Towards Reusability of Computational Experiments
Capturing and Sharing Research Objects from Knowledge Discovery Processes
Armel Lefebvre, Marco Spruit
and Wienand Omta
Department of Information and Computer Sciences, Utrecht University, Princetonplein 5, Utrecht, The Netherlands
Keywords: Knowledge Discovery, Reproducible Research, Bioinformatics, Research Objects, Software Development.
Abstract: Calls for more reproducible research by sharing code and data are released in a large number of fields from
biomedical science to signal processing. At the same time, the urge to solve data analysis bottlenecks in the
biomedical field generates the need for more interactive data analytics solutions. These interactive solutions
are oriented towards wet lab users whereas bioinformaticians favor custom analysis tools. In this position
paper we elaborate on why Reproducible Research, by presenting code and data sharing as a gold standard
for reproducibility misses important challenges in data analytics. We suggest new ways to design interactive
tools embedding constraints of reusability with data exploration. Finally, we seek to integrate our solution
with Research Objects as they are expected to bring promising advances in reusability and partial
reproducibility of computational work.
1 INTRODUCTION
Over the last few years, calls from researchers
defending better data and code sharing for
computational experiments (CE) are propagated in
high-ranked journals (McNutt, 2014; Peng,
2011).Usually grouped under Reproducible Research
(RR), these invitations elevate reproducibility or
replicability as a central key of the scientific method.
One of the interpretations presents reproduction as an
application, by independent researchers, of identical
methods on identical data to obtain similar results
whereas replication is similar except that different
data is selected. According to RR proponents,
benefits would be numerous.
First, for verifying results of a published study
(Peng, 2011). Second, for reusing previous work and
build new knowledge. While the latter brings a
constructive and enriching dimension to reproducible
science, the first one is clearly oriented to alleviating
scientific misconduct, particularly in Life Sciences
(Laine et al., 2007).
Despite the fact that RR proponents are focused
on suggesting to exchange code and data as a minimal
threshold for “good science”, they do not examine the
methods used or people participating in CEs.
Methods are not of interest to RR as the main focus
lays on getting similar results for verification. Hence,
the end product of a CE is seen as a script, or package
that should be made available by the authors of a
paper as supplementary material.
The issue investigated in this work emerged from
three phenomena: (1) the notorious increase of data
generation and resource intensive analytics. Here in
the biomedical domain, (2) ignorance about data
generation processes and their impact in terms of
modelling. For instance, the sequencing instruments
and custom bioinformatics pipelines producing
analytical data and how well they represent
underlying biological facts and (3) non-specialists,
not trained in data analytics, eager to participate in
computationally intensive experiments but preferably
via convenient end-user interfaces instead of custom
scripts or programs (Holzinger et al., 2014).
The phenomena described above were observed
during a design science research (DSR) (Hevner &
Chatterjee, 2010) we conducted in the domain of
biomedical genetics. Our research was focused on
designing an interactive data mining tool for
biologists to identify interesting outliers in RNA-Seq
count tables. Ultimately, the goal is to seek how to
facilitate access and how to reuse scripts and
packages for bioinformaticians and biologists at the
same time. After one design cycle of a technical
artifact and its evaluation by three focus groups
gathering biologists and bioinformaticians (n=15) we
collected evidence against some practices proposed
by RR and suggest potentially fruitful improvements.
456
Lefebvre, A., Spruit, M. and Omta, W..
Towards Reusability of Computational Experiments - Capturing and Sharing Research Objects from Knowledge Discovery Processes.
In Proceedings of the 7th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2015) - Volume 1: KDIR, pages 456-462
ISBN: 978-989-758-158-8
Copyright
c
2015 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
Indeed, reproducibility of CEs should not be
reduced to code and data sharing as it does not cover
fundamental characteristics of modern data analysis
in biology. We state that web resources and their
support for multiple representations that satisfy the
interest of both types of users involved will have a
positive impact on reproducibility by facilitating
reusability first.
2 BACKGROUND
Two aspects of knowledge creation and sharing are
presented. Together, they clarify what issues emerge
from code and data sharing when all stakeholders
involved in a CE are not considered. We make use of
a standard knowledge cycle, the Integrated
Knowledge Cycle (IKC) (Dalkir, 2005) to emphasize
the issues of codification implied by Reproducible
Research. In knowledge management, codification
aims at making implicit knowledge (from an
individual) available as an object that is separated
from the individual (Hislop, 2005). This can also be
seen as the goal of RR which distributes experiments
as packages.
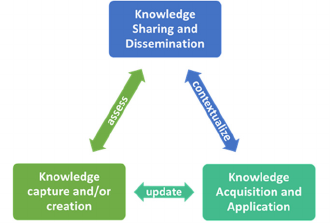
Figure 1: Integrated Knowledge Cycle with three stages
(Dalkir, 2005).
The IKC is illustrated in Figure 1. We focus our
discussion on the knowledge capture and creation
and knowledge sharing and dissemination phases.
The last phase acquisition is not discussed here as we
believe it to be the role of academia or industry in
general.
2.1 HCI-KDD
We start with Human-computer Interaction (HCI)
which is the “study of the way in which computer
technology influences human work and activities.”
(Dix, 2009). Knowledge discovery from databases
(KDD) is defined by Fayyad as “the nontrivial
process of identifying valid, novel, potentially useful,
and ultimately understandable patterns in data”
(Fayyad et al., 1996).
The first aspect is that an end-user should be able
to analyze data by using steps from the knowledge
discovery process but interactively. This combination
between KDD and human-computer interaction was
theorized by Holzinger (2013). Tailored to the
biomedical field, the process emphasizes that an end-
user needs powerful visualization tools as much as
data management and analytics capabilities.
Holzinger also stresses the fact that reproducibility
should be investigated further as it represents a major
problem with data intensive experiments (Holzinger,
2013).
The steps of the HCI-KDD are integration, pre-
processing and data mining. Integration is the
activity of merging structured or unstructured data
sets. Pre-processing applies normalization or
transformation techniques to make the data sets
suitable for data analysis. Data mining is the design
and application of algorithms to identify patterns,
associations or outliers.
2.2 Reproducible Research
The second aspect is the need for better
reproducibility of experiments which are conducted
with computers. Here we integrate notions belonging
to two approaches to reuse context and computational
material.
On the one hand, based on literate programming
(Knuth, 1984), dynamic documents (Pérez and
Granger, 2007) and compendiums (Gentleman and
Lang, 2007) constrain design choice to add human
and machine readable context to executable code.
Compendiums aggregate dynamic documents.
Dynamic documents are executable files that contain
code with descriptive information. They are currently
available with authoring packages in R (Knitr,
Sweave) or Python (IPython notebooks, Jupyter).
On the other hand, an ontology based approach
for dissemination of reusable components is assured
by semantically enriched objects aggregating
resources about the context of an experiment and its
material. These are called Research Objects (RO)
(Bechhofer et al., 2013).
3 DISCUSSION
As we noticed, the fact that one end-user deals with
each step is, at least, a very optimistic view on data
analytics. The HCI-KDD process implemented in our
prototype was discussed among participants (see
Towards Reusability of Computational Experiments - Capturing and Sharing Research Objects from Knowledge Discovery Processes
457
section 3.1). The questions were oriented to the flow
of analysis and presence or absence of components
(e.g. charts, packages, result tables, context…) in the
interface. Additionally, a survey was answered by 11
respondents (n=11) about how they are dealing with
data and Reproducible Research.
3.1 Focus Groups Result
Inside our three focus groups we divide participants
according to their main interests, i.e.
bioinformaticians and biologists.
For the first type of participants,
bioinformaticians, a friendly user interface is
rejected. Scripts are preferred for analyzing data.
Regarding methods applied, a participant indicated
that a method is sometimes selected because “it
works” and is not a matter of “hidden” assumptions.
By assumption we refer to prior knowledge of the
state of the world embedded in packages or statistical
models. Not being aware of them makes a package
acting as a “black-box” with unknown consequences
on the rest of the processing.
For the second type of participants, biologists,
they estimated the presence of such methods as
appropriate. The indications given on the website
(package name, version, reference paper, running
environment and online documentation) are sufficient
if kept up-to-date. The web interface offered the
possibility to apply different methods on the same
data set. This was judged as beneficial because the
influence of a choice could be assessed by the user
interactively. In that case, another concern raised by
bioinformaticians is about the interpretation of
results by users that would not be trained in statistics.
Regarding reproducibility, the lab part of an
experiment has strong influences on the rest of the
pipeline and it is perceived as challenging to integrate
in the tool. Efforts for improving reproducibility are
welcome but full reproducibility is impossible, as
indicated by participants in the third focus group.
3.2 Code and Data for Verification
It is the view of Peng (2011) that executable code and
data form a gold standard of reproducible research.
We argue that these elements are not of interest for
each important type of stakeholder involved in a
computational experiment. We may admit though that
what the author tries to achieve is a minimal level of
reproducibility for verification purposes. The idea is
that a reviewer would carefully inspect code shared
with a paper, e.g. as an R package on Bioconductor.
With that package, the entire computational workflow
is runnable and shows figures that are identical to
their online or printed counterpart.
But as even noticed by Peng (2011), papers
validating previous work are rarely acclaimed by
publishers which expect “new” knowledge to be
submitted. This may be an explanation while results
from our survey showed a poor interest in full
replication. On a scale from 1 (never) to 5 (always).
The need for full replication has a Mode of 2
(Median=2). Partial replication did slightly better
with a Mode of 3 (Median=3).
3.3 Reusability and Interactivity
Regarding Research Objects, they sometimes appear
to be developed as external solutions or repositories.
We would lose a major group of researchers if the
goal of an application is to purely manage research
objects. Instead, the software application should
produce resources that might be automatically
aggregated in a RO. This is a transparent manner for
users more interested in advanced visualization
capabilities.
Therefore, we claim that Research Objects could
be a hidden component of any interactive mining tool.
By doing this, we encourage RO generation and usage
without transforming such tools in a “reproducibility
manager” for users interested in getting precious
insights from their experiments. Exaggerating any
requirement of RO management for these
stakeholders will most probably result in a rejection
of the entire application. This could be achieved by
automatically extracting information from earlier
processing stages and intermediate data sets in the
analysis flow.
3.4 Resources and Representations
An interesting proposal in compendium design was
the notion of transformer. We present it in this work
as the creation of a representation (or view) from a
single resource. A resource is an object of interest
whereas a representation is a usable form of a
resource which corresponds to the consumer’s
interest. We designate by consumers both human and
machine readers or interpreters.
In the RO world, it implies to work on ontologies
and machine readable standards. For biologists, it
means that a chart resource has to render a dynamic
representation. We can imagine that after exchanging
a RO, we find a data object resource and a chart
resource. A chart shows the content of a data object
as, for instance, a scatterplot. We expect an end-user
to be willing to select parts of this scatterplot, zoom-
KDIR 2015 - 7th International Conference on Knowledge Discovery and Information Retrieval
458
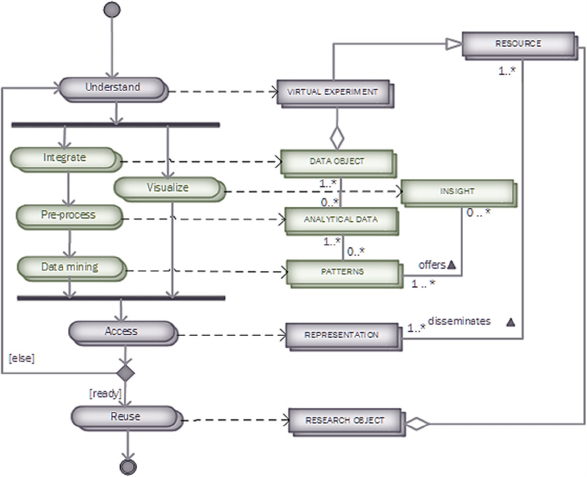
Figure 2: The Reproducible Research Oriented Knowledge Discovery in Databases (RRO-KDD) process.
in or display labels. We also expect that this chart
resource is identical to what was generated by a team
of researchers which created this RO.
As we show in the next section, open source
technologies for visualization “as a resource” exist
and are under heavy development. They are able to
create Json or html/JS serialization of a chart resource
while providing enough interactivity for end-users.
4 SOLUTION
The evaluation of our prototype yielded limitations of
both HCI-KDD and current practices defended by
Reproducible Research. Hence, we suggest an
improved knowledge discovery process embedding
the HCI-KDD in an extended process named
Reproducible Research Oriented Knowledge
Discovery in Databases (RRO-KDD).
When conducting a DSR, four stages appear at
each design cycle (Hevner and Chatterjee, 2010). The
problem specification resulted from a literature
review and meetings with experts in biomedical
genetics. The other steps found in design science
research are Intervention, Evaluation and Reflection.
Each of them are described in the next
subsections.
4.1 Specification
The problem addressed in this work encompasses
reproducibility and visualization for researchers in
biology who are collaborating with bioinforma-
ticians. As explained in the background section,
computational experiments are not only conducted on
the bioinformatics side of data analysis. Hence, an
application enabling self-service data analytics for
biologists has additional constraints. Self-service is
understood as letting users perform analytics tasks
without advanced knowledge of programming or
statistical modelization.
4.2 Intervention
As technical outcome of the DSR we conducted, a
prototype was developed and deployed in a research
lab for structural genomics at the University Medical
Centre Utrecht (UMCU) in the Netherlands.
The prototype started from the HCI-KDD process
by implementing interactive visualization capabilities
together with methods to pre-process and mine data
sets. Pre-processing consisted in normalization and
transformation of table of counts generated by RNA-
Seq technologies and tools. A table of counts has
samples of patients in columns and a list of genes as
rows (60 000 in the files used).
This table is the result of a bioinformatics
pipeline. Hence, analytical data is generated by
various levels of data processing from raw DNA
sequence quality checks to counting how many RNA
fragments found in a patient tissue overlap a gene.
Via the web interface, users start with these tables
Towards Reusability of Computational Experiments - Capturing and Sharing Research Objects from Knowledge Discovery Processes
459
in a virtual experiment (gathering data and contextual
information). Then a possibility is offered to
normalize or transform data sets by calling packages
from Bioconductor. Normalization is an important
pre-processing task to make samples comparable due
to the presence of (technical) biases in the raw data.
4.3 Evaluation
Exploratory focus groups with biologists and
bioinformaticians provided input for conducting
additional iterations, similar to an agile approach.
From requirements and discussions with specialists a
set of functionalities for KDD and visualization were
implemented. The facet of RR was imposed as it was
not a primary requirement from the field experts.
Hence, design choices for RR were inspired by
previously described literature about compendiums
and ROs.
Next, three confirmatory focus groups invited
bioinformaticians and biologists to discuss about the
prototype and judge the applicability of the KDD
steps implemented. We addressed results obtained
from the focus groups in section 3. These results are
further processed is section 4.4. We present a design
proposition which is an outcome of the evaluation of
the prototype. Furthermore, our design proposition
covers architectural choices which are mainly
grounded in the web architecture.
4.4 Reflection
The lessons learned from our DSR are described in
the RRO-KDD process. We processed the input of
three confirmatory focus groups with 15 participants.
We described the results earlier and elaborate on their
processing further in the next section.
4.5 RRO-KDD Process
In Figure 2, the RRO-KDD process is modeled with
its related “deliverables” in a so-called process-
deliverable diagram (PDD) (Weerd and
Brinkkemper, 2008). Here, the elements of the HCI-
KDD process are integrated with contextual and
technological outputs. These outputs are directed to
reusability of previous experiment code, data and
methods. Below, we shortly describe the steps and
deliverables:
1) Understand is an activity where sufficient
description of the data sets are provided. For instance,
information about instruments, sequencing platforms,
sample preparation. It builds a container for an
experiment which is denoted by virtual experiment.
Virtual experiments are uniquely identified
aggregation of resources and group data sets together
with context and methods.
2) Integrate, pre-process and data mining are the
steps elaborated by the HCI-KDD process.
Visualization is an activity that occurs in parallel to
KDD and enables to get insight of what happens at
each step. For instance, it helps the users to judge the
impact of pre-processing methods on the data set.
Activity Integrate results in data objects, and Pre-
process will normalize or transform these integrated
data sets into analytical data which are more easily
interpretable than raw data, e.g. from sequencing
instruments. Finally, data mining results find useful
patterns from data, according to Fayyad’s definition
(Fayyad et al., 1996). Visualization is here a subpart
of the whole HCI field of research as it was not
extensively investigated in this work.
3) Visualization has a deliverable called insight,
which informs researchers on patterns, scores or
relations in their data on an interactive manner.
Interactive plots were rendered with bokeh, a python
library for creating browser compatible
visualizations.
4) Access presents previous, interactively created
components of an experiment (like charts and new
data objects) as REST resources that might be
accessed without the user interface via RESTAPIs.
5) These resources, aggregated in a virtual
experiment can be semantically enriched for reuse as
ROs. This is made possible because each component
is uniquely identified and accessible via a
programming interface. As an example, a mining task
created by a biologist is reusable via a RO with its
unique identifier.
The code of the prototype is hosted on GitHub
under MIT license and is available here:
https://github.com/armell/RNASEqTool.
5 CONCLUSION
Results suggest that reproducibility cannot be
reduced to data and code sharing and that the field of
biomedical genetics suffers from a lack of software
solutions that are both satisfactory for
bioinformaticians and biologists who are mutually
engaged in CEs. There are overlapping data analytics
practices but also serious apprehensions from
bioinformaticians to offer such a type of application
to biologists if they exceed data visualization.
Despite these concerns, we found that there is gap
to fill both in terms of data analytics and reuse of
previous work.
KDIR 2015 - 7th International Conference on Knowledge Discovery and Information Retrieval
460
As we have seen biologists were more inclined to
ask more visualization capabilities whereas
bioinformaticians expect a solution where scripting or
custom data processing is allowed. Unique identifier
of resources and platform-independent information
exchange via REST enables this. Nevertheless, HCI
alone for biologists is not satisfactory as they want to
query data and compare the impact of different
methods. These comparisons require pre-processing
and mining.
Reusability of data, workflows or parts of
experiments seems to be more interesting for the two
types of end-users which evaluated the artifact than
reproducibility.
6 FUTURE WORK
The suggested RRO-KDD is still in a design
proposition phase that needs to be evaluated in other
settings and the interest in sharing Research Objects
must be assessed. For this assessment, the mining
tools have to be upgraded and provide more realistic
possibilities to exchange and reuse virtual
experiments and their components.
In addition, extending the RRO-KDD to
distributed systems will have similar problems
encountered in previous studies and known as
workflow decay. This issue still holds in the RRO-
KDD context which is built around web services and
URLs that may be inactive after some time.
Permanent Identifiers may moderate accessibility
issues but not the support of data objects or remote
implementations of analysis packages.
Recommendations to face these issues are an
integration with virtual environments or containers
(e.g. Docker), dynamic documents and proper data
management solutions. More research on integrating
virtual containers for reusability of computational
experiments for bioinformaticians and biologists is
needed. Dynamic documents generated by the tool
could also play a role for bioinformaticians to
understand what decisions were taken by biologists
processing data via a user-friendly interface.
These investigations should be made by
effectively combining HCI and KDD as suggested by
Holzinger. But the multiplicity of actors, analysis
tools and techniques remains a great challenge first
for reusability then for reproducibility.
Hence, reproducibility arguments in literature
should be replaced by better designs for reusability in
IT solutions, at least for enhancing collaboration
between bioinformatics and biologists. Reusability is
broader than reproducibility as it enables repurposing
of previous work and, in essence, reproducibility.
ACKNOWLEDGEMENTS
Our thanks go to Dr. Wigard Kloosterman (UMCU)
and his team for hosting us, providing any resource to
conduct our research and assisting at the demo
sessions.
REFERENCES
Bechhofer, S., Buchan, I., De Roure, D., Missier, P.,
Ainsworth, J., Bhagat, J., … Goble, C. (2013). Why
linked data is not enough for scientists. Future
Generation Computer Systems, 29(2), 599–611.
doi:10.1016/j.future.2011.08.004
Dalkir, K. (2005). Knowledge Management in Theory and
Practice. Knowledge Management (Vol. 4).
Dix, A. (2009). Human-Computer Interaction. In L. LIU &
M. T. ÖZSU (Eds.), Encyclopedia of Database Systems
SE - 192 (pp. 1327–1331). Springer US.
doi:10.1007/978-0-387-39940-9_192
Fayyad, U., Piatetsky-Shapiro, G., & Smyth, P. (1996).
Knowledge Discovery and Data Mining: Towards a
Unifying Framework. In Proc 2nd Int Conf on
Knowledge Discovery and Data Mining Portland OR
(pp. 82–88).
Gentleman, R., & Lang, D. (2007). Statistical analyses and
reproducible research. Journal of Computational and
…, 16(1), 1–23.
Hevner, A., & Chatterjee, S. (2010). Design research in
information systems. Springer New York.
Hislop, D. (2005). Knowledge management in
organizations: A critical introduction. Management
Learning (Vol. 36).
Holzinger, A. (2013). Human-Computer Interaction and
Knowledge Discovery (HCI-KDD): What is the benefit
of bringing those two fields to work together? In
Lecture Notes in Computer Science (including
subseries Lecture Notes in Artificial Intelligence and
Lecture Notes in Bioinformatics) (Vol. 8127 LNCS, pp.
319–328). doi:10.1007/978-3-642-40511-2_22
Holzinger, A., Dehmer, M., & Jurisica, I. (2014).
Knowledge Discovery and interactive Data Mining in
Bioinformatics - State-of-the-Art, future challenges and
research directions. BMC Bioinformatics, 15 Suppl
6(Suppl 6), I1. doi:10.1186/1471-2105-15-S6-I1
Knuth, D. E. (1984). Literate Programming. The Computer
Journal, 27(2), 97–111. doi:10.1093/comjnl/27.2.97
Laine, C., Goodman, S. N., Griswold, M. E., & Sox, H. C.
(2007). Reproducible Research : Moving toward
Research the Public Can Really Trust. Annals of
Internal Medicine, 146(6), 450–453. Retrieved from
http://annals.org/article.aspx?articleid=733696
Towards Reusability of Computational Experiments - Capturing and Sharing Research Objects from Knowledge Discovery Processes
461
McNutt, M. (2014). Journals unite for reproducibility.
Science, 346(6210), 679–679.
Peng, R. D. (2011). Reproducible research in computational
science. Science (New York, N.Y.), 334(6060), 1226–7.
doi:10.1126/science.1213847
Pérez, F., & Granger, B. E. (2007). IPython: A system for
interactive scientific computing. Computing in Science
and Engineering, 9, 21–29. doi: 10.1109/MCSE.
2007.53
Weerd, I. Van De, & Brinkkemper, S. (2008). Meta-
modeling for situational analysis and design methods.
Handbook of Research on Modern Systems Analysis
and Design Technologies and Applications, 38–58.
KDIR 2015 - 7th International Conference on Knowledge Discovery and Information Retrieval
462
