Clustering using Cellular Genetic Algorithms
Nuno Leite
1,3
, Fernando Mel
´
ıcio
3
and Agostinho C. Rosa
2,3
1
Instituto Superior de Engenharia de Lisboa/ADEETC, Polytechnic Institute of Lisbon, Rua Conselheiro Em
´
ıdio Navarro,
n.° 1, 1959-007, Lisboa, Portugal
2
Department of Bioengineering/Instituto Superior T
´
ecnico, Universidade de Lisboa, Av. Rovisco Pais, n.° 1, 1049-001,
Lisboa, Portugal
3
Institute for Systems and Robotics/LaSEEB, Instituto Superior T
´
ecnico, Universidade de Lisboa, Av. Rovisco Pais, n.° 1,
TN 6.21, 1049-001, Lisboa, Portugal
Keywords:
Cellular Genetic Algorithms, Clustering, Classification, Evolutionary Computation, Nature Inspired
Algorithms.
Abstract:
The goal of the clustering process is to find groups of similar patterns in multidimensional data. In this work,
the clustering problem is approached using cellular genetic algorithms. The population structure adopted in
the cellular genetic algorithm contributes to the population genetic diversity preventing the premature con-
vergence to local optima. The performance of the proposed algorithm is evaluated on 13 test databases. An
extension to the basic algorithm was also investigated to handle instances containing non-linearly separable
data. The algorithm is compared with nine non-evolutionary classification techniques from the literature, and
also compared with three nature inspired methodologies, namely Particle Swarm Optimization, Artificial Bee
Colony, and the Firefly Algorithm. The cellular genetic algorithm attains the best result on a test database. A
statistical ranking of the compared methods was made, and the proposed algorithm is ranked fifth overall.
1 INTRODUCTION
Clustering is an important unsupervised learning ap-
proach used in applications in areas such as data min-
ing, statistical data analysis, data compression, or vec-
tor quantization. The task of clustering is to discover
natural groupings of a set of patterns, points, or ob-
jects (Jain, 2010).
Clustering approaches are generally categorized
into two classes (Jain, 2010): hierarchical and par-
titional. In hierarchical clustering, nested clusters are
found recursively according to two modes, agglomer-
ative and divisive. In the agglomerative mode, the
clustering process starts with each data point in its
own cluster and then proceeds merging the most sim-
ilar pair of clusters successively to form a cluster hi-
erarchy. In the divisive (top-down) mode, the clus-
tering starts with all the data points in one cluster
and then proceeds by recursively dividing each cluster
into smaller clusters. Partitional clustering algorithms
work differently compared to hierarchical clustering
algorithms as they find all the clusters at the same
time as a partition of the data without imposing a hi-
erarchical structure. The prototype-based clustering
algorithms are the most popular partitional clustering
algorithms. In these, each cluster is represented by
its center and the objective function used is the sum
of squared distances of the data points to the cluster
centers they are assigned to (Mirkin, 1996; Borgelt,
2006). An example of a partitional clustering algo-
rithm is the popular k–means algorithm. Although
still widely used, the main drawback of the k–means
algorithm is that it converges to a local minima from
the starting position of the search (Selim and Ismail,
1984).
The clustering process can be done through two
ways: by unsupervised learning or by supervised
learning. In unsupervised clustering, also called au-
tomatic clustering, there’s no information about the
classes of the patterns, and the clustering is done by
finding the natural aggregations in the dataset. On
the other way, in supervised clustering, also known
as classification, a classifier is learned given a train-
ing set with known pattern classes. After the training
phase, a test set is classified based on the learned clas-
sifier. The learned classifier contains information of
the optimal cluster centers that characterize the parti-
tions in the training set. These cluster centers are used
to classify the patterns in the test set.
In the recent years, nature inspired algorithms
366
Leite, N., Melício, F. and Rosa, A..
Clustering using Cellular Genetic Algorithms.
In Proceedings of the 7th International Joint Conference on Computational Intelligence (IJCCI 2015) - Volume 1: ECTA, pages 366-373
ISBN: 978-989-758-157-1
Copyright
c
2015 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

have been proposed to solve clustering and classi-
fication problems. Examples include Genetic Al-
gorithms (Falkenauer, 1998; Maulik et al., 2000),
Memetic Algorithms (Ni et al., 2013), Artificial Bee
Colony (ABC) (Karaboga and Ozturk, 2011), Parti-
cle Swarm Optimization (PSO) (Falco et al., 2007),
Firefly Algorithm (FA) (Senthilnath et al., 2011), and
Cuckoo Search Algorithm (Zhao et al., 2014). Re-
cently, hybrid evolutionary algorithms which com-
bine evolutionary algorithms with the k–means algo-
rithm were also proposed (Niknam and Amiri, 2010).
In the study presented in this work, a novel ap-
proach of cellular genetic algorithms (cGA) (Alba
and Dorronsoro, 2008) is proposed for solving the
clustering problem. The cGA is based on the par-
allel cellular model which maintains a greater pop-
ulation diversity compared to the standard evolution-
ary algorithm, thus contributing to find solutions with
better quality (Alba and Dorronsoro, 2008). To the
extent of our knowledge, cellular evolutionary algo-
rithms have not yet been applied successfully to the
clustering problem. The cGA is applied to classifica-
tion benchmark problems on 13 typical test databases.
The cGA is compared with nine non-evolutionary
classification techniques from the literature, and also
compared with three nature inspired methodologies,
namely PSO (Falco et al., 2007), ABC (Karaboga and
Ozturk, 2011), and the FA (Senthilnath et al., 2011).
The remaining of the paper is organized as fol-
lows. In Section 2, the clustering problem is formally
defined. Section 3 presents the canonical cellular evo-
lutionary algorithm. In Section 4, the adaptation of
the cGA for classification is described. Section 5 re-
ports the experimental results. Some conclusions are
drawn in Section 6.
2 CLUSTERING AND
CLASSIFICATION PROBLEMS
Given a set of multidimensional data, the clustering
process consists in finding groups of patterns, or clus-
ters, in the data whose members are more similar to
each other than they are to other patterns (Duda et al.,
2000). The clustering is done based on some simi-
larity measure (Jain et al., 1999), where the measure
based on the distance is generally used.
The clustering problem can be stated as an opti-
mization problem, as in (Marinakis et al., 2008). In
order to formulate the clustering problem the follow-
ing symbols were defined:
• K, is the number of clusters.
• N, is the number of objects.
• x
i
∈ R
n
,(i = 1, . . . , N) is the location of the ith
pattern.
• z
j
∈ R
n
,( j = 1, . . . , K) is the center of the jth clus-
ter, computed as:
z
j
=
1
N
j
∑
x
i
∈C
j
x
i
(1)
where N
j
is the number of objects in the jth cluster
denoted by C
j
.
The optimization problem is formulated as:
Minimise J(w, z) =
N
∑
i=1
K
∑
j=1
w
i j
k x
i
− z
j
k
2
(2)
Subject to
K
∑
j=1
w
i j
= 1, i = 1,. . . , N (3)
w
i j
= 0 or 1, i = 1,. . . , N, j = 1,. . . , K. (4)
In Eq. (1), each z
j
cluster center is computed as the
centroid of the region formed by the cluster objects.
The goal of the clustering problem, as given in Eq. (2),
is to find the clusters centers z
j
, such that the sum of
the squared Euclidean distances between each pattern
and the center of its cluster is minimized.
As mentioned in the introduction, the related
problem known as classification is solved in this
work. To assess the quality of the classification, the
performance measure – classification error percentage
(CEP) – was used:
CEP = 100 ·
Number of misclassified instances
Total size of test set
.
(5)
3 CELLULAR GENETIC
ALGORITHMS
The cellular evolutionary algorithm (cGA) used to
solve the Clustering problem relies on the parallel
cellular model. In this model, the populations are
organized in a special structure defined as a con-
nected graph, in which each vertex is a solution that
communicates with its neighbours (Alba and Dor-
ronsoro, 2008). More specifically, individuals are set
in a toroidal mesh and are only allowed to recombine
with the close neighbours (Figure 1). The population
structure used in cellular evolutionary algorithms con-
tributes to the population genetic diversity thus avoid-
ing the premature convergence to a local optimum.
Algorithm 1 depicts the steps of the general cel-
lular genetic algorithm. First, the initial population is
Clustering using Cellular Genetic Algorithms
367
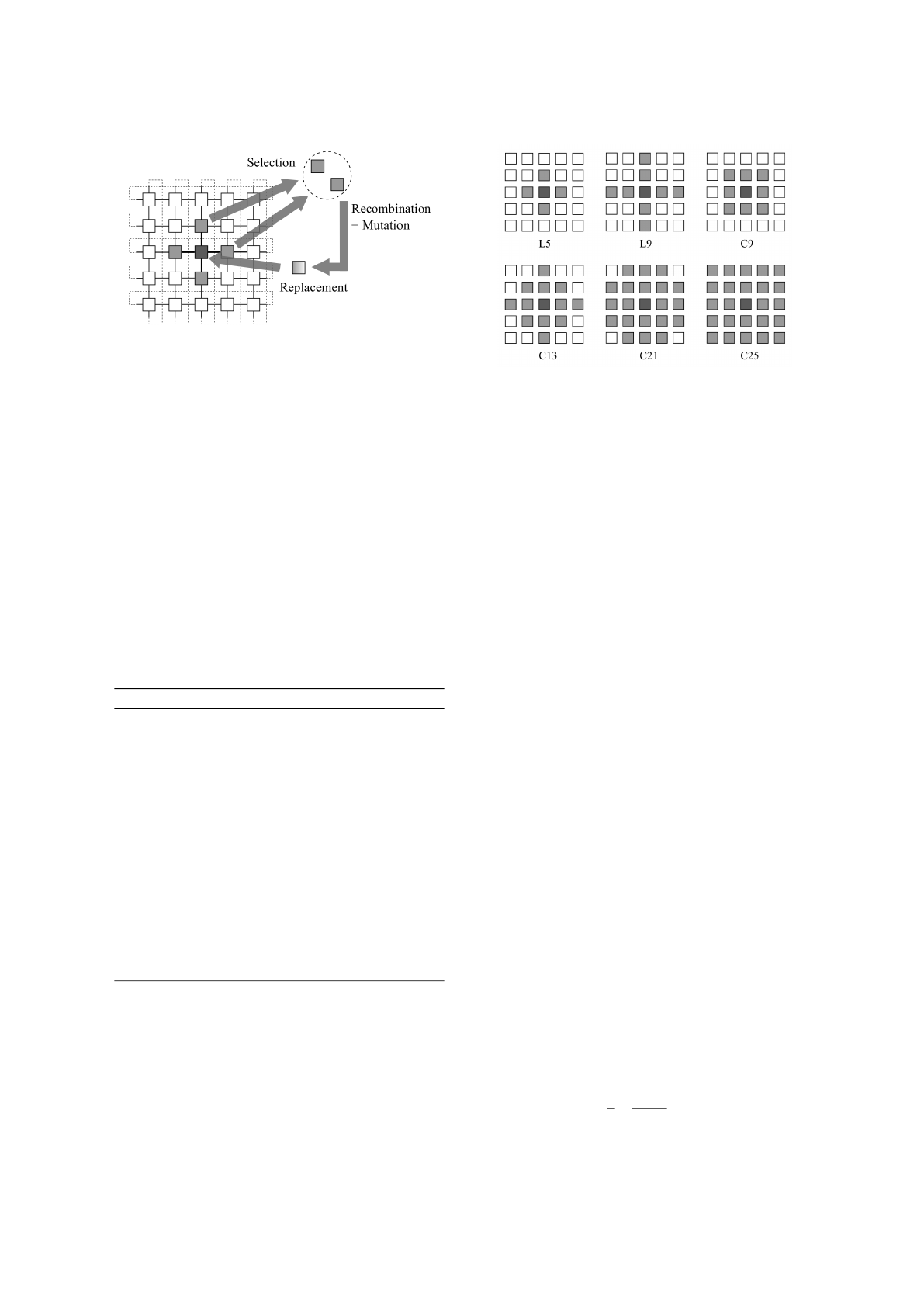
Figure 1: The parallel cellular model for evolutionary algo-
rithms.
generated and evaluated. Then, while the stop condi-
tion is not achieved (e.g., maximum number of itera-
tions is achieved, fitness value of the best individual
is within a desired interval), the following steps are
executed sequentially for each population’s individ-
ual: (1) obtain the individual’s neighbours according
to a specified neighbourhood, (2) select individuals
to recombine from the neighbours’ set, (3) recombine
(crossover and mutation) the selected offspring, and
(4) put the best offspring in an auxiliary population.
After doing steps (1) to (4) for all the elements of
the population, the auxiliary population replaces the
original population. In this version of the cGA, all the
individuals are updated at the same time, so the the
algorithm is called synchronous cGA. Figure 2 illus-
trates six typical neighbourhoods used in the cGA.
Algorithm 1: Pseudo-code of the canonical cGA.
1: procedure CGA(cga) // Parameters in ‘cga’
2: GenerateInitialPopulation(cga.pop);
3: Evaluation(cga.pop);
4: while !StopCondition() do
5: for indiv ← 1 to cga.popSize do
6: neighs ← GetNeighs(cga, pos(indiv));
7: par ← Selection(neighs);
8: offs ← Recombination(cga.P
c
, par);
9: offs ← Mutation(cga.P
m
, offs);
10: Evaluation(offs);
11: Replace(pos(indiv), offs, auxPop);
12: end for
13: cga.pop ← auxPop;
14: end while
15: end procedure
In the implemented approach, it was used the L5
neighbourhood type, also known as the von Neu-
mann neighbourhood. This neighbourhood promotes
a smother actualisation of the populations compared
with the other neighbourhoods. In the selection pro-
cedure (Figure 1), one of the four neighbours of the
central individual is selected with a binary tourna-
Figure 2: Typical neighbourhoods used in cellular evolu-
tionary algorithms.
ment. The chosen neighbour and the middle individ-
ual are then recombined producing two offspring so-
lutions. A mutation operator is then applied to these
two solutions and the best offspring is selected. The
offspring solution is compared with the original in-
dividual, and, if it is better, it will replace the origi-
nal solution. Otherwise, the original solution is main-
tained.
4 CGA APPLIED TO
CLASSIFICATION
In this section, the cGA adaptation to tackle classifi-
cation problems is described.
4.1 Encoding
Given an instance with C classes and N attributes, the
classification problem considered consists in finding
the optimal positions of the C centroids in the N-
dimensional space. The i-th individual of the popu-
lation is encoded as follows:
(~p
1
i
,...,~p
C
i
) (6)
where the j-th centroid position is formed by N
real numbers representing the coordinates in the N-
dimensional space (Falco et al., 2007):
~p
j
i
= {p
j
1,i
,..., p
j
N,i
}. (7)
So, each population individual is composed by C ∗ N
components, each represented by a real number.
4.2 Fitness Function
Each solution i is evaluated using the weighted fitness
function ψ
3
(i) from (Falco et al., 2007):
ψ
3
(i) =
1
2
ψ
1
(i)
100.0
+ ψ
2
(i)
, (8)
ECTA 2015 - 7th International Conference on Evolutionary Computation Theory and Applications
368

where ψ
1
(i) is given by
ψ
1
(i) =
100.0
D
Train
D
Train
∑
j=1
δ(~x
j
) (9)
δ(~x
j
) =
1 if CL(~x
j
) 6= CL
known
(~x
j
),
0 otherwise.
(10)
and ψ
2
(i) is given by
ψ
2
(i) =
1
D
Train
D
Train
∑
j=1
d
~x
j
,~p
CL
known
(~x
j
)
i
. (11)
As described in (Falco et al., 2007), the function ψ
1
(i)
is computed in two steps. In the first step, the train-
ing set patterns are assigned to the class of the closest
centroid in the N-dimensional space. Then, the fitness
of the i-th individual is computed as the percentage
of incorrectly assigned instances on the training set,
i.e., counts the number of patterns ~x
j
for which the
assigned class CL(~x
j
) is different from the true class
CL
known
(~x
j
). The function ψ
2
(i) is computed in one
step as the sum of the Euclidean distances between
each pattern ~x
j
in the training set and the centroid of
the true cluster (~p
CL
known
(~x
j
)
i
).
When computing the distance, the components of
the i-th individual are normalized using the Min-Max
Normalization.
4.3 Population Initialization
The components of each individual in the population
are initialized with random numbers in the range [0,1]
generated with uniform distribution. Notice that the
components could have been initialized with any ran-
dom numbers, or even with the components of ran-
dom chosen patterns. Empirical tests were undertaken
with different ranges ([0, 1], [0,5], [0, 50], and com-
ponents of random patterns). The results showed that
there are not significant differences between the meth-
ods.
4.4 Crossover
Given two parent solutions, the crossover recombina-
tion process exchanges, in a probabilistic fashion, the
information between the parent chromosomes pro-
ducing two offspring chromosomes. In this work,
single-point crossover with crossover probability P
c
was used.
4.5 Mutation
The mutation operator used was based on that pub-
lished in (Maulik et al., 2000). For a fixed probability
Table 1: Properties of the examined databases.
N D C miss
Balance 4 625 3 No
Credit 15 690 2 Yes
Dermatology 34 366 6 Yes
Diabetes 8 768 2 No
E.Coli 7 327 5 No
Glass 9 214 6 No
Heart 13 303 2 Yes
Horse Colic 27 368 2 Yes
Iris 4 150 3 No
Thyroid 5 215 3 No
WDBCancer 30 569 2 No
WDBCancer-Int 9 699 2 Yes
Wine 13 178 3 No
P
m
, a gene is selected randomly. A number δ in the
range [−1, 1] is generated with uniform distribution.
If the value at a gene position is v, after mutation it
becomes
v =
v · (1 + 2 · δ), v 6= 0,
v + 2 · δ, v = 0.
(12)
With this mutation, the value of the selected gene, v,
is increased or decreased in small random amounts.
4.6 Handling Non-linearly Separable
Problems
As pointed out by De Falco et al. (Falco et al., 2007)
in their work, the use of centroids to approach the
classification problem is not adequate in some situ-
ations. The authors list three cases in which an algo-
rithm based on the concept of centroids and distance
to separate the samples into the correct classes is very
difficult or even impossible. The first one is when in-
stances of a class are located in two distinct regions
of the N-dimensional space, and these instances are
separated by instances of a second class. The second
situation occurs when a region comprising instances
of a class are completely surrounded by a region con-
taining instances belonging to another class. The third
one is related to the existence of confusion areas in the
search space, i.e., parts of the search space containing
examples belonging to two or more classes. The first
two cases could be solved by considering two or more
centroids per class. The third case cannot be solved
effectively with the centroid approach.
To solve the first two cases mentioned in the pre-
vious paragraph, the cGA chromosome was extended
by considering K clusters per class (K ≥ 1).
Clustering using Cellular Genetic Algorithms
369

5 EXPERIMENTS AND RESULTS
5.1 Settings
The evaluation of the proposed cGA was carried out
on 13 well known databases taken from the UCI Ma-
chine Learning Repository (Lichman, 2013). The
characteristics of the 13 examined UCI databases are
depicted in Table 1. For each database it is indicated
the number N of attributes composing each instance,
the total instance number D, the number of classes
C into which is divided, and a flag miss indicating
if there is missing data in the database. More in-
formation about the test databases can be obtained
from (Falco et al., 2007).
The algorithm was programmed in the C++ lan-
guage and was based on the ParadisEO frame-
work (Cahon et al., 2004). The experiments were
made on an Intel Core i7-2630QM (CPU @ 2.00 GHz
with 8 GB RAM) PC running Ubuntu 14.04 LTS –
64 bit OS. The Linux kernel used was: 3.13.0-49-
generic. The compiler used was: GCC v. 4.8.2.
In this section, all the statistical tests were per-
formed with a 95% confidence level. The popula-
tion size was set to 100 individuals distributed on a
5 × 20 rectangular grid. The neighbourhood used was
the von Neumann neighbourhood. The crossover and
mutation probabilities, respectively, P
c
and P
m
, were
set equal to 0.6 and 0.1. The number of generations
of the cGA was set to 1000.
The parameter values were chosen empirically
taking into account a reasonable balance of obtained
solution quality versus total time taken. The cGA
was executed 20 times on each dataset. The execu-
tion time range from few seconds to about a minute
depending on the dataset size. These times are within
the times published by (Falco et al., 2007).
The source code and the datasets used are
publicly available on the following Git reposi-
tory: https://github.com/nunocsleite/cellularGA-
clustering.
The cGA results were compared with re-
sults produced by nine classical classification tech-
niques, and with Particle Swarm Optimization
(PSO) (Falco et al., 2007), Artificial Bee Colony
(ABC) (Karaboga and Ozturk, 2011), and Firefly Al-
gorithm (FA) (Senthilnath et al., 2011). In order to
perform the classification, the datasets were divided
into 75% for training and 25% for testing. The sam-
ples were selected randomly, and divided in a bal-
anced way, in order to guarantee that the percentage of
samples in each class for the testing and training par-
titions was approximately the same. The same trans-
formations to the tested databases as in (Falco et al.,
2007) were carried out (transformation on attributes
with missing data, data shuffling, and E.Coli database
class removal).
5.2 Comparison with Other
Classification Techniques
Table 2 compares the cGA against three nature based
metaheuristics. Table 3 shows the results of the cGA
on the tested datasets and comparison with the nine
classification methods from literature as published
in (Falco et al., 2007). The tables present the aver-
age percentages of incorrect classification on the test-
ing set for each algorithm. For every method except
for ABC, the number of runs reported is 20. For the
ABC, the authors report five runs. From the registered
results it can be observed that the cGA is competitive
with the other techniques, attaining the best value on
the Diabetes dataset.
To assess the effectiveness of the analysed tech-
niques, a statistical ranking was performed (Table 4).
The results of the statistical tests and analysis were
produced using the Java tool of (Garc
´
ıa and Herrera,
2008). The cGA is positioned in the fifth position.
Relating the metaheuristics, the cGA is superior to
the PSO approach. Relating the other classification
methods, only the Bayesian network (Bayes Net) and
the MultiLayer Perceptron Artificial Neural Network
(MLP ANN) are superior to the cGA.
Table 2: Average percentages of incorrect classification on
the testing set. For each test database, the best value ob-
tained among all the techniques is presented in bold.
cGA PSO ABC FA
Balance 12.02 13.12 15.38 14.10
Credit 13.34 18.77 13.37 12.79
Dermatology 4.95 6.08 5.43 5.43
Diabetes 21.61 21.77 22.39 21.88
E.Coli 14.32 13.90 13.41 8.54
Glass 37.36 38.67 41.50 37.74
Heart 18.60 15.73 14.47 13.16
Horse colic 33.91 35.16 38.26 32.97
Iris 1.62 5.26 0.00 0.00
Thyroid 3.49 3.88 3.77 0.00
WDBCancer 4.26 3.49 2.81 1.06
WDBCancer-Int 1.61 2.64 0.00 0.00
Wine 2.84 2.88 0.00 0.00
Figure 3 ilustrates the evolution of the train and
classification error as a function of the number of it-
erations for the Balance dataset. Observing Figure 3
it can be concluded that for the Balance dataset the
algorithm steadily improves the training and testing
ECTA 2015 - 7th International Conference on Evolutionary Computation Theory and Applications
370
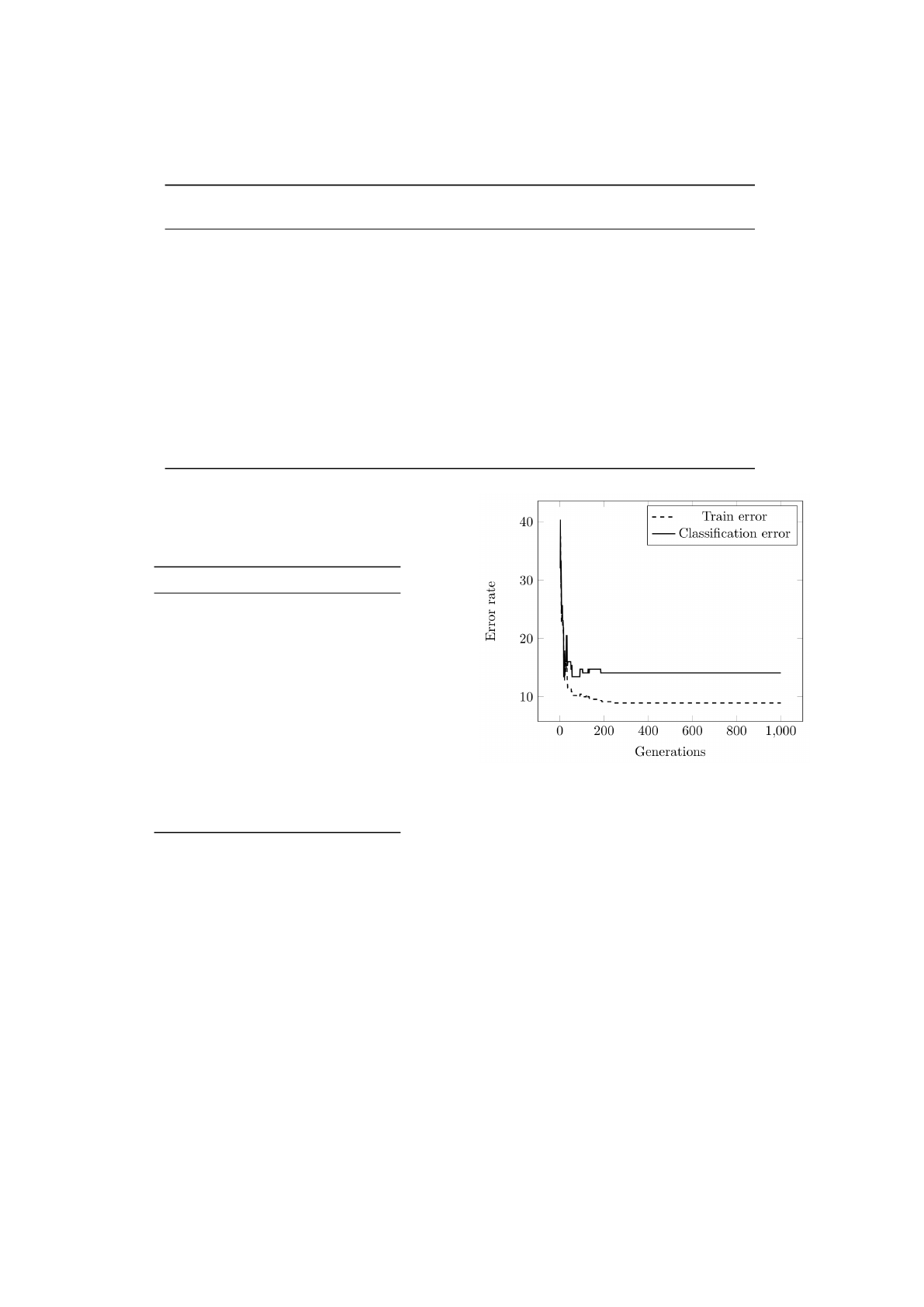
Table 3: Average percentages of incorrect classification on the testing set. For each test database, the best value obtained
among all the techniques is presented in bold.
cGA Bayes MLP RBF KStar Bagging Multi NBTree Ridor VFI
Net ANN Boost
Balance 12.02 19.74 9.29 33.61 10.25 14.77 24.20 19.74 20.63 38.85
Credit 13.34 12.13 13.81 43.29 19.18 10.68 12.71 16.18 12.65 16.47
Dermatology 4.95 1.08 3.26 34.66 4.66 3.47 53.26 1.08 7.92 7.60
Diabetes 21.61 25.52 29.16 39.16 34.05 26.87 27.08 25.52 29.31 34.37
E.Coli 14.32 17.07 13.53 24.38 18.29 15.36 31.70 20.73 17.07 17.07
Glass 37.36 29.62 28.51 44.44 17.58 25.36 53.70 24.07 31.66 41.11
Heart 18.60 18.42 19.46 45.25 26.70 20.25 18.42 22.36 22.89 18.42
Horse colic 33.91 30.76 32.19 38.46 35.71 30.32 38.46 31.86 31.86 41.75
Iris 1.62 2.63 0.00 9.99 0.52 0.26 2.63 2.63 0.52 0.00
Thyroid 3.49 6.66 1.85 5.55 13.32 14.62 7.40 11.11 8.51 11.11
WDBCancer 4.26 4.19 2.93 20.27 2.44 4.47 5.59 7.69 6.36 7.34
WDBCancer-Int 1.61 3.42 5.25 8.17 4.57 3.93 5.14 5.71 5.48 5.71
Wine 2.84 0.00 1.33 2.88 3.99 2.66 17.77 2.22 5.10 5.77
Table 4: Average Rankings of the algorithms (higher rank-
ing values are better). The Friedman test on these results
has a p-value of 2.002 × 10
−8
0.05 making these results
statistically significant.
Algorithm Average ranking value
FA 10.58
MLP ANN 9.19
ABC 9.12
Bayes Net 8.81
cGA 8.62
Bagging 8.23
PSO 7.50
KStar 6.35
NBTree 6.31
Ridor 5.62
MultiBoost 4.27
VFI 4.12
RBF 2.31
error in the first 30 generations. Then, it suffers from
a slight overfitting effect as the train error continues
to decrease and the test error increases.
5.3 Train with Cross-validation
To assess if the results of using a simple split were bi-
ased or not, the classifier was trained using the cross-
validation technique. The instances composing each
dataset were split in the following manner: 75% of
the instances for training the classifier, and 25% for
testing the learned classifier. The training set was fur-
ther divided into a training partition (84%), and a de-
velopment partition (16%). The results for the 13 UCI
Figure 3: Evolution of the train and classification error as a
function of the number of iterations for the Balance dataset.
datasets are presented in Table 5. The cross-validation
results are slightly different from those with simple
split but the deviation is not significant except for the
Balance and Glass datasets.
5.4 Non-linearly Separable Dataset
In this section, an experiment with a non-linearly
separable synthetic dataset was made. This dataset
(called ‘rings’ (Lourenc¸o et al., 2015)) has N = 450
samples classified into C = 3 classes (Figure 4). There
are 200 samples of class 1, 200 samples of class 2, and
the remaining 50 samples belong to class 3.
In order classify this dataset a variable number of
clusters per class, instead of a single one, was used.
Figures 5, 6, and 7, illustrate the use of one, three,
and four clusters per class, respectively. As can be ob-
Clustering using Cellular Genetic Algorithms
371
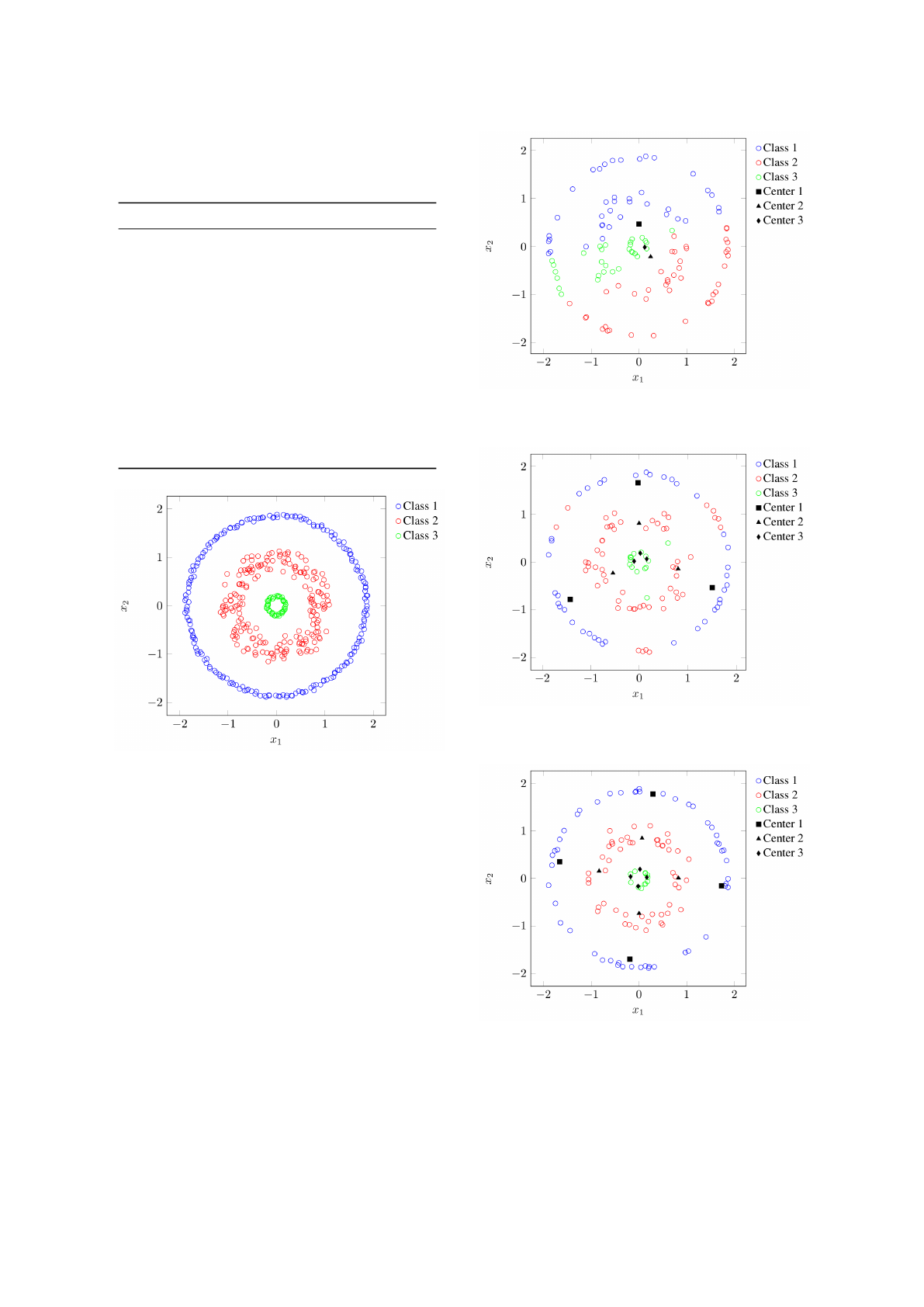
Table 5: Execution of cGA with simple split and using split
into three partitions (train, validation, and test partitions)
with cross-validation (CV) on the validation partition. 20
runs were executed.
Dataset Simple split Split with CV
Balance 12.02 15.77
Credit 13.34 12.62
Dermatology 4.95 5.60
Diabetes 21.61 21.41
E.Coli 14.32 13.09
Glass 37.36 33.68
Heart 18.60 21.93
Horse colic 33.91 32.88
Iris 1.62 1.35
Thyroid 3.49 3.58
WDBCancer 4.26 3.56
WDBCancer-Int 1.61 3.74
Wine 2.84 3.30
Figure 4: Distribution of samples into classes for the syn-
thetic ‘rings’ dataset.
served, the classification error is significantly reduced
with the addition of more clusters.
Although the addition of more clusters could solve
this specific clustering problem, it couldn’t improve
the classification accuracy on the UCI datasets. This
indicate that the UCI datasets probably belong to the
third type mentioned in Section 4.6, where the exam-
ples of each class are mixed into confusion regions,
that are difficult to tackle by a centroid based algo-
rithm.
6 CONCLUSIONS
In this work, a Cellular Genetic Algorithm has been
used to tackle the problem of classification of pat-
terns into clusters. The algorithm was tested on 13
Figure 5: Classification of the ‘rings’ dataset using one clus-
ter per class. The obtained classification error is equal to
54.4643% (61 misclassified examples out of 112).
Figure 6: Classification of the ‘rings’ dataset using three
clusters per class. The obtained classification error is equal
to 11.6071% (13 misclassified examples out of 112).
Figure 7: Classification of the ‘rings’ dataset using four
clusters per class. The obtained classification error is equal
to 0%.
ECTA 2015 - 7th International Conference on Evolutionary Computation Theory and Applications
372
diverse benchmarks and compared with nine classifi-
cation techniques and three metaheuristics.
Experiments show that the cGA is competitive
with the chosen special purpose classification tech-
niques and also with the general purpose metaheuris-
tics, attaining the best result on one problem instance.
The cGA was ranked in fifth place.
Regarding future developments, two points were
considered. The first is test other crossover and mu-
tation operators, and verify if the hybridization of the
cGA with local search metaheuristics is able to im-
prove the basic cGA. The second point aims at per-
forming a more rigorous study of the population grid
layout, size, and neighbours used.
ACKNOWLEDGEMENTS
This work was partially supported by national funds
through Fundac¸
˜
ao para a Ci
ˆ
encia e a Tecnologia
(FCT) under project [UID/EEA/50009/2013], and by
the PROTEC Program funds under the research grant
[SFRH/PROTEC/67953/2010].
REFERENCES
Alba, E. and Dorronsoro, B. (2008). Cellular Genetic Algo-
rithms. Springer Publishing Company, Incorporated,
1st edition.
Borgelt, C. (2006). Prototype-based classification and clus-
tering. PhD thesis, Otto-von-Guericke-Universit
¨
at
Magdeburg, Universit
¨
atsbibliothek.
Cahon, S., Melab, N., and Talbi, E.-G. (2004). Par-
adiseo: A framework for the reusable design of paral-
lel and distributed metaheuristics. Journal of Heuris-
tics, 10(3):357–380.
Duda, R. O., Hart, P. E., and Stork, D. G. (2000). Pattern
Classification (2Nd Edition). Wiley-Interscience.
Falco, I. D., Cioppa, A. D., and Tarantino, E. (2007). Facing
classification problems with particle swarm optimiza-
tion. Appl. Soft Comput., 7(3):652–658.
Falkenauer, E. (1998). Genetic Algorithms and Grouping
Problems. John Wiley & Sons, Inc., New York, NY,
USA.
Garc
´
ıa, S. and Herrera, F. (2008). An extension on “sta-
tistical comparisons of classifiers over multiple data
sets” for all pairwise comparisons. Journal of Ma-
chine Learning Research, 9:2677–2694.
Jain, A. K. (2010). Data clustering: 50 years beyond k-
means. Pattern Recognition Letters, 31(8):651 – 666.
Award winning papers from the 19th International
Conference on Pattern Recognition (ICPR)19th Inter-
national Conference in Pattern Recognition (ICPR).
Jain, A. K., Murty, M. N., and Flynn, P. J. (1999). Data
clustering: A review. ACM Comput. Surv., 31(3):264–
323.
Karaboga, D. and Ozturk, C. (2011). A novel clustering
approach: Artificial bee colony (abc) algorithm. Appl.
Soft Comput., 11(1):652–657.
Lichman, M. (2013). UCI machine learning repository.
University of California, Irvine, School of Informa-
tion and Computer Sciences.
Lourenc¸o, A., Bul
`
o, S. R., Rebagliati, N., Fred, A. L. N.,
Figueiredo, M. A. T., and Pelillo, M. (2015). Proba-
bilistic consensus clustering using evidence accumu-
lation. Machine Learning, 98(1-2):331–357.
Marinakis, Y., Marinaki, M., Doumpos, M., Matsatsinis, N.,
and Zopounidis, C. (2008). A hybrid stochastic genet-
icgrasp algorithm for clustering analysis. Operational
Research, 8(1):33–46.
Maulik, U., Bandyopadhyay, S., and B, S. (2000). Genetic
algorithm-based clustering technique. Pattern Recog-
nition, 33:1455–1465.
Mirkin, B. (1996). Mathematical Classification and Clus-
tering. Kluwer Academic Publishers, Dordrecht, The
Netherlands.
Ni, J., Li, L., Qiao, F., and Wu, Q. (2013). A novel
memetic algorithm and its application to data cluster-
ing. Memetic Computing, 5(1):65–78.
Niknam, T. and Amiri, B. (2010). An efficient hybrid ap-
proach based on pso, aco and k-means for cluster anal-
ysis. Appl. Soft Comput., 10(1):183–197.
Selim, S. Z. and Ismail, M. A. (1984). K-means-type algo-
rithms: A generalized convergence theorem and char-
acterization of local optimality. IEEE Trans. Pattern
Anal. Mach. Intell., 6(1):81–87.
Senthilnath, J., Omkar, S., and Mani, V. (2011). Clustering
using firefly algorithm: Performance study. Swarm
and Evolutionary Computation, 1(3):164 – 171.
Zhao, J., Lei, X., Wu, Z., and Tan, Y. (2014). Clustering
using improved cuckoo search algorithm. In Tan, Y.,
Shi, Y., and Coello, C., editors, Advances in Swarm
Intelligence, volume 8794 of Lecture Notes in Com-
puter Science, pages 479–488. Springer International
Publishing.
Clustering using Cellular Genetic Algorithms
373
