
Managing Fragmented Personal Data: Going beyond the Limits of
Personal Health Records
Juha Puustjärvi
1
and Leena Puustjärvi
2
1
Department of Computer Science, University of Helsinki, P.O. Box 68, Helsinki, Finland
2
The Pharmacy of Kaivopuisto, Neitsytpolku 10, Helsinki, Finland
Keywords: Personal Health Record, Personal Data, Data Integration, Smart Home, Semantic Web, RDF, SPARQL.
Abstract: A personal health record (PHR) is a record of a consumer that includes data gathered from different sources
such as from health care providers, pharmacies, insures, the consumer, and third parties. Gathering data is
technically complicated and error-prone due to the heterogeneities of the data sources. Further, due to failed
or missed transmissions patients’ PHRs are often incomplete. However, a consumer should have easy access
to their own health information as well as to any relevant information they need in order to make decisions
about their own heath care. Nevertheless, no holistic approach for managing personal data beyond PHRs has
been developed. Satisfying this challenge requires a means to capture and interconnect information from a
variety of personal data sources and from public data sources. In order to achieve this goal, we have
designed a Personal Record (PR). It is virtually a single record in the sense that it gives an illusion of a
traditional standalone tool, such as a traditional PHR, although its content may locate in a variety of sources,
e.g., in systems storing data of health, gyms, smart homes, or personal notes. By means of PR we can also
achieve synergy, e.g., in using health data together with welfare and smart home data we can produce
outcomes that could not be achieved by functioning independently with single data sources. Moreover,
using personal data together with public data sources we can also achieve more informal outcomes. The
only requirement is that the data sources are in RDF-format, i.e., in the form of subject–predicate–object
expressions. Then the SPARQL processor has the ability to process the data as well as to find the
connections between triples from separate sources.
1 INTRODUCTION
A personal health record (PHR) is a record of a
consumer that includes data gathered from different
sources such as from health care providers,
pharmacies, insures, the consumer, and third parties
(Raisinghani and Young, 2008). It includes
information about medications, allergies,
vaccinations, illnesses, laboratory and other test
results, and surgeries and other procedures. An ideal
PHR would provide a complete and accurate
summary of the health and medical history of a
consumer (Angst et al., 2008).
PHRs have the potential to dramatically change
healthcare in the near future as they enable patients
to become more involved and engaged in their care
and allow other authorized stakeholders to access
information about patients that was previously not
available. The changes effected by PHR systems
could have a significant, positive impact on the
efficiency of healthcare sector and thus resulting
considerable cost savings to the healthcare systems.
In order to avoid the compatibility problems in
importing data to PHRs various standardization
efforts on PHRs have been done. In particular, the
use of the Continuity of Care Record of ASTM
(CCR, 2011) and HL7’s Continuity of Care
Document (CCD, 2009) has been proposed for using
in standardizing the structure PHRs. From
technology’s point of view CCR and CCD-standards
represent two different XML schemas designed to
store patient clinical summaries. However, both
schemas are identical in their scope in the sense that
they contain the same data elements.
A problem of current PHRs is that they assume
all its content to be in one source although patient
may have lived in many places and used various
healthcare specialities. This in turn requires moving
records from a variety of sources into PHR. Such
transmissions are technically complicated and error-
Puustjärvi, J. and Puustjärvi, L.
Managing Fragmented Personal Data: Going beyond the Limits of Personal Health Records.
DOI: 10.5220/0005626101450150
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 5: HEALTHINF, pages 145-150
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
145

prone. As a result patients’ PHRs are incomplete in
the sense that lot of relevant data is often missing.
Our argument is that by exploiting Semantic web
technologies it is easier to ensure the consistency of
personal data by retrieving it from its original
sources instead of first gathering it into one source,
Another problem of current PHRs is that their
content is restricted on health oriented data.
However, there are a lot of related data that are
stored in other systems, and which use together with
PHR data would produce outcomes that could not be
achieved by functioning independently.
Examples of PHR-related data sources include
gyms, smart homes and personal note books: gyms
store data that is gathered by sensor and training
equipment, smart homes store a lot of data related to
heating, air conditioning as well as with personal
well fare such as weight measurements, and personal
note books may include a variety of useful
information concerning working hours, meals and
location data.
By connecting these data sources we can achieve
new outcomes: For example, a person can query his
or her blood pressures when his or her weigh had
maximal and minimum values. Also a person can
query his or her cholesterol values when his or her
training hours had maximal or minimum values.
There are also a lot of public data sources, which
use together with personal data would produce
outcomes that could not be achieved by using only
personal data. For example, personal data may
indicate the vaccinations of a person while public
data source can augment this information by more
informal descriptions of the vaccinations.
In this paper, we introduce a personal record, or
shortly PR. Although its data may locate in one or
more sources, it is virtually a standalone record in
the sense that it gives an illusion of a standalone
record. The only requirements of a PR are that its
data sources have a Unified Resource Locator
(URL), and the data is in RDF-format, i.e., in the
form of subject–predicate–object expressions (RDF,
2011). Then the SPARQL processor (SPARQL,
2008) has the ability to process the data as wells as
to find connections between triples from different
sources. Hence, the synergy of accessing data from
different sources can be achieved.
The rest of the paper is organized as follows.
First, in Section 2, we consider related work by
focusing on the way patients´ scattered clinical
documents are managed and gathered in the context
of the IXE XDS (IHE, 2005). Then, in Section 3, we
present the architecture of our designed PR-system.
In Section 4, we consider PR-system’s data sets. In
particular, we present our developed Welfare
ontology and illustrate its use in RDF-statements. In
Section 5, we present how traditional XML-based
PHRs, as well as any XML-document, can be
transformed into RDF-format. Finally, Section 6
concludes the paper by discussing the gains and the
challenges of the PR-system.
2 RELATED WORK
The idea of gathering patients’ clinical documents
dynamically from a variety of sources is not new: in
the context of electronic health record (EHR) the
problem of patients’ scattered clinical documents is
studied (Boone, 2011). EHRs differ from PHRs in
that former is a record of healthcare provider while
the latter is a record of consumer (Puustjärvi and
Puustjärvi, 2015). In particular, in IHE XDS
architecture original documents are dynamically
retrieved by exploiting relevant registries. That is,
the idea behind the IHE XDS is to build virtual
patient records on the fly from a variety of clinical
documents created by different healthcare
organizations (Benson, 2010).
In IHE XDS terminology healthcare enterprises
that agree to work together for clinical document
sharing is called clinical affinity domain (IHE,
2015). Its enterprises agree on a common set of
policies such as how the patients are identified, the
access is controlled, and the common set of coding
terms to represent the metadata of the documents.
Further, patients expect their records to follow them
as they move from one clinical affinity domain to
another (Dogac et al., 2002).
Examples of XDS clinical affinity domains
include: nationwide and regional EHRs, federations
of enterprises, regional federations made up of
several local hospitals, healthcare providers, and
insurance provider supported communities
(Puustjärvi and Puustjärvi, 2014).
IHE XDS has proven to be useful and workable
innovation, and hence we could adopt the ideas of
IHE XDS into PHRs. However, we argue that by
exploiting modern information technology we can
avoid many of the drawbacks of the IHE XDS. In
particular, we have addressed the following two
problems of the IHE XDS.
The main problem with its used ebXML
registries is that searches can only be based on the
keywords and folders. Although the keywords are
taken from a taxonomy only a very limited amount
of semantics can be provided (Dogac et al., 2007).
Folders group the related documents together (e.g.,
HEALTHINF 2016 - 9th International Conference on Health Informatics
146
based on a period of time, episode, or
immunizations). However, there are numerous cases
where retrieving predefined folders are not
appropriate but rather dynamic grouping of
documents should be possible.
Another problem with the IHE XDS is that it
expects patients’ records to follow then when they
move from one affinity domain to another. The
problem here are twofold: First, moving records
between affinity domains is technically complicated
and error-prone due to the heterogeneities of affinity
domains. Second, due to the failed or missed
transmissions patients’ EHRs are incomplete.
In the PR-system these kinds of problems can be
avoided by the solutions presented in the next
sections.
3 PR-SYSTEM
3.1 Searching Multiple Datasets by
SPARQL
SPARQL allows users to write queries against data
that follows the RDF specification of the W3C. The
name SPARQL is a recursive acronym for SPARQL
Protocol and RDF Query Language, which is
described by a set of specifications from the W3C
(DuCharme, 2011). SPARQL Protocol refers to the
rules for how a client program and a SPARQL
processing server exchange SPARQL queries and
results.
A typical SPARQL query specifies the pieces of
information that meets the stated conditions. The
conditions are described with triple patterns, which
are similar to RDF triples but may include variables
to add flexibility in how they match against the data.
There is a variety of SPARQL processors (also
called SPARQL engines) available for running
queries against data both locally and remotely.
SPARQL provides two ways for querying remotely:
using FROM keyword or using SERVICE keyword.
In the former way, the FROM keyword names a
dataset to query that may be local or remote file. In
the latter way, instead of pointing at an RDF file
somewhere, a SPARQL endpoint is pointed. A
SPARQL endpoint is a web service that accepts
SPARQL queries, runs the queries, and then returns
the result.
In addition, SPARQL allows searching multiple
datasets with one query. This enables a variety of
useful applications. To illustrate this, assume that
user’s blood pressure values, weight measurements,
and medication information are stored in different
data sources such that each data source has a URL.
Then, by using SPARQL Federated queries and
MAX-function, a user may query for example:
Give me my ongoing medication when my
weight had a minimum value.
Give me my training ours of the day when my
blood pressure had maximal value.
We next consider the architecture where this
kind of queries can be processed.
3.2 The Architecture of the
PR - System
In our PR-related terminology organizations that
produce and maintain RDF-formatted personal data
and agree on a common set of policies make up a
collaboration domain. The policies specify how the
personal data sets are identified, the access is
controlled, and the common set of coding terms to
represent the RDF-files or SPARQL endpoints.
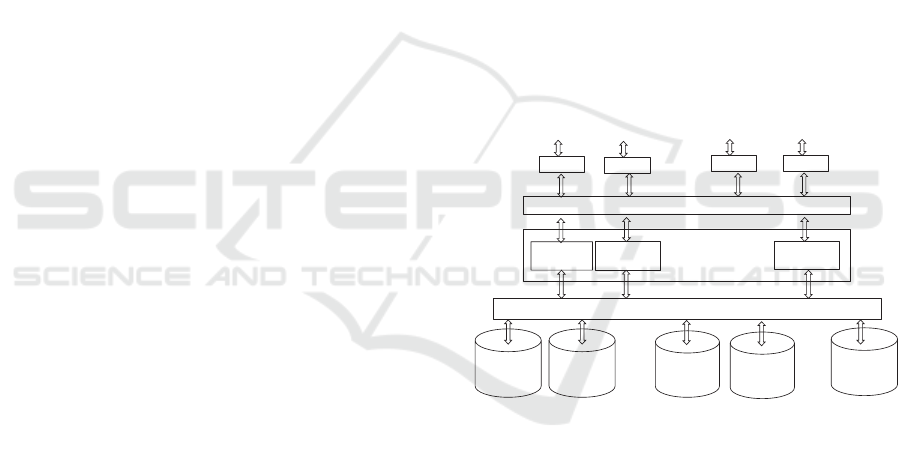
The architecture of a collaboration domain is
presented in Figure 1.
Browser
PR-Portal
Application 1
. . .
Community applications
SPARQL Processor
User of a
virtual PHR
Application 2
Application n
A dataset of a
healthcare
organization A
A dataset of
gym A
Browser
User of a
virtual PHR
Browser
User of a
virtual PHR
Browser
User of a
virtual PHR
A dataset of a
smart home
. . .
. . .
A dataset of
gym B
A dataset of a
healthcare
organization B
. . .
Figure 1: The architecture of the virtual PR-system.
The users of the collaboration domain access (by
their browsers) their personal datasets through the
PR-Portal. The portal provides connections to
relevant applications. For example there may be
separate applications for a traditional PHR, welfare
data, and smart home. However, the key point here
is that the applications use data from a variety of
data sets and thus enables outcomes that could not
be achieved by single systems, e.g., by smart home
system or PHR system.
In addition, a property of the PR-system is that
each user may have one or more predefined profiles.
Each profile specifies a set of user’s data sources,
i.e., URLs. So, the system can provide an illusion of
the traditional tools such as a PHR-system or a note
book.
Managing Fragmented Personal Data: Going beyond the Limits of Personal Health Records
147
The applications are based on the use cases of
various user groups, and they may interoperate
through accessing the same datasets. Also new
applications can be easily inserted when new needs
arise. Even new tools can be easily added, e.g.,
inserting a personal photo album requires adding a
relevant application and a data set for the album.
Note that in this this architecture some of the
users may have all their data in a data source. In
such a case their PR can behave like a traditional
Internet-based PHR. Such a case can also be
considered as the first step towards the use of the
PR-system. If the user already has a traditional
XML-based PHR, it must be transformed into RDF-
format. The way such a transformation can be
automatically done is presented in Section 5. The
transformation exploits the ontology presented in
Section 4.
4 THE ROLE OF OTOLOGIES IN
PR-SYSTEM
4.1 Welfare Ontology and OWL
In the context of computer science, an ontology is a
general vocabulary of a certain domain, and it can be
defined as “an explicit specification of a
conceptualization” (Antoniou and Harmelen, 2004).
It tries to characterize that meaning in terms of
concepts and their relationships (Daconta et al.,
2003). It is typically represented as classes,
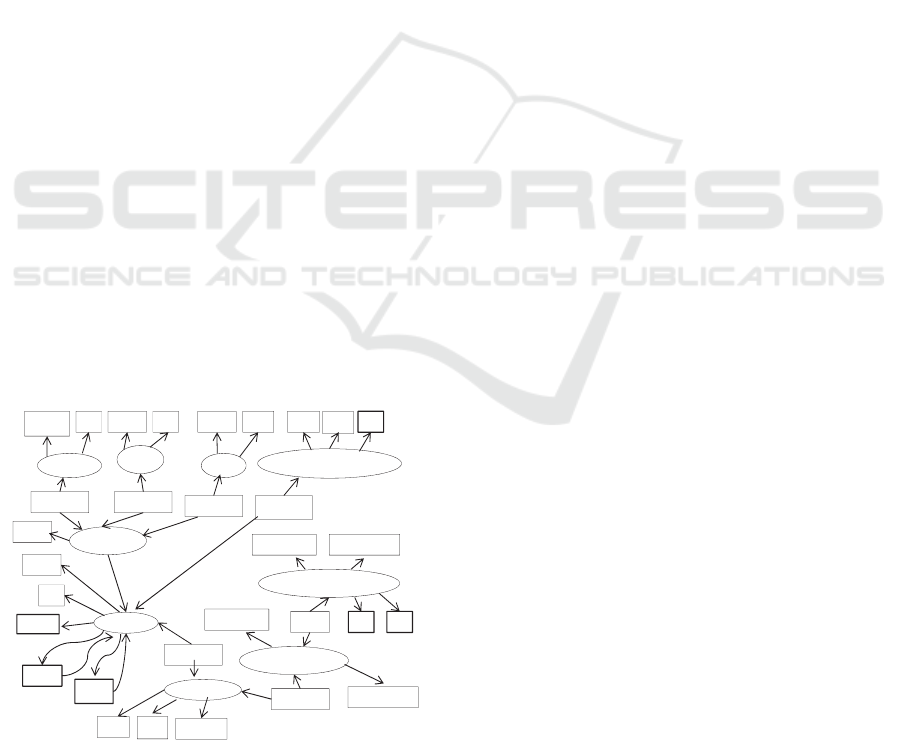
properties, attributes and values. As an example,
consider a subset of our designed Welfare Ontology,
which is presented in a graphical way in Figure 2.
MedicalTest
BloodPressureLimits
SystolicLimit
DiastolicLimit
SSN
Measures
SubclassOf
Performed
ExcerciseTest
Stored
SubclassOf
SubclassOf
Swimming
Duration Da te
Walk
Length
Date
Mother
Father
Hobby
SubclassOf
Run
Length
Date
TestName
Person
Name
WeightMeasurement
Value Date
Time
BloodPressureTestValue
SystolicValue DiastolicValue
TimeTypeOf Date
Value Date
Figure 2: Graphical presentation of a portion of the
Welfare Ontology.
In this graphical representation ellipses represent
classes and subclasses while rectangles represent
data type and object properties. Classes, subclasses,
data properties and object properties are modeling
primitives in OWL (Web Ontology Language)
(OWL, 2011). Object properties (e.g., Measures)
relate objects to other objects while data type
properties (e.g., Name) relate objects to datatype
values. In Figure 2, we have presented only a few of
objects’ datatype properties.
Fundamentally the Welfare Ontology comprises
the vocabulary that a person can use in describing
his or her personal welfare information. Hence we
do not assume that a person uses all the terms of the
vocabulary (ontology). For example, datatype
properties Father and Mother are included in the
vocabulary, but the person does not have to give
values for these properties. Neither the person needs
class Swimming, if swimming is not included in his
or her hobbies.
On the other hand, a person may use whatever
ontology (vocabulary) or ontologies in describing
his or her personal welfare information.
Respectively a person may use a variety of
ontologies in his or her smart home or notebook
data.
We next illustrate the use of ontologies in RDF
descriptions.
4.2 Using Welfare Ontology in
RDF-Formatted Datasets
RDF itself is a data model. Its modeling primitive is
an object-attribute-value triple, which is called a
statement (Antoniou and Harmelen, 2004). In order
that RDF data can be represented and transmitted it
needs a concrete syntax, which is given in XML,
i.e., RDF statements are usually coded in XML.
Hence, RDF inherits the benefits associated with
XML. However, other syntactic representations
(e.g., Turtle) are also possible, meaning that XML-
based syntax is not a necessary component of the
RDF model.
<rdf:RDF
xmlns :rdf=”http://www.w3.org /1999/02/22‐rdf‐syntax‐ns#”
xmlns :po=http://www.helsinki.fi/WelfareOntology#>
<rdf:Description
rdf:about=”weightmeasurement100820151028”>
<rdf:type rdf:resource=“&po;WeightMeasurement”/>
<po :Measures>RitaSmith</po :Measures>
<po :Date>10:08:2015</po:Uses>
<po :Time>10:28</po:Time>
<po :Value>68.7</po:Value>
</rdf :Description>
</rdf:RDF>
Figure 3: An instance of the Welfare Ontology in RDF.
HEALTHINF 2016 - 9th International Conference on Health Informatics
148
One RDF description may contain one or more
RDF statements about an object (Daconta et al.,
2003). For example, in Figure 3, the description
concerning Rita Smith’s weight measurement
(identified by “weightmeasurement100820151028”)
contains five RDF statements: the first states that its
type in the Welfare Ontology is Weight-
Measurement, and the second states that it measures
Rita Smith. Subsequent statements specify the date,
the time, and the value of the weight measurement.
5 TRANSFORMING AN
XML-BASED PHR INTO RDF
In our model, the organizations of the collaboration
domain maintain personal datasets, which are in
RDF. The personal data sets that are not initially in
XML (e.g., most PHRs) have to be transformed into
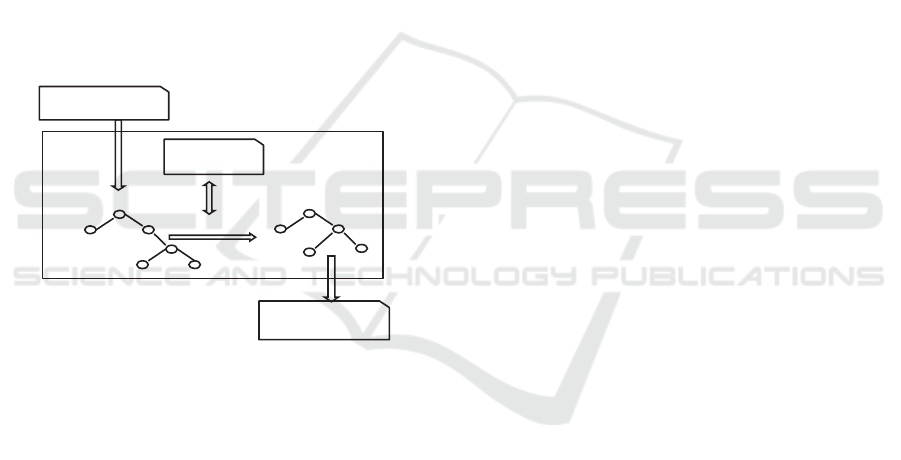
RDF. Such a transformation is illustrated in Figure
4.
XML tree
RDF/XML tree
Stylesheet Engine
Transformation
CCD document
in RDF/XML
Stylesheet
in XSLT
CCD document
in XML
Figure 4: Transforming a CDD document into RDF/XML
format.
The Stylesheet Engine takes an XML document
(e.g., a PHR based on the CDD-standard), loads it
into a DOM (Document Object Model) (Daconta et
al., 2003) source tree, and transforms that document
with the instructions given in the stylesheet into
RDF/XML format. The instructions use XPath
expressions (Daconta et al., 2003) in referencing to
the source tree and in placing it into the result tree.
The result tree is then formatted, and the resulting
element in RDF/XML format is returned.
To illustrate the transformation an input
document (a CDD document) is presented in Figure
5, and its output document is presented in Figure 6.
<SimplifiedCCDfile>
<DocumentID>DOC_123</DocumentID>
<Patient>
<PatientID>AB‐12345></PatientID>
<PatientName>RitaSmith></PatientName>
</Patient>
<Medications>
<Medication>
<MedicationID>Medication.567</MedicationID>
<DateTime>
<ExactDateTime>2012‐03‐ 01TO12:00</ExactDateTime>
</DateTime>
<Source>
<Actor>
<ActorID>Pharmacy ofKaivopuisto</ActorID>
<ActorRole>Pharmacy</ActorRole>
</Actor>
</Source>
<Description>
<Text>Onetablet three times aday</Text>
</Description>
<Product>
<ProductName>Voltaren</ProductName>
<BrandName>Diclofenac</BrandName>
</Product>
<Strenght>
<Value>50</Value>
<Unit>milligram</Unit>
</Strenght>
<Quantity>
<Value>30</Value>
<Unit>Tabs</Unit>
</Quantity>
</Medication>
</Medications>
</SimplifiedCCDfile>
Figure 5: A simplified example of a CCD document.
<rdf:RDF
xmlns :rdf=”http://www.w3.org/1999/02/22‐rdf‐syntax‐ns#”
xmlns :xsd=”http://www.w3.org/2001/XMLSchema#”
xmlns:po=http://www.lut.fi/ontologies/EHR‐Ontology#>
<rdf:Description rdf:about=”AB‐12345”>
<rdf:type rdf:resource=“&po;Patient”/>
<po :PatientName>RitaSmith</po :PatientName>
<po :Us es>MO‐5481</po:Uses>
<po :Performed>H‐257L</po :Performed>
</rdf :Description>
<rdf:Description rdf:about=”MO‐5481”>
<rdf:type rdf:resource=“&po;Medication”/>
<po :Contains>Voltaren</po :Contains>
<po:ExactDateTime>2012‐03‐01TO12:00
</po: ExactDateTime>
<po :StrenghtV alue rdf:datatype=
”&xsd;integer”>30</po :StrenghtValue>
<po :StrenghtUnit>Tabs</po :StrenghtUnit>
</rdf :Description>
<rdf:Description rdf:about=”211708‐8”>
<rdf:type rdf:resource=“&po;Source/>
<po :ActorID>Pharmacy</po :ActorID>
<po :ActorRole>Pharmacy</po :ActorRole>
</rdf :Description>
<rdf:Description rdf:about=”Voltaren”>
<rdf:type rdf:resource=“&po;ProductName”/>
<po :BrandName>Diclofenac</po :Contains>
</rdf :Description>
</rdf:RDF>
Figure 6: A transformed CCD document in RDF/XML-
format.
Managing Fragmented Personal Data: Going beyond the Limits of Personal Health Records
149

6 CONCLUSIONS
RDF-based data formats have not yet achieved the
mainstream status that XML and relational databases
have. However an increasing number of
professionals are discovering that tools using the
RDF data model let them expose diverse sets of data
with a common, standardized interface. The data sets
may be public or private. Private data sets include a
variety of personal data including health data,
welfare data, and smart home data.
By connecting personal data among themselves,
or with public data we can achieve synergy. For
example, connecting person’s vaccinations data with
public informal data dealing with vaccinations gives
outcomes that could not be achieved by functioning
independently with personal or public data.
However, achieved synergy is not the only gain of
our designed PR-system: by integrating a variety of
personal tools we can also significantly improve
their usability.
The SPARQL processor is a corner stone of our
approach. It has the ability to process the data and to
find the connections between RDF-triples from
separate data sources. Especially we have exploited
this feature in developing the PR-system.
We have also presented our developed Welfare
Ontology, which can be used in data sets concerning
individual’s welfare data. However, it is just an
alternative: in RDF-based data sets we can use any
ontology (vocabulary). Even each RDF-statement in
a dataset may be based on different ontology. The
possibility of using existing public ontologies as
well as user specific ontologies makes this approach
very flexible
On the other hand, to succeed PR-system should
not be considered just as a technical infrastructure
but rather as ecosystems having many
interconnected parts. So far we have considered the
technical infrastructure and the services of our
designed PR-system. The other key parts of the e-
health ecosystem are governance regulations,
financing and stakeholders. In our future research
we will focus on these issues.
In addition, there are many other challenges.
The introduction of new technology is also an
investment. Also a consequence of introducing new
healthcare model is that it significantly changes the
daily duties of the employees in the organizations,
which produce personal digital data. Thus the most
challenging aspect will not be the technology but
rather changing the mind-set of the employees of
these organizations.
REFERENCES
Angst, C.M., Agarwal, R, Downing, J., 2008. An
empirical examination of the importance of defining
the PHR for research and for practice, Proceedings of
the 41st Annual Hawaii International Conference on
System Sciences.
Antoniou, G., Harmelen, F., 2004. A Semantic Web
Primer. The Mitt Press.
Benson, T., 2010. Principles of Health Interoperability
HL7 and SNOMED. Springer.
Boone, K., 2011. The CDA Book. Springer.
CCD, 2009. What Is the HL7 Continuity of Care
Document? Available at: http://www.neotool.com/
blog/2007/02/15/ what-is-hl7-continuity-of-care-docu
ment/
CCR, 2011. Continuity of Care Record (CCR) Standard.
Available at: http://www.ccrstandard.com/
Daconta, M., Obrst, L., Smith, K., 2003. The semantic
web: A Guide to the Future of XML, Web Services,
and Knowledge Management, John Wiley & Sons.
Dogac, A., Laleci, G., Kabak, Y., Cingil, I., 2002.
Exploiting web service semantics: Taxonomies vs
ontologies, IEEE Data Eng. Bull, Vol. 25, No. 4,
pp.10-16.
Dogac, A., Gokce, B., Aden, T., Laleci, T., Eichelberg,
M., 2007. Enhancing IHE XDS for Federated Clinical
Affinity Domain Support. IEEE Transactions on
Information Technology in Biomedicine, Vol.11, No.
2.
DuCharme, B., 2011. Learning SPARQL. O’Reilly Media.
IHE, 2005. IT Infrastructure Technical Framework
Volume 1 (ITI TF-1). Available at:
www.ihe.net/Technical_Framework/upload/ihe_iti_tf_2.0
_vol1_FT_2005-08-15.pdf.
OWL, 2011. Web Ontology Language. Available at:
http://www.w3.org/TR/owl-features/
Puustjärvi, J., Puustjärvi.L., 2014. Using Ontology-Based
Registry and SPARQL Engine in Searching Patient’s
Clinical Documents. In the proc. of the International
Conference on Health Informatics. Pages 151-158.
Puustjärvi, J., Puustjärvi. L., 2015. Maintaining the
consistency of Electronic Health Record’s Medication
List. In the proc. of the International Conference on
Health Informatics.
RDF, 2011. Resource Description Language. Available at:
http://www.w3.org/RDF/
Raisinghani M.S., Young, E., 2008. Personal health
records: key adoption issues and implications for
management, International Journal of Electronic
Healthcare. Vol. 4, No.1 pp.67-77.
SPARQL, 2008. SPARQL Query Language for RDF.
Available at: http://www.w3.org/TR/rdf-sparql-query/
HEALTHINF 2016 - 9th International Conference on Health Informatics
150
