
Multiobjective Bacterial Foraging Optimization using Archive Strategy
Cuicui Yang and Junzhong Ji
College of Computer, Beijing University of Technology, Beijing Municipal Key Laboratory of Multimedia and
Intelligent Software, Pingleyuan 100, Chaoyang District, Beijing, China
Keywords:
Bacterial Foraging Optimization, Archive Strategy, Conjugation, Multiobjective Optimization.
Abstract:
Multiobjective optimization problems widely exist in engineering application and science research. This paper
presents an archive bacterial foraging optimizer to deal with multiobjective optimization problems. Under
the concept of Pareto dominance, the proposed algorithm uses chemotaxis, conjugation, reproduction and
elimination-and-dispersal mechanisms to approximate to the true Pareto fronts in multiobjective optimization
problems. In the optimization process, the proposed algorithm incorporates an external archive to save the
nondominated solutions previously found and utilizes the crowding distance to maintain the diversity of the
obtained nondominated solutions. The proposed algorithm is compared with two state-of-the-art algorithms
on four standard test problems. The experimental results indicate that our approach is a promising algorithm
to deal with multiobjective optimization problems.
1 INTRODUCTION
The multiobjective optimization problem (MOP) usu-
ally involves more than one conflicting objectives that
need to be optimized simultaneously and is an im-
portant class of scientific and engineering problems
in real-world (Deb, 2001). The solution to a MOP
is a set of trade-off solutions known as Pareto opti-
mal solutions or non-dominated solutions which can-
not improve all the objectives simultaneously. Evolu-
tionary computation methods based on Darwin’s bi-
ological evolution theory deal with a group of candi-
date solutions, which makes that they are natural to
be used to handle multiobjective optimization prob-
lems (MOPs). In the past two decades, researchers
have proposed a variety of evolutionary computation-
based approaches for solving MOPs (Deb, 2001;
Coello Coello CA, Van Veldhuizen DA, Lamont GB,
2007), such as the well-known algorithms PESA-
II (Corne D W, Jerram N R, Knowles J D, et al,
2001), NSGA-II (Deb K, Pratap A, Agarwal S, et
al, 2002) and SPEA2 (E Zitzler, M Laumanns, L
Thiele, 2002). Evolutionary multiobjective optimiza-
tion (EMO) that uses evolutionary computation meth-
ods to solve MOPs has become a relatively hot re-
search area.
In recent years, some new bio-inspired optimiza-
tion technologies have been successfully introduced
to deal with MOPs, where Particle swarm optimiza-
tion (PSO) is a prominent example. So far, there
have been a number of methods based on PSO for
solving MOPs (X Li, 2003; Coello C A C, Pulido
G T, Lechuga M S, 2004; Tripathi P K, Bandyopad-
hyay S, Pal S K, 2007). Bacterial foraging opti-
mization (BFO) is another popular bio-inspired op-
timization technology which simulates the foraging
behavior of E. coli bacteria (K.M. Passino, 2002).
BFO has been proved to be an efficient optimiza-
tion method for single objective optimization prob-
lems (Agrawal V, Sharma H, Bansal J C, 2011), and
more recently researchers have also shown promising
results for MOPs (Panigrahi B K, Pandi V R, Das
S, et al, 2010; Guzm ´an M A, Delgado A, De Car-
valho J, 2010; Niu B, Wang H, Tan L, et al, 2012;
Niu B, Wang H, Wang J, et al, 2013). However, to
the best of our knowledge, none of these algorithms
incorporate an external archive to preserve the elitism
solutions, namely, there is no way to keep the non-
dominated solutions found previously in the optimiza-
tion process. Elitism-preservation is an importance
strategy in multiobjective search, which has been rec-
ognized and supported experimentally (Parks G T,
Miller I, 1998; Zitzler E, Deb K, Thiele L, 2000).
To further explore the potential of BFO algorithm in
finding Pareto optimal solutions for MOPs, this pa-
per presents an archive bacterial foraging optimizer
for multiobjective optimization, called as MABFO.
MABFO mainly includes four optimization mech-
anisms: chemotaxis, conjugation, reproduction and
elimination-and-dispersal. The implements of these
Yang, C. and Ji, J.
Multiobjective Bacterial Foraging Optimization using Archive Strategy.
DOI: 10.5220/0005668601850192
In Proceedings of the 5th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2016), pages 185-192
ISBN: 978-989-758-173-1
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
185

mechanisms are different from other current meth-
ods based on BFO. More improtant, MABFO incor-
porates an external archive to save the nondominated
solutions previously found and uses the crowding dis-
tance to maintain the diversity of the nondominated
solutions. These strategies are conducive to produce
well-distributed and high-quality solutions.
MABFO is validated on four standard test prob-
lems and compared against two start-of-the-art EMO
approaches NSGA-II (Deb K, Pratap A, Agarwal S,
et al, 2002) and SPEA2 (E Zitzler, M Laumanns, L
Thiele, 2002). The experimental results indicate that
MABFO is a promising algorithm and can be consid-
ered an alternative to deal with MOPs.
The remainder of the paper is structured as fol-
lows. Section II briefly introduces basic concepts in-
volving MOPs. Section III presents the details of the
MABFO algorithm. We then present experiments in
section IV, and finally, section V concludes the paper.
2 BASIC CONCEPTS
A MOP can be described as the problem of finding
a vector of decision variables that optimizes a vector
function and satisfies some restrictions. Without loss
of generality, a MOP is formulated as follows (Deb,
2001):
min y = F(x) = ( f
1
(x), f
2
(x),· ·· , f
m
(x))
T
s.t. g
i
(x) ≤ 0, i = 1, 2,·· · , p
h
j
(x) = 0, j = 1,2, ··· , q
x
L
i
≤ x
i
≤ x
U
i
, (1)
where x = (x
1
,x
2
,··· ,x
n
) ∈ X ⊂ R
n
is a n-dimensional
decision vector, X represents a n-dimensional deci-
sion space, x
L
i
and x
U
i
are the upper and lower bound-
ary values of x
i
, respectively. y = (y
1
,y
2
,··· ,x
m
) ∈
Y ⊂ R
m
is a m-dimensional objective vector, Y rep-
resents a m-dimensional objective space. F(x) is a
mapping function from n-dimensional decision space
to m-dimensional objective space. g
i
(x) ≤ 0 (i =
1,2,· ·· , p) and h
j
(x) = 0 ( j = 1, 2,··· , q) defines p
inequalities and q equalities, respectively.
In the following, we will list four definitions in-
volving MOPs.
Pareto Dominant: x
α
, x
β
are two feasible solu-
tions for problem (1), x
α
is Pareto dominant compared
with x
β
if and only if:
∀i = 1, 2,··· , m, f
i
(x
α
) ≤ f
i
(x
β
) ∧
∃ j = 1,2,··· ,m, f
i
(x
α
) < f
i
(x
β
)
. (2)
We call this relationship x
α
≻ x
β
, x
α
dominate x
β
, or
x
β
is dominated by x
α
.
Pareto Optimal Solution: Ω is the feasible solu-
tion set of problem (1), x
∗
∈ Ω, x
∗
is a Pareto optimal
solution if and only if:
¬∃x ∈ Ω : x ≻ x
∗
. (3)
Pareto Optimal Set: The Pareto optimal set of
problem (1) includes all the Pareto optimal solutions
and is given as follows:
X
∗
= {x
∗
|¬∃x ∈ Ω : x ≻ x
∗
}. (4)
Pareto Front: The Pareto front of problem (1)
includes all the objective vectors corresponding to X
∗
and is given as follows:
PF = { F(x
∗
) = ( f
1
(x
∗
), f
2
(x
∗
),· ·· , f
m
(x
∗
))
T
|x
∗
∈ X
∗
}.
(5)
3 THE MABFO ALGORITHM
In this section, we will introduce MABFO algorithm.
Algorithm 1 is the framework of MABFO. In the fol-
lowing, we will describe its five important operators
chemotaxis, archive updating, conjugation, reproduc-
tion and elimination-and-dispersal as Algorithm 1
shown.
Algorithm 1: MABFO algorithm (main loop).
Input: Different parameters:
N
c
: maximum number of chemotaxis,
N
re
: maximum number of reproduction,
N
ed
: maximum number of elimination-and-dispersal,
N1: the size of population P,
N2: the maximum size of external archive A.
Output: A (Pareto optimal set)
1: Initialization: Generate an initial population P and an
empty archive A.
2: for l = 1 to N
ed
do
3: for k = 1 to N
re
do
4: for j = 1 to N
c
do
a). Each bacterium in P takes a chemotaxis
process.
b). Archive updating: copy all nondominated
individuals in joint population of A and P to A.
If the size of A exceeds N2, then reduce it
based on the crowding distance.
c). Each bacterium takes a conjugation process.
5: end for
6: The population P perform a reproduction process
which selects N1 superior individuals in the joint
population P and A and then copy them to P.
7: end for
8: The population P perform an elimination-and-
dispersal process and update the archive A as step
4 b).
9: end for
10: Return A.
ICPRAM 2016 - International Conference on Pattern Recognition Applications and Methods
186

3.1 Chemotaxis
Chemotaxis simulates the foraging movement of
E.coli bacteria through tumbling and swimming. To
absorb more nutrients, each bacterium tries to find
food in two ways: tumbling and swimming. A bac-
terium tumbles in a random direction to exploratively
search for food. If the food is rich in the selected di-
rection, the bacterium will swim along this direction,
till the food gets bad or the bacterium has swum the
fixed steps.
A random direction is a random unit vector in n-
dimensional space. In the original BFO algorithm, it
is very cumbersome to calculate a random unit vector
for each bacterium in every chemotaxis. In the pro-
posed MABFO algorithm, we use a simple unit vec-
tor e
m
with n dimensions as the chemotaxis direction,
in which only one component randomly selected (i.e.
the mth component) is either -1 or 1 and all the others
are 0. The method of updating a solution is given as
follows:
x
i
( j + 1, k, l) = x
i
( j,k,l) + c(i) e
m
, (6)
where x
i
( j,k,l) represents the ith bacterium at jth
chemotaxis, kth reproduction, lth elimination-and-
dispersal step. c(i) is the step size along the direc-
tion e
m
. The original BFO algorithm uses a fixed
step size which would lead to bad convergence per-
formances (S Dasgipta, S Das, A Abraham and A
Biswas, 2009). In the proposed MABFO algorithm, a
dynamic step size is adopted and is given as follows:
c(i) = r (x
i
′
m
( j,k,l) − x
i
m
( j,k,l)), (7)
where r is a random number uniformly distributed
between −1 and 1, x
i
′
m
( j,k,l) denotes the mth com-
ponent of another different bacterium i
′
which is se-
lected randomly in P.
When bacterium i carries out a chemotaxis oper-
ator, it first generates a direction e
m
and a step size
c(i) as described above, and then swims a step along
the direction e
m
according to Eq.(6). If the new solu-
tion x
i
( j+1,k,l) dominates the old solution x
i
( j,k,l),
bacterium i will swim another step in the direction e
m
according to Eq.(6). This process is continued until
bacterium i has swum the maximum steps N
s
or the
obtained new solution x
i
( j + 1,k,l) is dominated by
the old solution x
i
( j,k,l). Such chemotaxis mecha-
nism is an important driving force for locally opti-
mizing each candidate solution, where each bacterium
ties its best to search for non-dominated solutions.
3.2 Archive Updating
The main goal of the external archive is to keep a his-
torical record of the nondominated solutions obtained
in the search process. In MABFO, we use an exter-
nal archive A with a fixed number N2. Whenever the
populationP carries out a chemotaxis process, we will
reselect all the nondominated solutions from the joint
population of P and A, and then update archive A by
copying them to A. If the size of A exceeds N2, an
archive truncation procedure is invoked, which itera-
tively remove individuals from A based on the crowd-
ing distance until the size of A is N2. At each itera-
tion, the individual which has the minimum distance
to another individual is removed according to the for-
mula:
dis(x
i
) < dis(x
j
) ⇔
∀ 0 < k < |Q| : σ
k
i
= σ
k
j
∨
∃ 0 < k < |Q| : [(∀0 < l < k : σ
l
i
= σ
l
j
) ∧ σ
k
i
< σ
k
j
]
(8)
where Q is a group of individuals, |Q| denotes the size
of Q and here Q = A. σ
k
i
is the distance of ith solution
x
i
to its kth nearest neighbor in Q.
The external archive A promises not to miss the
nondominated solutions found in the search process.
The crowding distance eliminates the individuals in
dense area, which can ensure the obtained solutions
distributed evenly. The using of external archive
and crowding distance is beneficial to find well-
distributed Pareto solutions.
3.3 Conjugation
Conjugation, as well as chemotaxis, is an important
biological behavior of bacteria. Bacterial conjugation
is the transfer of part of plasmid (genetic material)
from donor bacteria to recipient bacteria by directly
physical contact and is often regarded as the sex-
ual reproduction or mating between bacteria. Some
researchers have taken the bacterial conjugation as
a message passaging mechanism in their work (C
Perales-Grav´an, R Lahoz-Beltra, 2008; A Balassub-
ramaniam, Memeber, IEEE, P Lio, 2013).
In this paper, we also simulate the bacterial con-
jugation behavior as an information exchange mech-
anism between bacteria in population P and archive
A. To model this biological behavior, we first define
conjugation length L (L < n) measured by the number
of decision variables. Then each bacterium i in pop-
ulation P will randomly select another bacterium i
′
in
A and a conjugation point Bt (Bt < n − L) to take a
conjugation step. In this step, the way of updating the
solution is given as follows:
x
i
new
( j,k,l) = x
i
( j,k,l) + ω ◦ (x
i
′
( j,k,l) − x
i
( j,k,l)),
(9)
where x
i
new
( j,k,l) denotes the new solution after bac-
terium i performing the conjugation operator. ω is
Multiobjective Bacterial Foraging Optimization using Archive Strategy
187
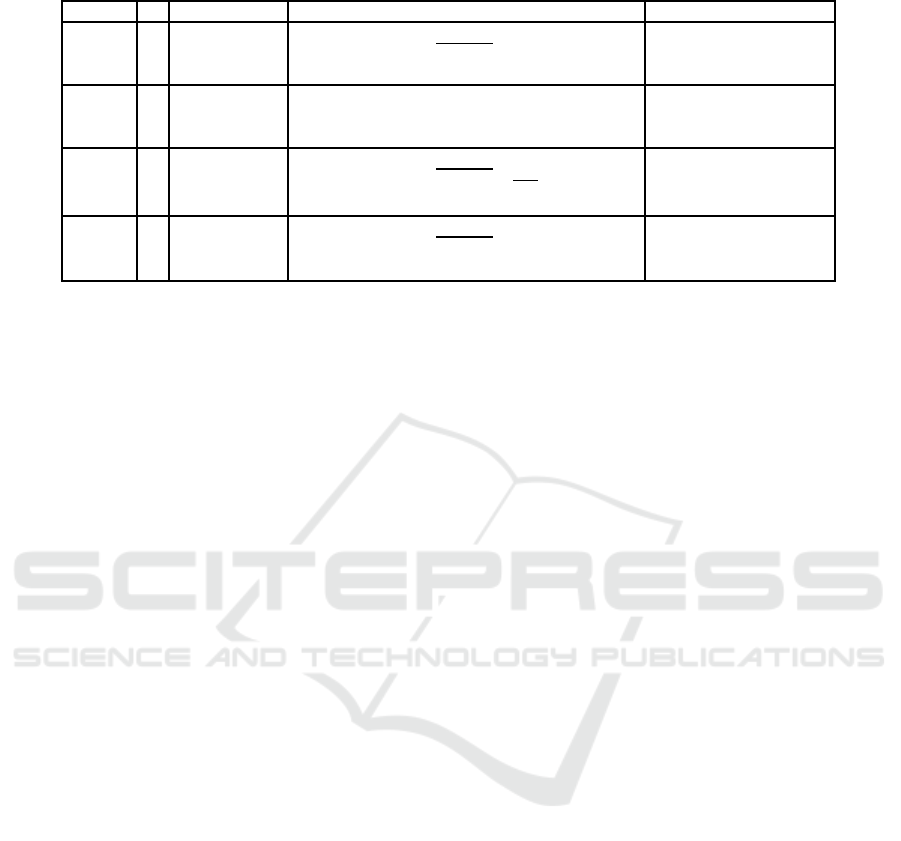
Table 1: Test problems used in this paper.
Problem n Variable range Objective functions Pareto-optimal solutions
ZDT1 30 [0,1] f
1
(x) = x
1
x
1
∈ [0,1]
f
2
(x) = g(x)[1−
p
x
1
/g(x)] x
i
= 0,i = 2,··· ,n
g(x) = 1+9(
∑
n
i=2
x
i
)/(n−1)
ZDT2 30 [0,1] f
1
(x) = x
1
x
1
∈ [0,1]
f
2
(x) = g(x)[1− (x
1
/g(x))
2
] x
i
= 0,i = 2,··· ,n
g(x) = 1+9(
∑
n
i=2
x
i
)/(n−1)
ZDT3 30 [0,1] f
1
(x) = x
1
x
1
∈ [0,1]
f
2
(x) = g(x)[1−
p
x
1
/g(x) −
x
1
g(x)
sin(10πx
1
)]
x
i
= 0,i = 2,··· ,n
g(x) = 1+9(
∑
n
i=2
x
i
)/(n−1)
ZDT4 10 x
1
∈ [0,1] f
1
(x) = x
1
x
1
∈ [0,1]
x
i
∈ [−5,5], f
2
(x) = g(x)[1−
p
x
1
/g(x)] x
i
= 0,i = 2,··· ,n
i = 2, ··· ,n g(x) = 1+ 10(n− 1) +
∑
n
i=2
x
2
i
− 10cos(4πx
i
)
a n-dimensional random vector in which the values
of components Bt to Bt + L − 1 are uniformly dis-
tributed random numbers between −1 and 1, and
those of other components are 0. “◦” is an opera-
tor, called Hadamard product, which represents mul-
tiplying the corresponding elements of two vectors.
Take 3-dimensional vectors for an example, if a =
(a
1
,a
2
,a
3
), b = (b
1
,b
2
,b
3
), then D = A ◦ B = (a
1
×
b
1
,a
2
× b
2
,a
3
× b
3
). If the new solution x
i
new
( j,k,l)
is dominated by the old solution x
i
( j,k,l), x
i
( j,k,l) is
kept; otherwise, x
i
new
( j,k,l) replaces x
i
( j,k,l). With
this communication mechanism, each bacterium in
the population P searches for the nondominated so-
lutions under the guidance of the superior individuals
in the archive A. Thus, the bacterial population are
likely to quickly converge to the global Pareto front.
3.4 Reproduction
The bacteria grow longer with the increasing of the
nutrients absorbed. The more nutrients a bacterium
gets, the healthier it is. Under appropriate conditions,
some of bacteria in a population who are healthy
enough will asexually split into two bacteria, and
the other ones will die. Essentially, the reproduction
mechanism is to generate newpopulation based on the
superior individuals in current population. In the pro-
posed MABFO algorithm, the reproduction operator
selects N1 superior individuals from the joint popu-
lation of the current population P (|P| = N1) and the
external archive A and produces a new population P
by copying these selected N1 superior individuals to
P. The way to select N1 superior individuals is as fol-
lows: first sort the joint population of P and A into
different nondominated levels as reference (?) did.
The first nondominated level F
1
contains all the cur-
rent nondominated solutions. Then we pick out solu-
tions in ascending order of the nondominated level to
form a new group R until the size of R would be equal
or larger than N1 if we incorporate the tth nondomi-
nated level F
t
. In the first case, the size of R is exactly
equal to N1 if putting F
t
into R, just put it into R and
this select step is completed. In the second case, the
size of R is larger than N1 if F
t
is loaded into R. In
this case, we need to first pick out N1 − |R| solutions
from F
r
based on the crowding distance according to
(8) and then put them into R. In this way, |R| is also
exactly the same with N1. Next, R is used to update
the old population P by copying it to P. Thus, the
new population P which contains the best individuals
is obtained. Such reproduction mechanism follows
the rule of survival of the fittest, which plays a role of
transmitting good information among the whole pop-
ulation and speeding up the convergence.
3.5 Elimination-and-dispersal
With changes to the local environment in which a
population of bacteria lives, all of the bacteria in
this region may be killed, or a group of bacteria
may be dispersed into a new environment to find bet-
ter food sources. To simulate this phenomenon, an
elimination-and-dispersal step is taken in BFO after
N
re
reproduction steps. Each bacterium in the popula-
tion may be eliminated or dispersed to a new location
with a given probability P
ed
. The rule is shown in the
following:
x =
x
′
, if r < P
ed
x, otherwise
, (10)
where r is a random number uniformly distributed in
[0,1], x is the current solution associated with a bac-
terium, x
′
is a new solution generated at random in n-
dimension search space. That is, for each bacterium,
if the number generated randomly is smaller than P
ed
,
it will move to a new random solution, otherwise, it
will keep the original solution unchanged. This mech-
anism is helpful to escape from local Pareto optima
ICPRAM 2016 - International Conference on Pattern Recognition Applications and Methods
188
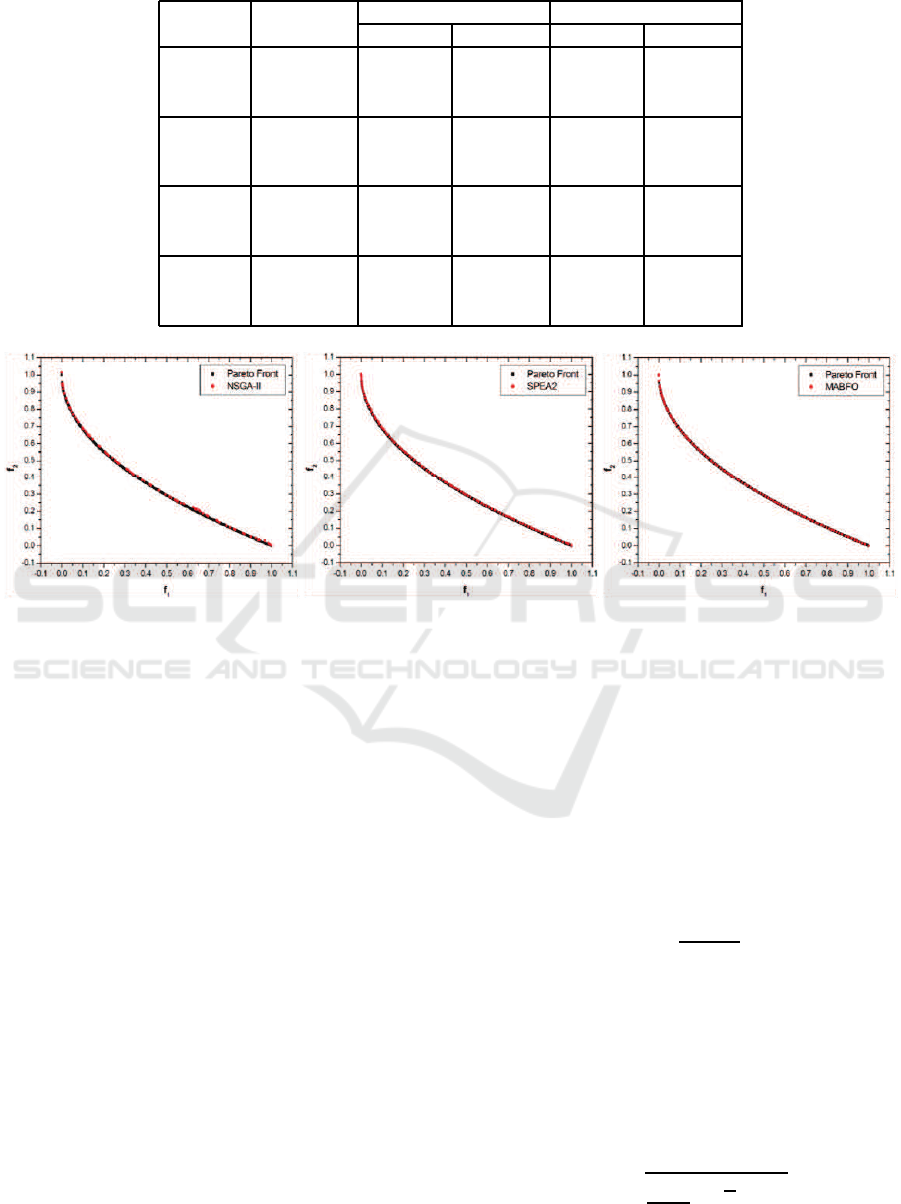
Table 2: Performance comparisons of NSGA-II, SPEA2 and MABFO on four test problems.
Problem Algorithm
GD SP
Average Std. Dev. Average Std. Dev.
NSGA-II 5.10e-04 1.24e-04 9.14e-03 1.13e-03
ZDT1 SPEA2 3.76e-04 6.36e-05 3.28e-03 3.80e-04
MABFO 1.96e-04 4.55e-05 3.24e-03 5.48e-04
NSGA-II 3.66e-04 2.80e-04 1.02e-02 3.09e-03
ZDT2 SPEA2 1.74e-04 2.83e-05 3.15e-03 3.62e-04
MABFO 9.26e-05 4.42e-06 3.04e-03 2.98e-04
NSGA-II 5.22e-04 2.46e-04 1.02e-02 1.50e-03
ZDT3 SPEA2 3.73e-04 1.10e-04 4.42e-03 5.79e-04
MABFO 1.59e-04 1.24e-05 4.85e-03 1.11e-03
NSGA-II 2.45e-03 9.01e-04 1.33e-02 5.07e-03
ZDT4 SPEA2 1.88e-03 7.50e-04 4.17e-03 1.25e-03
MABFO 2.34e-04 2.20e-04 2.40e-03 1.52e-03
Figure 1: Pareto fronts produced by NSGA-II (left), SPEA2 (middle) and MABFO (right) for the ZDT1 test problem.
and to explore the global Pareto optima in the search
space.
4 EXPERIMENTS
In this section, we will compare the proposed
MABFO algorithm with two state-of-the-art algo-
rithms NSGA-II and SPEA2. The experimental plat-
form is a PC with Inter(R) Core(TM) i5- 3470 CPU
3.20GHz, 4GB RAM and Windows 7, and all the al-
gorithms are implemented using C++ language.
4.1 Test Functions and Evaluation
Metrics
We choose four test problems ZDT1, ZDT2, ZDT3
and ZDT4 which are suggested by Zizler and com-
monly used in a number of significant past studies.
All the test problems have two objective functions and
have not any constraint. Table 1 lists these test prob-
lems and also provides the number of variables, their
ranges, the Pareto-optimal solutions for each prob-
lem.
In general, there are two issues to consider for as-
sessing a method in multiobjective optimization area:
(1) the convergence of the obtained Pareto front to-
ward the true Pareto front and (2) spread of the ob-
tained solutions. Based on this notion, we adopt two
common metrics: Generational distance (GD) (Van
Veldhuizen D A, Lamont G B, 1998) and Space
(SP) (Schott J R, 1995).
GD measures the distance between the Pareto
front found so far and the true Pareto front. It is de-
fined as:
GD =
∑
n
i=1
d
2
i
n
, (11)
where n is the number of members in the Pareto front
found so far, d
i
is the Euclidean distance between the
ith member of the Pareto front found and the nearest
member of the true Pareto front. The smaller the value
of this metric, the nearer the Pareto front found so far
to the true Pareto front.
SP judges how well the Pareto front found so far
distributed and is formulated as follows:
SP =
s
1
n− 1
n
∑
i=1
(
d − d
i
)
2
, (12)
Multiobjective Bacterial Foraging Optimization using Archive Strategy
189
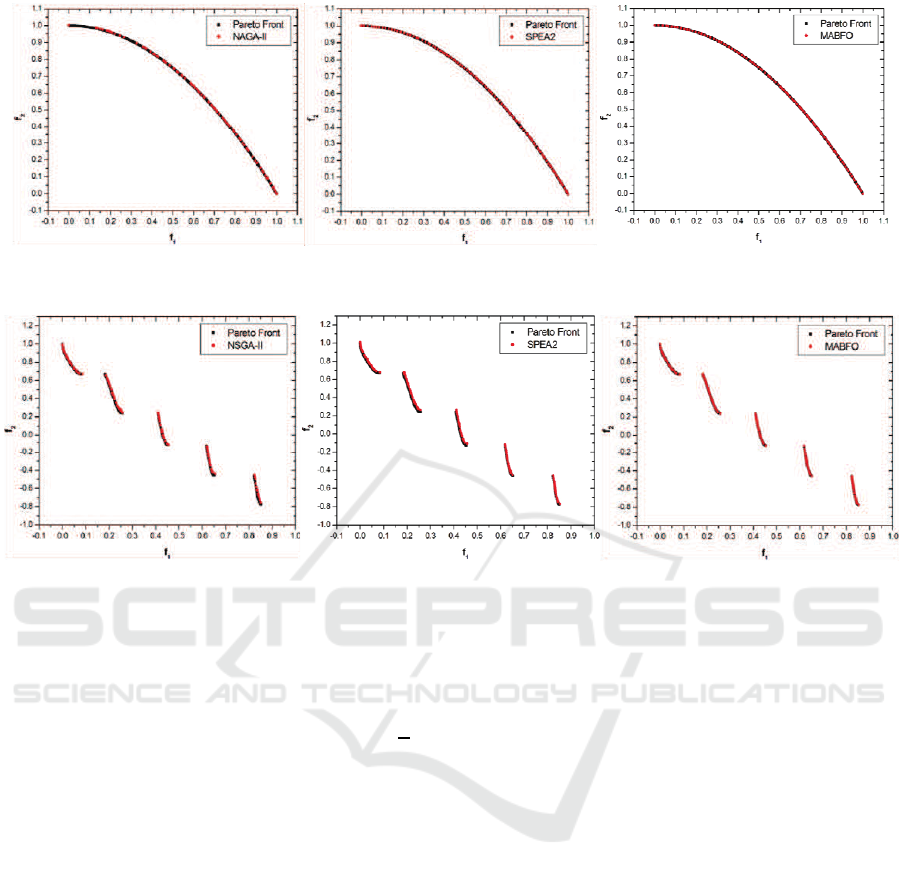
Figure 2: Pareto fronts produced by NSGA-II (left), SPEA2 (middle) and MABFO (right) for the ZDT2 test problem.
Figure 3: Pareto fronts produced by NSGA-II (left), SPEA2 (middle) and MABFO (right) for the ZDT3 test problem.
where n is the number of members in the Pareto
front found so far, d
i
is the minimum Manhatton dis-
tance between the ith member and other members
in the Pareto front found and d
i
= min
j
(| f
1
(x
i
) −
f
1
(x
j
)| + | f
2
(x
i
) − f
2
(x
j
)| + ··· + | f
m
(x
i
) − f
m
(x
j
)|),
j = 1,2,· ·· ,n. m is the number of the objectives.
d is
the average value of all d
i
. The smaller the value of
this metric, the more uniform the Pareto front found
is distributed.
4.2 Results and Analysis
In the experiments, each algorithm was tested 30 in-
dependent runs on each test problem. To be fair,
the population sizes and the elite archive sizes of the
three algorithms were set to 100. The generations of
NSGA-II and SPEA2 were set to 500. For the other
parameters in NSGA-II and SPEA2 algorithms, we
tried to use identical settings as suggested in the orig-
inal studies. For the left parameters in the proposed
MABFO algorithm, we didn’t make any serious at-
tempt to find the best settings and only chose a reason-
able set of values: N
s
= 4, N
c
= 10, N
re
= 25, N
ed
= 2,
P
ed
= 0.2 and L = 0.4× n.
Table 2 provides the average results (Average) and
standard deviations (Std. Dev.) with respect to the
two metrics GD and SP. The best average results with
respect to each metric are shown in bold. It can
be seen from the table, the average performance of
MABFO is the best with respect to the GD and SP
metrics on ZDT1, ZDT3 and ZDT4. As for ZDT2,
MABFO achieves the best result in term of GD and is
only slightly worse than SPEA2 in term of SP, but it
has the smallest deviation with respect to SP.
Figures 1-4 show the Pareto fronts obtained by
NSGA-II, SPEA2 and our MABFO algorithm on the
four test problems–ZDT1, ZDT2, ZDT3 and ZDT4,
respectively. The Pareto fronts displayed correspond
to the median results over 30 runs with respect to
the GD metric. From these figures, we can see
that the three algorithms are able to cover the entire
Pareto fronts on ZDT1, ZDT2 and ZDT3. But our
MABFO algorithm produces better-distributed and
higher-quality Pareto front on these three test prob-
lems, especially on the ZDT3 problem. For the ZDT4
problem, both NSGA-II and SPEA2 fail to cover the
true Pareto front, whereas our MABFO algorithm
successfully does it. Through the comparison with
two best EMO algorithms NSGA-II and SPEA2, our
MABFO algorithm is a viable alternative to solve
MOPs.
ICPRAM 2016 - International Conference on Pattern Recognition Applications and Methods
190
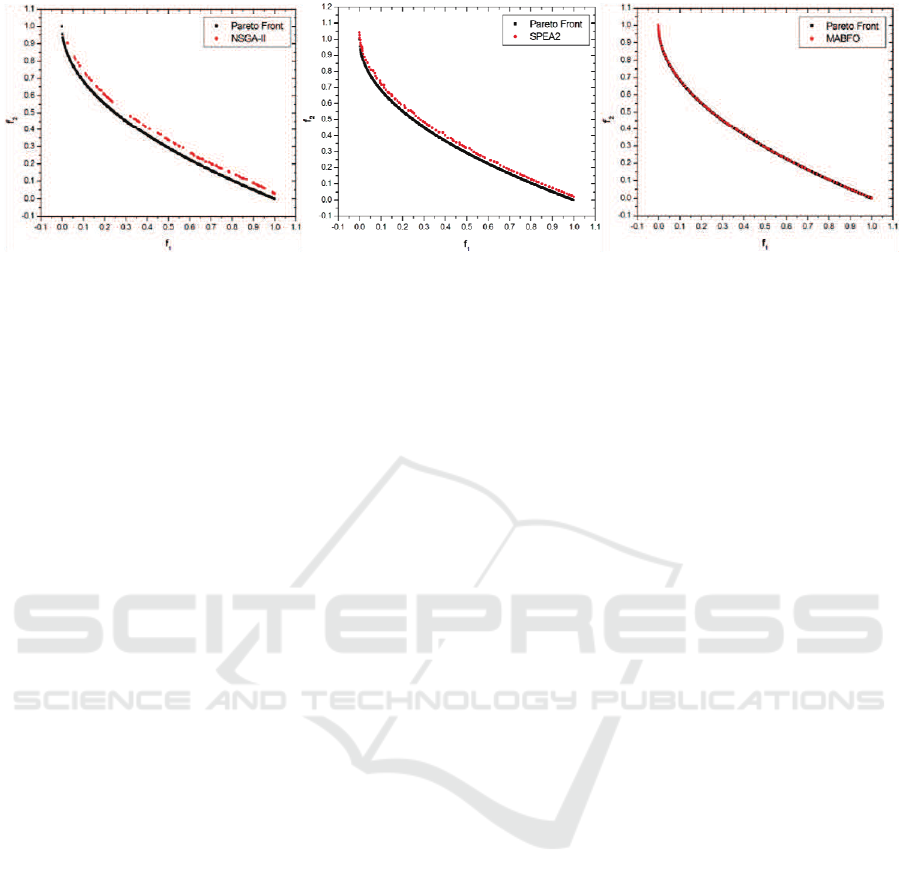
Figure 4: Pareto fronts produced by NSGA-II (left), SPEA2 (middle) and MABFO (right) for the ZDT4 test problem
5 CONCLUSIONS
The MOP is a very important research topic in both
science and engineering communities. In recent
years, researchers are interested in using some new
bio-inspired optimization models to solve MOPs. Al-
though several methods based on BFO have been
proposed for handling MOPs. But to the best of
our knowledge, the current methods based on BFO
havenot incorporated the elitist strategy which always
plays an important role in getting global Pareto front.
In this paper, we present a new algorithm based on
archive bacterial foraging optimization for multiob-
jective optimization (called MABFO). MABFO sim-
ulates four biological mechanisms: chemotaxis, con-
jugation, reproduction and elimination-and-dispersal.
The implements of these mechanisms are different
from other methods based on BFO. More impor-
tant, MABFO incorporates an external archive to
save the nondominated solutions previously found
and maintains the diversity of the nondominated so-
lutions found based on the crowding distance. To
demonstrate the performance of MABFO algorithm,
we compared it with two best methods known to date
(NSGA-II and SPEA2) on four test problems, the re-
sults indicate that MABFO is a promising alternative
since it has the best average performance with respect
to two metrics in most cases.
In the future, we will give an detailed analysis
of the algorithm, make a comprehensive testing on
more test problems and continue to explore more ef-
fective diversity preservation strategies to better cover
the global Pareto front. We also hope to extend this
algorithm so that it can handle dynamic functions.
ACKNOWLEDGEMENTS
This work is partly supported by the NSFC
Research Program (61375059, 61332016), the
National 973 Key Basic Research Program of
China (2014CB744601), Specialized Research Fund
for the Doctoral Program of Higher Education
(20121103110031), and the Beijing Municipal Edu-
cation Research Plan key project (Beijing Municipal
Fund Class B) (KZ201410005004).
REFERENCES
A Balassubramaniam, Memeber, IEEE, P Lio (2013).
Multi-hop conjugation based bacteria nanonetworks.
In IEEE Trans. on Nanobiosci., volume 12, pages 47–
59.
Agrawal V, Sharma H, Bansal J C (2011). Bacterial for-
aging optimization: A survey. In SocProS’2011, Pro-
ceedings of the International Conference on Soft Com-
puting for Problem Solving, pages 227–242, India.
C Perales-Grav´an, R Lahoz-Beltra (2008). An am radio
receiver designed with a genetic algorithm algorithm
based on a bacterial conjugation genetic operator. In
IEEE Trans. Evol. Comput., volume 12, pages 129–
142.
Coello C A C, Pulido G T, Lechuga M S (2004). Handling
multiple objectives with particle swarm optimization.
In IEEE Transactions on Evolutionary Computation,
volume 8, pages 256–279.
Coello Coello CA, Van Veldhuizen DA, Lamont GB (2007).
Evolutionary Algorithms for Solving Multi-Objective
Problems. New York, springer-verlag edition.
Corne D W, Jerram N R, Knowles J D, et al (2001). PESA-
II: Region-based selection in evolutionary multiobjec-
tive optimization. In GECCO’2001, Proceedings of
the Genetic and Evolutionary Computation Confer-
ence, pages 283–290.
Deb (2001). Multi-objective Optimization using Evoulu-
tionary Algorithms. Chichester, john wiley & sons
edition.
Multiobjective Bacterial Foraging Optimization using Archive Strategy
191

Deb K, Pratap A, Agarwal S, et al (2002). A fast and
elitist multiobjective genetic algorithm: NSGA-II.
In IEEE Transactions on Evolutionary Computation,
volume 6, pages 182–197.
E Zitzler, M Laumanns, L Thiele (2002). SPEA2: Improv-
ing the strength pareto evolutionary algorithm. In Evo-
lutionary Methods for Design, Optimization and Con-
trol with Applications to Industrial Problems, pages
95–100, Berlin.
Guzm´an M A, Delgado A, De Carvalho J (2010). A novel
multiobjective optimization algorithm based on bacte-
rial chemotaxis. In Engineering Applications of Arti-
ficial Intelligence, volume 23.
K.M. Passino (2002). Biomimicry of bacterial foraging for
distributed optimization and control. In Control Sys-
tems, volume 22, pages 52–67.
Niu B, Wang H, Tan L, et al (2012). Multi-objective opti-
mization using bfo algorithm. In Bio-Inspired Com-
puting and Applications, pages 582–587.
Niu B, Wang H, Wang J, et al (2013). Multi-objective bac-
terial foraging optimization. In Neurocomputing, vol-
ume 116, pages 336–345.
Panigrahi B K, Pandi V R, Das S, et al (2010). Multiobjec-
tive fuzzy dominance based bacterial foraging algo-
rithm to solve economic emission dispatch problem.
In Energy, volume 35, pages 227–242.
Parks G T, Miller I (1998). Selective breeding in a multiob-
jective genetic algorithm. In Parallel Problem Solving
From NaturełPPSN V, pages 250–259.
S Dasgipta, S Das, A Abraham and A Biswas (2009). Adap-
tive computational chemotaxis in bacterial foraging
optimiztion: An analysis. In IEEE Transactions on
Evolutionary Computation, volume 13, pages 919–
941.
Schott J R (1995). Fault tolerant design using single and
multicriteria genetic algorithm optimization. In MS
Thesis, Massachusetts institute of Technology, Cam-
bridge.
Tripathi P K, Bandyopadhyay S, Pal S K (2007). Multi-
objective particle swarm optimization with time vari-
ant inertia and acceleration coefficients. In Informa-
tion Sciences, volume 177, pages 5033–5049.
Van Veldhuizen D A, Lamont G B (1998). Multiobjec-
tive evolutionary algorithm research: A history and
analysis. In Technical Report TR-98-03, Department
of Electrical and Computer Engineering, Graduate
School of Engineering, Air Force Institute of Technol-
ogy, Ohio.
X Li (2003). A non-dominated sorting particle swarm
optimizer for multiobjective optimization. In
GECCO’2003, Proceedings of Genetic and Evolu-
tionary Computation, pages 37–48.
Zitzler E, Deb K, Thiele L (2000). Comparison of multiob-
jective evolutionary algorithms: Empirical results. In
Evolutionary computation, volume 8, pages 173–195.
ICPRAM 2016 - International Conference on Pattern Recognition Applications and Methods
192
