTs. For example, the authors don’t take
into account the What type of questions that
considered one of the most complicate questions to
classify (Li et al., 2008).
Weissenborn et al. (2013) have presented a QA
System for factoid questions in the biomedical
domain. Their system is able to answer only factoid
questions (e.g. “Where in the cell do we find the
protein Cep135?”). The authors have not taken into
account other QTs and have not addressed the
Biomedical QTs Classification problem.
Yang et al. (2015) have described a Biomedical
QA System deals with just factoid questions.
However, the authors have not addressed the
Biomedical QTs Classification challenge. They have
clearly noted that the Biomedical QTs Classification
is a big challenge for building an extensible
Biomedical QA System.
To our knowledge, the literature has not
discussed the QTs Classification problem in the
biomedical domain, whereas our study addresses this
problem of Biomedical QTs Classification and takes
into account all Biomedical QTs in order to build an
extensible Biomedical QA System.
3 PROPOSED METHOD
Our main goal is to classify the Biomedical
Questions into three broad categories: Yes/No,
Factoid and Summary Questions. To achieve this
goal, we propose several Syntactic Patterns of each
Biomedical QTs. We first run a POS Tagger on each
question of Benchmark dataset using the Stanford’s
POS Tagger (Toutanova and Manning, 2000) and
manually analyze these questions and constructing
Syntactic Patterns for QTs. On the basis of that we
have classified each QTs into three categories.
Table 1 show that Which, How and What Types
of questions could belongs to Factoid and Summary
Questions, while Why Type of questions belongs to
only Summary Questions, etc.
Table 1: Question Categories and their Excepted Answer
Types.
Question Category Answer(s)
How
Factoid
Biomedical Entity Names,
Number(s), short expression
Summary
Phrase, Paragraph, short text
summarization
Why Summary
Phrase, Paragraph, short text
summarization
Where Factoid
Biomedical Entity Names,
Number(s), short expression
Which
Factoid
Biomedical Entity Names,
Number(s), short expression
Summary
Phrase, Paragraph, short text
summarization
What
Factoid
Biomedical Entity Names,
Number(s), short expression
Summary
Phrase, Paragraph, short text
summarization
Yes/No Yes/No Yes or No
Biomedical Question Types Classification using Syntactic and Rule based Approach
267

To detect Yes/No Questions we used the regular
expression (see pattern (1)), where the questions
should start with three types of words. We found
that this method is significantly efficient.
[Be verbs |Modal verbs |Auxiliary
verbs]+[.*]+?
(1)
Where:
Be verbs = «am, is, are, been, being, was,
were»
Modal verbs= «can, could, shall, should, will,
would, may, might»
Auxiliary verbs= «do, did, does, have, had,
has»
Examples:
Is intense physical activity associated with
longevity?
Does Serca2a bind PLN in the heart?
As it is difficult to distinguish Factoid Type of
Question Wh-word, from Summary Type of
Question Wh-word (Biswas et al., 2014). In this
paper, we have used WordNet in order to improve
the performance of classification. Indeed, The
WordNet (Fellbaum, 1998) is a large English
lexicon in which meaningfully related words are
connected via cognitive synonyms (synsets). The
WordNet is a useful tool for word semantics analysis
and has been widely used in Question Classification
as semantic feature for machine learning approach
such as (Schlaefer et al., 2007).
Additionally, we have used MetaMap (Aronson,
2001) for mapping terms in questions to UMLS in
order to extract the Biomedical Entity Names
(BENs). The UMLS (Bodenreider, 2004) is a
repository of biomedical vocabularies developed by
the US National Library of Medicine.
To overcome this issue, we list all regular
expression patterns of each QTs that are used in our
experiments as follow (NOUN is a noun, JJ is an
Adjective):
What ─ Question category: Summary or Factoid
Question Pattern:
What+[is|are]+
(2)
Examples:
What is the definition of autophagy?
What is the function of the yeast protein Aft1?
What+[is|are]+ (3)
Examples:
What is clathrin?
What is the ubiquitin proteome?
What+[Modal: does]+
«
Summary»
(4)
What+
(5)
Example:
What types of cancers and inherited diseases have
been associated to mutations in the Notch pathway?
What+[is|are]+
(6)
Examples:
What is the indication of Daonil (Glibenclamide)?
What is the prevalence of short QT syndrome?
Which ─ Question category: Summary or Factoid
Question Pattern:
Which+
(7)
Example:
Which is the phosphorylated residue in the promoter
paused form of RNA polymerase II?
Which+[is|are]+
(8)
Example:
Which is the prevalence of cystic fibrosis in the
human population?
definition
role «Summary»
treatment
aim+[of]+[NOUN|JJ+NOUN]+?
effect
mechanism
Synonyms of those words
Where NOUN is BEN
numbe
r
name «Factoid»
indication
value
+[of]+[NOUN|JJ+NOUN]+?
frequency
prevalence
frequency
Synonyms of those words
Where NOUN is BEN
[NOUN]+[.*]+?
Or «
Factoid»
[Modal (does| do)] + [.*] + [stand for
|
bind to
]
+?
numbe
r
name «Factoid»
indication
value
+[of]+[NOUN|JJ+NOUN]+?
frequency
prevalence
frequency
Synonyms of those words
Where NOUN is BEN
[NOUN]+ ? «Summary»
Where NOUN is BEN
[NOUN]+[.*]+[do]+?
Where NOUN is BEN
[NOUN] where NOUN is BEN
Or
[Verb]+[.*]
+[NOUN]+ [.*]+[NOUN]+?
Or
[JJ+NOUN]
«Factoid»
KDIR 2015 - 7th International Conference on Knowledge Discovery and Information Retrieval
268

Which+[is|are]+
(9)
Example:
Which is the definition of pyknons in DNA?
Where ─ Question category: Always Factoid
Question Pattern:
Where + [.*] +? «Always Factoid» (10)
Example:
Where in the cell do we find the protein Cep135?
Why ─ Question category: Always Factoid
Question Pattern:
Why + [.*] +? «Always Summary» (11)
Examples:
Why does the prodrug amifostine (ethyol) create
hypoxia?
How ─ Question category: Factoid or Summary
Question Pattern:
How + (12)
Example:
How many recombination hotspots have been found
in the yeast genome?
How +
(13)
Example:
How does adrenergic signaling affect thyroid
hormone receptors?
H
ow are thyroid hormones involved in the
development of diabetic cardiomyopathy?
Define| Synonyms+
(14)
Example:
Define marine metaproteomics.
In addition, we have proposed an algorithm (see
algorithm (1)) to classify the given Biomedical
Question into any one of the predefined categories
using the questions patterns presented above. Then,
the appropriate Answer Extraction Algorithms can
be applied for extracting the precise and most
appropriate answer for that Biomedical Question.
Algorithm 1: Biomedical QTs Classification.
Input: Biomedical Questions
Output: Biomedical QTs /*Yes/No, Factoid or
Summary*/
1: If Biomedical Question matched to [Be verbs |
Modal verbs | Auxiliary verbs] + [.*] +? then
2: Return “Yes/No Question”
3: Else
4: Case 1: Wh-word = “How”
If wh-word+ [Adjective| Adverb] then
5: Return “Factoid”
6: Else if wh-word+ [Modal| Verb] then
Return “Summary”
7: Else if wh-word+[Noun] && Noun =
BEN then Return “Summary”
8: End if
9: Case 2: Wh-word = “Why” Return “Summary”
10: Case 3: Wh-word = “Where” Return “Factoid”
11: Case 4: Wh-word = “Which”
12: If wh-word+ [Noun| Verb|JJ+Noun]&&
Noun = BEN then Return “Factoid”
13: Else if wh-word+ [is| are]+ [[indication of]|
[number of]|…| [synonym of those words]
]+ [NOUN|JJ+NOUN] && Noun= BEN
then Return “Factoid”
14: Else if wh-word+ [is| are] + [[definition
of]| [role of]|…| [synonym of those
words] ]+ [NOUN|JJ+NOUN] &&
Noun= BEN then Return “Summary”
15: End if
16: Case 5: Wh-word = “What”
17: If wh-word+ [is| are] + [Noun] && Noun =
BEN | wh-word+ [Modal: does] + [NOUN]
+ [.*] + [do] +? Then Return “Summary”
18: Else if wh-word+ [Noun] | wh-
word+[Modal]+[.*]+[ stand for| bind to]+?
Then Return “Factoid”
19: Else if wh-word+ [is| are]+ [[indication of]|
[number of]|…| [synonym of those words]
]+ [NOUN|JJ+NOUN] && Noun= BEN
then Return “Factoid”
20: Else if wh-word+ [is| are] + [[definition
of]| [role of]|…| [synonym of those
words] ]+ [NOUN|JJ+NOUN] &&
Noun= BEN then Return “Summary”
21: End if
22: Case 6: Biomedical Question matched to [Define|
Synonyms] + [NOUN] +? && NOUN=BEN
23: Return “Summary”
24: End if
4 EXPERIMENTS AND RESULTS
In this section, we evaluate our proposed method for
definition
role «Summary»
treatment
aim
+[of]+[NOUN|JJ+NOUN]+?
effect
mechanism
Synonyms of those words
Where NOUN is BEN
[Adverb]
Or + [.*] +? «Factoid»
[Adjective]
[Modal]
Or
[Verb] + [.*] +? «Summary»
Or
[NOUN] where NOUN is BEN
[
N
OUN] +? «Summary»
Where NOUN is BEN
Biomedical Question Types Classification using Syntactic and Rule based Approach
269
Biomedical QTs Classification through the study of
the performance of Biomedical Syntactic Patterns
for each type of questions.
4.1 Datasets
The 1433 Biomedical Questions of the Benchmark
datasets are the evaluation text collection of our
study. Those over one thousand Biomedical
Questions were obtained from BioAsk challenges
(Tsatsaronis et al., 2012). Each type of question was
assigned to one category. For example, the question
“What is the role of the Tsix gene during X
chromosome inactivation?” was assigned to
Summary Questions.
Table 2 shows the 3 categories and the number
of Questions Types assigned to each one. For
example, 198 What Type of questions were assigned
to Factoid Questions, 234 What Type of questions
were assigned to Summary Questions, etc.
Table 2: Question Types and their Question Categories.
Question
Types
Question Categories
Yes/No Factoid Summary
Total
How 0 29 37 66
Why 0 2 12 14
Where 0 15 0 15
Which 0 394 55 439
What 0 198 234 432
Yes/No 467 0 0 467
Total 467 638 328 1433
4.2 Results and Discussion
To conduct the experiments, we have used the
Benchmark datasets (Tsatsaronis et al., 2012) that
have been presented above. We have applied
Stanford’s POS Tagger (Toutanova and Manning,
2000) for finding the syntactic structure of
questions. We proceeded by passing the questions
one by one to Stanford’s POS Tagger in order to
capture their syntactic structure. Additionally, We
have applied MetaMap (Aronson, 2001) to extract
the Biomedical Entity Names of each Biomedical
Questions. Moreover, we have used WordNet in
order to generate synonyms of words (e.g. treatment,
effect, etc.) that have been presented in patterns
[2,6,8,9].We have exploited the Syntactic patterns
that have been presented in section 3.
For evaluation, accuracy performance have been
widely used to evaluate Question Types
Classification methods (Li and Roth, 2002; Zhang et
al., 2003; Metzler et al., 2004; Biswas et al., 2014).
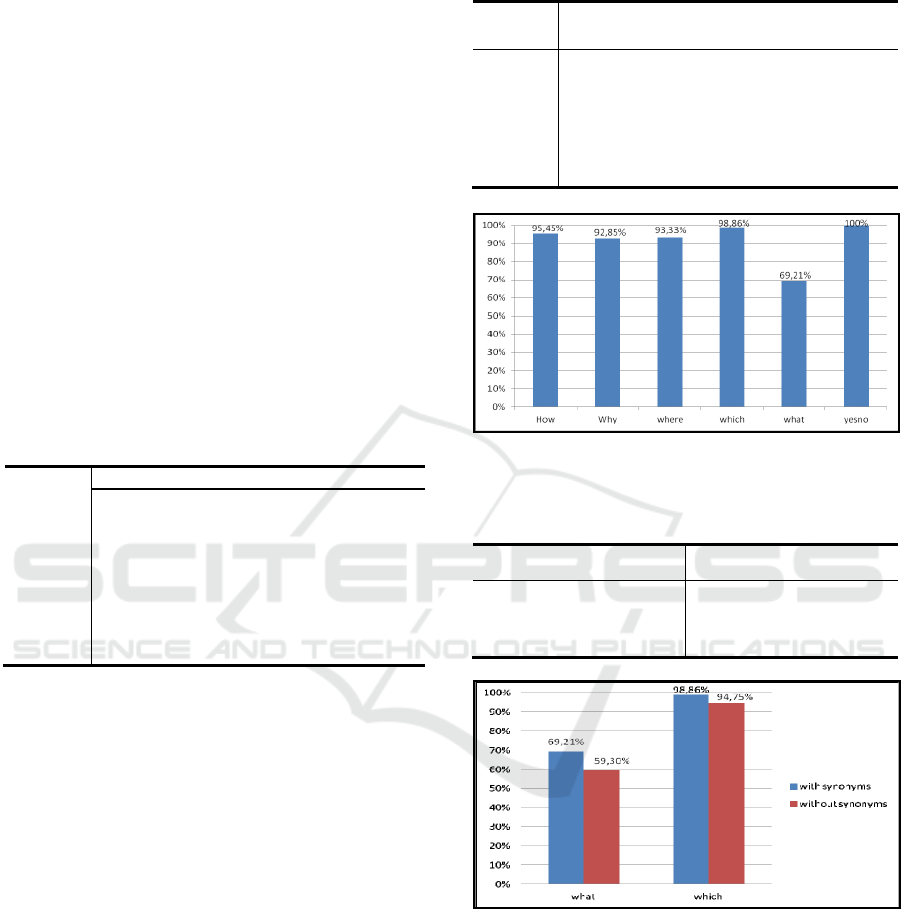
Table 3: Accuracy Performance of Biomedical QTs
Questions.
Question
Types
Total Matched Mismatched Accuracy
How 66 63 3 95.45%
Why 14 13 1 92.85%
Where 15 14 1 93.33%
Which 439 434 5 98.86%
What 432 299 133 69.21%
Yes/No 467 467 0 100%
Figure 2: Performance Measurement.
Table 4: Increase performance of what and which types of
questions with/without synonyms.
Question Types What Which
Without Synonyms 59.30% 94.75%
With Synonyms 69.21% 98.86%
Increase Performance +10% +4%
Figure 3: Accuracy performance of what and which types
of questions with/without synonyms.
Overall, from the results presented in Table 3
and Figure 2, it can clearly be seen that the overall
accuracy of our proposed method for Biomedical
QTs Classification is very interesting. The average
performance for automatically assigning a category
to a question was accuracy of 91.62%.
In fact, the proposed method led to the highest
accuracy of 100% and 98.68% for classifying the
Yes/No Biomedical Questions and Which Type of
KDIR 2015 - 7th International Conference on Knowledge Discovery and Information Retrieval
270

questions. As it is difficult to distinguish Factoid
Type from Summary Type of What Type of
questions, nevertheless, our proposed have achieved
an accuracy of 69.21%. Indeed, we can see from
Table 4 and Figure 3 that synonyms extracted from
WordNet of words (e.g. definition, indication, etc.)
that have been presented in section 3 (see pattern (2)
and pattern (6)) enhanced the performance (+10%)
for classifying What Type of questions. In addition,
using synonyms in pattern (8) and pattern (9)
enhanced the performance (+4%) for classifying
Which Type of questions.
We have shown that the proposed method can
classify the Biomedical Questions into three broad
categories: Yes/No, Factoid and Summary
Questions. Indeed those three categories can
summarize all Question Type that can be posed by
user (Tsatsaronis et al., 2012). In other words, these
categories can summarize all possible cases of the
Expected Answer Types. For example: “Which
diseases are caused by mutations in Calsequestrin 2
(CASQ2) gene? “ is a Factoid Questions and the
answer is a Biomedical Named Entity (BNE)
"CPVT", “catecholaminergic polymorphic ventricu-
lar tachycardia”. In addition, one of the most
advantages of our proposed method is that it does
not require a learning phase and therefore it can be
easily integrated in a Biomedical QA System in
order to build an extensible Biomedical QA System.
As mentioned in the Introduction, so far no one
appears to have discussed QTs Classification in the
biomedical domain. Therefore, the importance of
our results using our proposed method thus lies both
in their generality and their relative ease of
application to Biomedical QA System.
In addition, maybe one limitation of our research
was the patterns used for classifying What Type of
question if we take into account its difficulty to
distinguish Factoid Type from Summary Type of
What Type of question. Clearly these patterns that
have been used for classifying What Type question
are not enough to make generalizations about this
one. However, The 69.21% for the What Type of
question is quite low compared to the rest of the
results. The 69.21% stay satisfactory and it could be
improved with more patterns. Indeed the presence of
large numbers of such rules does not introduce any
problems because the rules are represented by a set
of patterns, and Question Types Classification is
conducted by pattern matching.
Remarkably,
the comparative studies that have
been done have focused on QA Systems and due to
the lack of baseline of Biomedical QTs Classification
methods we could not have comparative study.
The main goal of our project is to develop an
extensible Biomedical QA System capable to answer
all Biomedical QTs that is Yes/No, Factoid and
Summary Questions which is composed by three
main components: Questions Processing, Documents
Processing and Answers Processing. Significantly,
we believe that QT Classification may impact
positively or negatively on the reset of the QA
process. Indeed, the output of QTs Classification
will be used to determine the appropriate Answer
Extraction Algorithm.
5 CONCLUSION
In this paper we have presented a novel method for
classifying the Biomedical Questions into three
broad categories that summarize all possible cases of
the Expected Answer Types: Yes/No, Factoid and
Summary Questions. We first defined the syntactic
structure of the Biomedical Questions using
Stanford’s POS Tagger (Toutanova and Manning,
2000) as well as MetaMap for Biomedical Named
Entities Recognition (BNER). We presented the
rules that are represented by a set of patterns for
each Biomedical QTs. Our experimental results
using our proposed algorithm prove that the
Biomedical QTs classification problem can be
solved quite accurately using our proposed method.
The proposed method will allow selecting the
appropriate Answer Extraction Algorithm and
therefore the Biomedical QA System will be able to
treat all types of questions.
In future work we plan to improve the
performance for classifying What Type of questions
by presenting more patterns. We will pay more
attention to integrate our Biomedical QTs
classification system in a Biomedical QA System.
ACKNOWLEDGEMENTS
We thank BioASK challenges for providing us with
Benchmark datasets.
REFERENCES
Aronson, a R., 2001. Effective mapping of biomedical text
to the UMLS Metathesaurus: the MetaMap program.
Proc. AMIA Symp. 17–21.
Athenikos, S.J., Han, H., 2009. Biomedical question
answering : A survey. Comput. Methods Programs
Biomed. 99, 1–24.
Biomedical Question Types Classification using Syntactic and Rule based Approach
271

Biswas, P., Sharan, A., Kumar, R., Sciences, S., 2014.
Question Classification Using Syntactic And Rule
Based Approach. Int. Conf. Adv. Comput. Commun.
Informatics 1033–1038.
Bodenreider, O., 2004. The Unified Medical Language
System (UMLS): integrating biomedical terminology.
Nucleic Acids Res. 32, D267–D270.
Ely, J.W., Osheroff, J. a, Ebell, M.H., Bergus, G.R., Levy,
B.T., Chambliss, M.L., Evans, E.R., 1999. Analysis of
questions asked by family doctors regarding patient
care. BMJ 319, 358–361.
Ely, J.W., Osheroff, J. a, Ebell, M.H., Chambliss, M.L., C,
D., Stevermer, J.J., Pifer, E. a, Vinson, D.C., 2002.
Obstacles to answering doctors’ questions about
patient care with evidence: qualitative study. BMJ 324,
1–7.
Ely, J.W., Osheroff, J. a, Gorman, P.N., Ebell, M.H.,
Chambliss, M.L., Pifer, E. a, Stavri, P.Z., 2000. A
taxonomy of generic clinical questions: classification
study. BMJ 321, 429–432.
Fellbaum, C., 1998. WordNet. Wiley Online Library.
Gupta, P., Gupta, V., 2012. A Survey of Text Question
Answering Techniques. Int. J. Comput. Appl. 53, 1–8.
Haris, S.S., Omar, N., 2012. A rule-based approach in
Bloom’s Taxonomy question classification through
natural language processing, in: Computing and
Convergence Technology (ICCCT), 2012 7th
International Conference on. pp. 410–414.
Jacquemart, P., Zweigenbaum, P., 2003. Towards a
medical question-answering system: A feasibility
study. Stud. Health Technol. Inform. 95, 463–468.
Khoury, R., 2011. Question Type Classification Using a
Part-of-Speech Hierarchy. Lect. Notes Comput. Sci.
212–221.
Li, F., Zhang, X., Yuan, J., Zhu, X., 2008. Classifying
what-type questions by head noun tagging. 22nd Int.
Conf.
Li, X., Roth, D., 2002. Learning question classifiers. Proc.
19th Int. Conf. Comput. Linguist. 1 1–7.
Metzler, D., Metzler, D., Croft, W.B., Croft, W.B., 2005.
Analysis of Statistical Question Classi cation for Fact-
based Questions. Inf. Retr. Boston. 8, 1–30.
Niu, Y., Hirst, G., 2004. Analysis of semantic classes in
medical text for question answering, in: Proceedings
of the ACL 2004 Workshop on Question Answering in
Restricted Domains. pp. 54–61.
Prager, J., Radev, D., Brown, E., Coden, A., Samn, V.,
1999. The Use of Predictive Annotation for Question
Answering in TREC8. Proc. 8th Text Retr. Conf. 399–
411.
Schlaefer, N., Ko, J., Betteridge, J., Pathak, M.A., Nyberg,
E., Sautter, G., 2007. Semantic Extensions of the
Ephyra QA System for TREC 2007., in: TREC.
Sun, J., Cai, D., Lv, D., DONG, Y., 2007. HowNet based
Chinese question automatic classification. J. Chinese
Inf. Process. 21, 90–95.
Sung, C.L., Day, M.Y., Yen, H.C., Hsu, W.L., 2008. A
template alignment algorithm for question
classification. IEEE Int. Conf. Intell. Secur.
Informatics, 2008, IEEE ISI 2008 197–199.
Tomuro, N., 2004. Question terminology and
representation for question type classification.
Terminology 10, 153–168.
Toutanova, K., Manning, C.D., 2000. Enriching the
Knowledge Sources Used in a Maximum Entropy
Part-of-Speech Tagger. Proc. Jt. SIGDAT Conf.
Empir. Methods Nat. Lang. Process. Very Large
Corpora 63–70.
Tsatsaronis, G., Schroeder, M., Paliouras, G., Almirantis,
Y., Androutsopoulos, I., Gaussier, E., Gallinari, P.,
Artieres, T., Alvers, M.R., Zschunke, M., others, 2012.
BioASQ: A Challenge on Large-Scale Biomedical
Semantic Indexing and Question Answering. AAAI
Fall Symp. Inf. Retr. Knowl. Discov. Biomed. Text.
Weissenborn, D., Tsatsaronis, G., Schroeder, M., 2013.
Answering Factoid Questions in the Biomedical
Domain. BioASQ@ CLEF 1094.
Xu, W.E.N., ZHANG, Y., Ting, L.I.U., Jin-Shan, M.A.,
others, 2006. Syntactic Structure Parsing Based
Chinese Question Classification [J]. J. Chinese Inf.
Process. 2, 4.
Yang, Z., Gupta, N., Sun, X., Xu, D., Zhang, C., Nyberg,
E., 2015. Learning to Answer Biomedical Factoid and
List Questions OAQA at BioASQ 3B, in: Working
Notes for the Conference and Labs of the Evaluation
Forum (CLEF), Toulouse, France.
Yu, H., 2006. System and methods for automatically
identifying answerable questions.
Yu, H., Lee, M., Kaufman, D., Ely, J., Osheroff, J. a.,
Hripcsak, G., Cimino, J., 2007. Development,
implementation, and a cognitive evaluation of a
definitional question answering system for physicians.
J. Biomed. Inform. 40, 236–251.
Yu, Z., Ting, L., Xu, W., 2005. Modified Bayesian model
based question classification. J. Chinese Inf. Process.
19, 100–105.
Yu, Z.-T., Fan, X.-Z., Guo, J.-Y., 2005. Chinese question
classification based on support vector machine.
Huanan Ligong Daxue Xuebai (Ziran Kexue Ban)/ J.
South China Univ. Technol. Sci. Ed. 33, 25–29.
Zhang, D., Zhang, D., Lee, W.S., Lee, W.S., 2003.
Question classification using Support Vector
Machines. Proc. 26th Annu. Int. ACM SIGIR Conf.
Res. Dev. Inf. Retr. (SIGIR ’03), July 28 - August 1,
2003, Toronto, Canada 26–32.
Zhang, L., Zhou, L., Chen, J.J., 2007. Structure analysis
and computation-based chinese question classification.
Proc. - ALPIT 2007 6th Int. Conf. Adv. Lang. Process.
Web Inf. Technol. 39–44.
KDIR 2015 - 7th International Conference on Knowledge Discovery and Information Retrieval
272
