Template-based Affine Registration of Autistic Brain Images
Porawat Visutsak
Dept. of Computer and Information Science, Faculty of Applied Science,
King Mongkut’s University of Technology North Bangkok (KMUTNB), Bangkok, Thailand
Keywords: Autism, Image Registration, Medical Imaging.
Abstract: This paper presents a new method for the study of autistic brain image called “Template-based affine
registration”, based on the transformation of the grid-line from a source image to a target image. By using
the locations of grid of both source and target images as the control structure, together with a smart
transition of grid computed by bilinear and affine transformations. Besides, the new locations of grid of a
target image corresponding to a source image are the best-move of all feature points translated from a
source to a target. The template named after the point set extracted from source image, the simple idea is to
use the affine transformation for mapping the target point set to the template. The transformation process is
used effectively by using the incorporating transition of grid to maintain geometric alignment throughout
the process; the proposed method achieves a smooth transformation for image registration.
1 INTRODUCTION
Autism is a neurodevelopmental disorder
characterized by marked deficits in communication,
social interaction, and interests. Various studies of
autism have suggested abnormalities in several brain
regions, with an increasing agreement on the
abnormal anatomy of the white matter (WM) and the
unused brain cells, called gray matter (GM).
(Rachid, et al., 2007), (Fisher, 2011). The WM
connections between brain regions are important for
language and social skills. Normally, as children
grow into teenagers, in order to understand and
respond to the world, the brain undergoes 2 major
changes — the creation of new connections in WM,
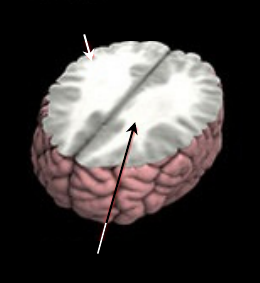
and the elimination, or “pruning,” of GM. Figure 1
shows the region of WM and GM. The brain-
imaging scan called a T1-weighted MRI (Magnetic
Resonance Imaging), which can map structural
changes during brain development (Fisher, 2011).
To study how the brains of autism changed over
time, the researchers captured the brain images of
children with autism before the treatment and they
did this again approximately three years later. By
doing this twice, the scientists were able to create a
detailed picture of how the brain changes. Thus, this
new knowledge may help to explain some of the
symptoms of autism and could improve future
treatment options later on. Unfortunately, only MRI
modality does not provide the brain activity analysis,
which is essential for understanding how the brain
works associated with the difficulties that many
autistic children have with — social impairment,
communication deficits and repetitive behaviour.
Figure 1: The connections between WM and GM.
A new methodology for analyzing fMRI scans
has been proposed by (Wei, et al., 2015). The
method called Brain-Wide Association Analysis
(BWAS), can analyze over 1 billion pieces of data
for creating panoramic views of the whole brain and
provides scientists with the 3D model to study the
brain connections. The ability to analyze the entire
data set from an fMRI scan provided the researchers
the opportunity to compile, compare and contrast
accurate imaging modalities for both autistic and
non-autistic brains. The major drawback of BWAS
Gray Matter
White Matter
188
Visutsak, P..
Template-based Affine Registration of Autistic Brain Images.
In Proceedings of the 7th International Joint Conference on Computational Intelligence (IJCCI 2015) - Volume 2: FCTA, pages 188-192
ISBN: 978-989-758-157-1
Copyright
c
2015 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
is that the computational cost of the big data analysis
should be concerned. A multi-modality of brain
imaging methodology has been introduced in
(Hughes, 2012). The autistic brain was scanned
using three different methods: high-resolution MRI,
which captures the structure of the brain; Diffusion
Tensor Imaging (DTI), a method to trace the
connections between brain regions; and functional
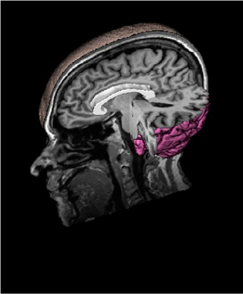
MRI, which indicates brain activity. Figure 2 shows
the vertical MRI scanning; the future work of
(Hughes, 2012) is how to combine the various types
of images into one common format.
Figure 2: Vertical MRI scanning.
2 PURPOSE OF PROPOSED
METHOD
This work aims to study the abnormalities in autistic
brain using image processing technique called Image
registration. Image registration is the process of
aligning the different sets of data of the same object
into a common format thus aligning them in order to
analyze subtle changes among each other. A
fundamental problem in medical image registration
is the integration of information from multiple
images of the same subject, acquired using the same
or different imaging modalities and possibly at
different time points (Maes, et al., 2013), i.e.,
recovering the geometric relationship between
corresponding points in multiple images of the same
scene.
As mentioned earlier in previous section; the
current research trends for analysis the autistic brain
image have been focused in 2 major fields:
1. To study the brain activities extraction using
fMRI modality, and
2. To study the brain active regions using DTI
modality.
The similarities between fMRI and DTI
modalities include the study of anomaly in WM and
deficits in the size of the corpus callosum (the white
crescent in figure 2.). (Lynn, et al., 2014) have been
supported the hypothesis that the disruption of the
corpus callosum constitutes a major risk factor for
developing autism, resulting in the difficulties that
many autistic people have with words and social
interaction. Unfortunately, to diagnose the
symptoms of autism, the doctor need to do it at least
twice; the first one concerns the observation of
fMRI, the second one the question of how to explain
the observed brain active region in DTI.
Therefore, the major contribution of this
proposed method is to integrate the fMRI and DTI
modalities into a common coordinate system, thus
the doctor can take the benefits of both fMRI and
DTI by observing the brain images at the same time.
This study proposes a new approach of registration
for Autistic brain images called Template-based
affine registration. The novelty of the method is that
the correlation of functional brain image data
obtained from different individuals can be achieved
by registration of the corresponding anatomical
brain images with a fixed template image (Visutsak,
2014). The brain image has been normalized to the
new coordinate system, such that after registration
process, functional measurements from different
individuals can be compared using the new
coordinates.
The term “Template” means the point set
extracted from source image (in this case; fMRI will
be chosen as source image and DTI will be chosen
as target image), the goal is to estimate the affine
transformation for source and target images using
two point sets extracted from these two images. By
performing the manual deformation to get source
and target image, the point sets of source and target
images (as well as the area of WM and corpus
callosum included in both images) will be extracted
respectively. In order to register two point sets of
images, two problems are needed to be solved
simultaneously, the first one is to estimate the
transformation between two point sets and the
second one is to concern with the mapped positions
of points using an appropriate transformation.
3 IMAGE REGISTRATION
The general term of image registration can be
defined as the evolution of source to target images;
this evolution refers to as what the proper mapping
function is used to spatially transform two images
Template-based Affine Registration of Autistic Brain Images
189

with respect to their intensities (Visutsak, 2014).
Given two images denoted by I
1
and I
2
, the mapping
between images can be expressed as:
I
2
(x,y)= g(I
1
(f(x,y)) (1)
Where, f() is the 2D spatial transformation
g() is the 1D intensity transformation
By assuming that the correspondences are
known, the goal of image registration is to find such
f() and g(), such that two images are best matched.
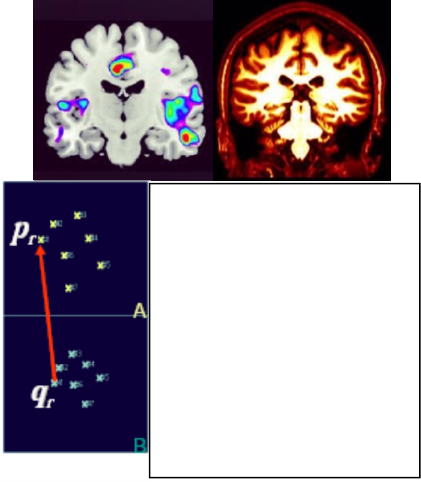
Figure 3 illustrates the concept of spatial
transformation that maps from arbitrary point P in
fMRI image to homologous point Q in DTI image.
Figure 3: Image registration finds a spatial transform
mapping one image into another.
Two images are involved when registration is
carried out. One image is taken as source image or
fixed image (fMRI), and the other as target image
(DTI). Registration is the determination of a one-to-
one mapping between the coordinates in fixed image
and those in target image, such that points in the two
images that correspond to the same anatomical point
are mapped to each other (Maurer, 1993). Referring
to equation 1, the simple task of image registration is
to establish correspondence between features in sets
of images, and using a transformation model to infer
correspondence away from those features (Crum,
2005).
Figure 4: 2D rigid transformation of skull radiographs.
Transformation represents the spatial mapping of
points in the floating image to points in the target
image (Porawat, 2014). In 2D to 2D image
transformation e.g. the transformation of fMRI to
DTI, 2 translations (up-down/left-right) and 1
rotation may be needed. Figure 4 shows the parallel
projection of skull radiographs, rigid 2D
transformation controlled by a rotation Ө and two
translation parameters T
1
and T
2
, respectively.
This is a Linear mapping from (x
1
, x
2
) to (y
1
, y
2
),
therefore,
y
1
= cosӨ.x
1
– sinӨ.x
2
+ T
1
y
2
= sinӨ.x
1
– cosӨ.x
2
+ T
2
(2)
Equation 2 can be derived into matrix form y =Ax,
such that,
y
1
= a
11
.x
1
+ a
12
.x
2
+ a
13
y
2
= a
21
.x
1
+ a
22
.x
2
+ a
23
(3)
This is so called the homogeneous coordinate
transformation of 2 images. In most cases of affine
transformations on images, the rotation around the
given location and the scaling with respect to a fixed
point are also needed to be considered, the
transformation function of these cases are
T(x,y).R(Ө).T(-x,-y),
T(x,y).S(S
x
,S
y
).T(-x,-y), respectively. (4)
There are many well-known techniques for
brain image registration, such as surface matching,
surface extraction, registration using external
landmark points set, and intensity-based registration.
Surface matching is a popular choice because of the
rigidity of the brain shape. Surface extraction is also
successful registration for the multi-modality of CT,
MR and PET brain images (Fitzgibbon et al., 2012).
External landmarks attached to the brain have also
been used to assist the registration (Wirth, et al.,
2002). Since the introduction of mutual into medical
image registration (Maes et al., 2013), intensity-
based registration methods have been widely used
(Pluim et al., 2003). The goal of these methods is to
choose the transformation types (e.g. rigid, non-
rigid, affine), and treat it as an optimization problem
with how to find the spatial mapping of images.
4 THE PROPOSED METHOD
The objectives of this study can be summarized as
to:
1. Purpose a new method of registration for
Autistic brain images called Template-based affine
registration.
FCTA 2015 - 7th International Conference on Fuzzy Computation Theory and Applications
190
2. Test the proposed method with brain images
data set, and compare the results with the well-
known registration methods.
The novel method of brain image registration has
been investigated. The method will be applied for
image analysis of autistic patients. The new method
involves integrating the images to create a composite
view, extracting information that would be
impossible to obtain from a single brain image.
The method will be started with manually
identify landmark points set (around 6-12 points) in
fMRI and DTI. There are two major concerns of this
selection: 1) the accuracy of the selection should be
1mm at center, and around 2 mm at the edge, 2) all
landmark points should be related to the soft tissue
structures in GM and WM such as enhancing the
brain development after taking some treatments.
Figure 5 illustrates the proposed method.
Figure 5: Template-Based affine registration.
Supposing that we have two lists of landmark
points of two images:
A = [p
1
, p
2
…p
N
] and
B = [q
1
, q
2
…q
N
]
It is the optimization problem of solving the
transformation T(q) that minimizes squared distance
between corresponding points in A and B.
E is the extrapolation function for all image
pixels:
E = ∑
r
|| p
r
– T(q
r
) ||
2
(5)
Where one set of points, q, is transformed by T().
5 CONCLUSIONS
This position paper presents a very simple but
important matter in image registration. The expected
result of this study is useful to register between two
multimodal brain images of autism (fMRI and DTI).
The expected benefit is the new method of
registration for autistic brain images which has many
potential applications in clinical diagnosis.
ACKNOWLEDGEMENTS
“This research was funded by the King Mongkut’s
University of Technology North Bangkok. Contract
no. KMUTNB-GOV-59-xx”.
REFERENCES
Cheng, Wei, et al. “Autism: Reduced Connectivity
between Cortical Areas Involved with Face
Expression, Theory of Mind, and the Sense of Self.”
Brain, A Journal of Neurology, Oxford University
Press, 2015.
Christopher Fisher, “Autistic Brains Develop More Slowly
Than Healthy Brains.” BMED Report, October 28,
2011.
Crum, William R., et al. “Generalised overlap measures
for assessment of pairwise and groupwise image
registration and segmentation.” Medical Image
Computing and Computer-Assisted Intervention–
MICCAI 2005. Springer Berlin Heidelberg, 2005. 99-
106.
A. Fitzgibbon et al. (Eds.): ECCV 2012, Part II, LNCS
7573, pp. 30–44, 2012.
Fahmi, Rachid, et al. “Structural MRI-Based
Discrimination between Autistic and Typically
Developing Brain.” Proc. of Computer Assisted
Radiology and Surgery (CARS’07), Berlin, Germany
(2007): 24-26.
Frederik Maes, Dirk Vandermeulen, and Paul Suetens,
“Medical Image Registration using Mutual
Information,” In Proc. of the IEEE, Vol. 91, No. 10,
Oct. 2003, pp. 1699-1722.
Maurer, Calvin R., and J. Michael Fitzpatrick. “A review
of medical image registration.” Interactive image-
guided neurosurgery 17 (1993).
Paul, Lynn K., et al. "Agenesis of the corpus callosum and
autism: a comprehensive comparison." Brain 137.6
(2014): 1813-1829.
J. P. W. Pluim, J. B. A. Maintz, and M. A. Viergever.
Mutual information based registration of medical
images: a survey. IEEE Trans. Medical Imaging,
22(8):986{1004, 2003.
Porawat Visutsak, "Grid Transformation for Image
Registration and Morphing", The 6th International
The Algorithm:
1. Assuming that A and B are
two lists of corresponding
feature locations:
[p
1
, p
2
…p
N
] and
[q
1
, q
2
…q
N
]
2. Find:
Transformation T(q) that
minimizes squared distance
between corresponding points:
E = ∑
r
|| p
r
– T(q
r
) ||
2
1
X
1
X
2
X
2
X
3
X
3
X
Template-based Affine Registration of Autistic Brain Images
191
Conference on Science, Technology and Innovation
for Sustainable Well-Being (STISWB VI), 28-30
August 2014, Apsara Angkor Resort & Conference
Siem Reap, Kingdom of Cambodia.
Porawat Visutsak, “Multi-Grid Transformation for
Medical Image Registration”, 2014 International
Conference on Advanced Computer Science and
Information Systems, pages 52-56, 18-19 Oct. 2014,
Ambhara Hotel, Blok M, Jakarta, Indonesia.
Virginia Hughes, “Researchers reveals first brain study of
Temple Grandin.” The 2012 Society for Neuroscience
annual meeting, Simons Foundation Autism Research
Initiative (SFARI), October 14, 2012.
M. A. Wirth, J. Narhan, and D. Gray. Nonrigid
mammogram registration using mutual information. In
SPIE: Medical Imaging: Image Processing, volume
4684, San Diego, CA, Feb. 2002.
FCTA 2015 - 7th International Conference on Fuzzy Computation Theory and Applications
192
