
Sparse-Reduced Computation
Enabling Mining of Massively-large Data Sets
Philipp Baumann, Dorit S. Hochbaum and Quico Spaen
IEOR Department, University of California, Berkeley, Etcheverry Hall, CA 94720, U.S.A.
Keywords:
Large-Scale Data Mining, Classification, Data Reduction, Supervised Normalized Cut.
Abstract:
Machine learning techniques that rely on pairwise similarities have proven to be leading algorithms for clas-
sification. Despite their good and robust performance, similarity-based techniques are rarely chosen for large-
scale data mining because the time required to compute all pairwise similarities grows quadratically with the
size of the data set. To address this issue of scalability, we introduced a method called sparse computation,
which efficiently generates a sparse similarity matrix that contains only significant similarities. Sparse compu-
tation achieves significant reductions in running time with minimal and often no loss in accuracy. However, for
massively-large data sets even such a sparse similarity matrix may lead to considerable running times. In this
paper, we propose an extension of sparse computation called sparse-reduced computation that not only avoids
computing very low similarities but also avoids computing similarities between highly-similar or identical ob-
jects by compressing them to a single object. Our computational results show that sparse-reduced computation
allows highly-accurate classification of data sets with millions of objects in seconds.
1 INTRODUCTION
In a recent computational comparison (Baumann
et al., 2015), the K-nearest neighbor algorithm, two
variants of supervised normalized cut, and support
vector machines (SVMs) with RBF kernels were the
leading classification techniques in terms of accuracy
and robustness. The success of these techniques is
attributed, in part, to their reliance on pairwise simi-
larities between objects in the data set. Pairwise simi-
larities can be defined flexibly and are able to capture
non-linear and transitive relationships in the data set.
Considering transitive relationships improves the per-
formance of supervised learning algorithms in cases
where an unlabeled object is only similar to a labeled
object through a transitive chain of similarities that in-
volves other unlabeled objects (Kawaji et al., 2004).
Computing pairwise similarities, however, poses a
challenge in terms of scalability, as the number of
pairwise similarities between objects grows quadrat-
ically in the number of objects in the data set. For
massively-large data sets, it is prohibitive to compute
and store all pairwise similarities.
Various methods have been proposed that sparsify
a complete similarity matrix while preserving specific
matrix properties (Arora et al., 2006; Spielman and
Teng, 2011; Jhurani, 2013). A sparse similarity ma-
trix requires less memory and allows faster classifi-
cation as the running time of the algorithms depends
on the number of non-zero entries in the similar-
ity matrix. However, existing sparsification methods
require as input the complete similarity matrix and
are thus not applicable to large-scale data sets. Re-
cently, (Hochbaum and Baumann, 2014) introduced
a methodology called sparse computation that gen-
erates a sparse similarity matrix without having to
compute the complete similarity matrix first. Sparse
computation significantly reduces the running time of
similarity-based classifiers without affecting their ac-
curacy. In sparse computation the data is efficiently
projected onto a low-dimensional space using a sam-
pling variant of principal component analysis. The
low-dimensional space is then subdivided into grid
blocks and similarities are only computed between
objects in the same or in neighboring grid blocks. The
density of the similarity matrix can be controlled by
varying the grid resolution. A higher grid resolution
leads to a sparser similarity matrix.
Although sparse computation works well in gen-
eral, it may occur that large groups of highly-similar
or identical objects project to the same grid block even
when the grid resolution is high. The computation of
similarities between these objects is unnecessary as
they often belong to the same class. In massively-
large data sets, large numbers of highly-similar ob-
jects are particularly common. The grid block struc-
ture created in sparse computation reveals the exis-
tence of such highly-similar objects in the data set.
224
Baumann, P., Hochbaum, D. and Spaen, Q.
Sparse-Reduced Computation - Enabling Mining of Massively-large Data Sets.
DOI: 10.5220/0005690402240231
In Proceedings of the 5th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2016), pages 224-231
ISBN: 978-989-758-173-1
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

In this paper we propose an extension of sparse
computation called sparse-reduced computation that
avoids the computation of similarities between
highly-similar and identical objects. The method
builds on sparse computation by using the grid block
structure to identify highly-similar and identical ob-
jects efficiently. In each grid block, the objects are
replaced by a small number of representatives. The
similarities are then computed only between represen-
tatives in the same and in neighboring blocks. The
resulting similarity matrix is not only sparse but also
smaller in size due to the consolidation of objects.
Sparse-reduced computation can be used to speed
up any machine learning algorithm. Although our
focus here is on similarity-based methods, sparse-
reduced computation also applies to non-similarity
based methods such as artificial neural networks, lo-
gistic regression, and K-means algorithms. In ad-
dition to enabling classification and clustering in
massively-large data sets, the method can be used as a
data compression method to represent data sets com-
pactly with minor loss of relevant information.
We evaluate sparse-reduced computation on four
real-world and one artificial benchmark data sets con-
taining up to 10 million objects. Sparse-reduced com-
putation delivers highly-accurate classification at very
low computational cost for most of the data sets.
The paper is structured as follows. In Section 2,
we review related data-reduction techniques. In Sec-
tion 3, we present the method of sparse-reduced com-
putation. In Section 4, we describe two similarity-
based machine learning techniques. In Section 5, we
explain the design and present the results of our com-
putational analysis. In Section 6, we conclude the pa-
per and provide directions for future research.
2 EXISTING SCALABLE DATA
MINING APPROACHES
Existing scalable data mining approaches can be
broadly divided into three categories: Algorithmic
modification approaches, problem decomposition ap-
proaches, and problem reduction approaches.
Algorithmic Modification Approaches: These ap-
proaches are designed to speed up a specific algo-
rithm. In particular, various algorithmic improve-
ments have been developed for SVM as they show
good performance for small data sets but suffer from
limited scalability. Some of these improvements
speed up the algorithms for solving the SVM opti-
mization problem (Shalev-Shwartz et al., 2011; Hsieh
et al., 2008). Another approach is to solve the
SVM optimization problem approximately as done by
(Tsang et al., 2005) in core vector machines. These
approaches are tailored to SVM only and lack broad
applicability.
Problem Decomposition Approaches: These ap-
proaches divide the data set into smaller subsets, train
a classifier on each of these subsets separately, and
combine the predictions of the local classifiers to ob-
tain an aggregated prediction. (Rida et al., 1999; Col-
lobert et al., 2002; Chang et al., 2010) propose gen-
eral strategies for decomposing the data set and com-
bining the predictions of the local classifiers. Other
approaches are tailored to specific algorithms. (Graf
et al., 2004) train multiple SVMs on random subsets
to create a cascade of SVMs. (Segata and Blanzieri,
2010) proposes to use local SVMs for each training
object based on its k-nearest neighbors. None of these
problem decomposition approaches is applicable to
massively-large data sets because either the decompo-
sition of the data sets or the combination of the local
classifiers takes too much time.
Problem Reduction Approaches: These ap-
proaches transform a large-scale problem instance
into an instance that can be solved quickly. They can
be further divided into two subcategories: sampling
approaches and representative approaches.
The idea of sampling approaches is to randomly
select a small subset of the training objects and per-
form the tuning of the algorithm on this subset.
This bears the risk of excluding valuable information
which may negatively affect the performance (Provost
and Kolluri, 1999).
Representative approaches discern the main struc-
ture in the data set by identifying a small set of repre-
sentatives, so that the machine learning task can then
be performed on the set of representatives. (Andrews
and Fox, 2007) propose to cluster the data set with
the k-means algorithm and select a representative for
each cluster. However, applying K-means on a large
data set is itself computationally expensive. (Yu et al.,
2003) propose a tree-based clustering model to itera-
tively train the SVM on the cluster representatives.
Although this approach is fast, it requires a specific
implementation for any algorithm it is combined with.
We next introduce a novel scalable data mining
approach that is based on representatives. In contrast
to existing scalable data mining approaches our ap-
proach is fast and can be used with any machine learn-
ing technique.
Sparse-Reduced Computation - Enabling Mining of Massively-large Data Sets
225

3 SPARSE-REDUCED
COMPUTATION
Sparse-reduced computation is an extension of sparse
computation introduced in (Hochbaum and Baumann,
2014). The idea of sparse computation is to avoid
the computation of very small similarities as they are
unlikely to affect the classification result. Sparse-
reduced computation not only avoids the computation
of very small similarities, but also avoids the com-
putation of similarities between highly-similar and
identical objects. Intuitively, sparse-reduced compu-
tation not only “rounds” very small similarities to zero
but it also “rounds” very large similarities to one.
Sparse-reduced computation consists of the four steps
of data projection, space partitioning, data reduction,
and similarity computation.
Data Projection: The first step in sparse-reduced
computation coincides with what is done in sparse
computation. The d-dimensional data set is projected
onto a p-dimensional space, where p d. This is im-
plemented using a sampling variant of principal com-
ponent analysis called Approximate PCA (Hochbaum
and Baumann, 2014). Approximate PCA computes
principal components that are very similar to those
computed by exact PCA as shown in (Drineas et al.,
2006), yet requires drastically reduced running time.
Approximate PCA is based on a technique devised by
(Drineas et al., 2006). The idea is to compute ex-
act PCA on a submatrix W of the original matrix A.
The submatrix W is generated by random selection
of columns and rows of A with probabilities propor-
tional to the `
2
norms of the respective column or row
vectors.
Space Partitioning: Once all objects are mapped
into the p-dimensional space, the next step is to subdi-
vide, in each dimension, the subspace occupied by the
objects into k intervals of equal length. This partitions
the p-dimensional space into k
p
grid blocks. Using
a uniform value of k for all dimensions allows us to
control the total number of grid blocks with a single
parameter. In the following, parameter k is referred
to as the grid resolution. Each object is assigned to a
single block based on the respective intervals in which
its p coordinates fall.
Data Reduction: The grid is then used to reduce
the size of the data set by replacing so-called δ-
identical objects by a single representative. Let δ =
1
κ
for κ ∈ N. In order to identify δ-identical objects
we subdivide each block into κ
p
sub-blocks. The
x
y
z
Figure 1: Data reduction with resolution k = 3 and κ = 1.
As shown in the magnified block, the negative training ob-
jects (gray), the positive training object (white fill), and the
testing objects (black) are each replaced by a single repre-
sentative.
sub-blocks are obtained by partitioning the grid block
along each dimension into κ intervals of equal length.
For each sub-block, we replace objects of the same
type (negative training objects, positive training ob-
jects and testing objects) by a single representative.
For example, if κ = 1, then all objects of the same
type that fall in the same grid block are considered
δ-identical and are thus grouped together. If κ = 2
for a three-dimensional space, then the grid block is
split into 2
3
= 8 sub-blocks and the replacement of
objects by representatives is done for each sub-block
separately. Different values for κ can be selected for
different blocks to account for an unequal distribu-
tion of the data. The representatives are computed as
the center of gravity of the corresponding objects and
have a multiplicity weight equivalent to the number of
objects they represent. Note that a representative may
represent a single object. Figure 1 illustrates the data
reduction step for a three-dimensional grid and κ = 1.
Similarity Computation: The sparse similarity
matrix on the representatives is computed based on
the concept of grid neighborhoods. This concept
is borrowed from image segmentation where simi-
larities are computed only between adjacent pixels
(Hochbaum et al., 2013). Here, we use the space par-
titioning and first consider all representatives in the
same block to be adjacent and thus have their similar-
ities computed. Then, adjacent blocks are identified
and the similarities between representatives in those
blocks are computed. Two blocks are adjacent if they
are within a one-interval distance from each other in
each dimension (the `
max
metric). Hence, for each
block, there are up to 3
p
−1 adjacent blocks. The sim-
ilarities are computed in the original d-dimensional
space. For very high-dimensional data sets, the
similarities could also be computed in the low p-
dimensional space. A finer grid resolution (higher
value of k) generally leads to a lower density of the
ICPRAM 2016 - International Conference on Pattern Recognition Applications and Methods
226

similarity matrix. Notice that for k = 2 all representa-
tives are neighbors of each other, and thus we get the
complete similarity matrix. The set of representatives
and the generated similarity matrix constitutes the in-
put to the classification algorithms which we describe
in the next section. The class labels that are assigned
to representatives will be passed on to each of the ob-
jects that they represent.
4 SIMILARITY-BASED MACHINE
LEARNING
In this section, we present two similarity-based ma-
chine learning techniques as binary classifiers that as-
sign a set of testing objects to either the positive or
the negative class based on a set of training objects.
The two techniques are the K-nearest neighbor al-
gorithm and the supervised normalized cut algorithm
(Hochbaum, 2010).
K-nearest Neighbor Algorithm (KNN): The KNN
algorithm (Fix and Hodges, 1951) finds the K nearest
training objects to a testing object and then assigns the
predominant class among those K neighbors. We use
the euclidean metric to compute distances between
objects and the nearest training object is used to break
ties. When KNN is applied with sparse-reduced data,
we consider the multiplicity weight of the representa-
tives to determine the K nearest training representa-
tives. For example, if K = 3 and the nearest training
representative is positive and has a multiplicity weight
of 2 and the second nearest training representative is
negative and has a multiplicity weight of 5, the test-
ing object is assigned to the positive class. A testing
object is assigned to the negative class if all similar-
ities to training objects are zero. In the experimental
analysis we treat K as a tuning parameter.
Supervised Normalized Cut (SNC): SNC is a su-
pervised version of HNC (Hochbaum’s Normalized
Cut), which is a variant of normalized cut that is
solved efficiently (Hochbaum, 2010). HNC is defined
on an undirected graph G = (V,E), where V denotes
the set of nodes and E the set of edges. A weight w
i j
is
associated with each edge [i, j] ∈ E. A bi-partition of
a graph is called a cut, (S,
¯
S) = {[i, j] |i ∈ S, j ∈
¯
S},
where
¯
S = V \ S. The capacity of a cut (S,
¯
S) is the
sum of weights of edges with one endpoint in S and
the other in
¯
S:
C(S,
¯
S) =
∑
i∈S, j∈
¯
S,[i, j]∈E
w
i j
.
In particular, the capacity of a set, S ⊂ V , is the sum
of edge weights within the set S,
C(S,S) =
∑
i, j∈S,[i, j]∈E
w
i j
.
In the context of classification the nodes of the
graph correspond to objects and the edge weights w
i j
quantify the similarity between the respective vec-
tors of attribute values associated with nodes i and j.
Higher similarity is associated with higher weights.
The goal of one variant of HNC is to find a cluster
that minimizes a linear combination of two criteria.
One criterion is to maximize the total similarity of the
objects within the cluster (the intra-similarity). The
second criterion is to minimize the similarity between
the cluster and its complement (the inter-similarity).
A linear combination of the two criteria is minimized:
min
/
0⊂S⊂V
C(S,
¯
S) − λC(S, S). (1)
The relative weighting parameter λ is one of the tun-
ing parameters. The input graph contains classified
nodes (training data) that refer to objects for which the
class label (either positive or negative) is known and
unclassified nodes that refer to objects for which the
class label is unknown. SNC is derived from HNC by
assigning all classified nodes with a positive label to
the set S and all classified nodes with a negative label
to the set
¯
S. The goal is then to assign the unclassified
nodes to either the set S or the set
¯
S. For a given value
of λ, the optimization problem is solved with a min-
imum cut procedure in polynomial time (Hochbaum,
2010; Hochbaum et al., 2013).
We implement SNC with exponential similarity
weights. The exponential similarity between object
i and j is quantified based on the respective vectors of
attribute values x
i
and x
j
by:
w
i j
= e
−ε||x
i
−x
j
||
2
,
where parameter ε represents a scaling factor. The
exponential similarity function is commonly used in
image segmentation, spectral clustering, and classi-
fication. When SNC is applied with sparse-reduced
computation, we multiply each similarity value by the
product of the weights of the two corresponding rep-
resentatives. There are two tuning parameters: the
relative weighting parameter of the two objectives,
λ, and the scaling factor of the exponential weights,
ε. The minimum cut problems are solved with the
MATLAB implementation of the HPF pseudoflow al-
gorithm version 3.23 of (Chandran and Hochbaum,
2009) that was presented in (Hochbaum, 2008).
Sparse-Reduced Computation - Enabling Mining of Massively-large Data Sets
227

5 EXPERIMENTAL ANALYSIS
In this section, we apply both sparse computation
and sparse-reduced computation with two similarity-
based classifiers to five data sets. The results, of com-
parable quality, for two additional data sets and a vari-
ant of SNC are not reported here due to a lack of
space. A comparison with existing sparsification ap-
proaches is not possible because these approaches re-
quire to first compute the full similarity matrix, which
would exceed the memory capacity of our machine.
In Sections 5.1–5.3, we describe the data sets, explain
the experimental design and report the numerical re-
sults, respectively.
5.1 Data Sets
We select four real-world data sets from the UCI Ma-
chine Learning Repository (Asuncion and Newman,
2007) and one artificial data sets from previous stud-
ies (Breiman, 1996; Dong et al., 2005). We substi-
tuted categorical attributes by a binary attribute for
each category. In the following, we briefly describe
each data set and mention further modifications that
are made. The characteristics of the adjusted data sets
are summarized in Table 1.
In the Bag of Words (BOW2) data set, the objects
are text documents from two different sources (New
York Times articles and PubMed abstracts). A docu-
ment is represented as a so-called bag of words, i.e. a
set of vocabulary words. For each document, an vec-
tor indicates the number of occurrences of each word
in the document. We treated New York Times articles
as positives.
The data set Covertype (COV) contains carto-
graphic characteristics of forest cells in northern Col-
orado. There are seven different cover types which
are labeled 1 to 7. Following (Caruana and Niculescu-
Mizil, 2006), we treat type 1 as the positive class and
types 2 to 7 as the negative class.
The data set KDDCup99 (KDD) is the full data
set from the KDD Cup 1999 which contains close to 5
million records of connections to a computer network.
Each connection is labeled as either normal, or as an
attack. We treat attacks as the positive class.
In the data set Record Linkage Comparison Pat-
terns (RLC), the objects are comparison patterns of
pairs of patient records. We substitute the missing
values with value 0 and introduce an additional bi-
nary attribute to indicate missing values. The goal
is to classify the comparison patterns as matches (the
corresponding records refer to same patient) or non-
matches. Matches are treated as positive class.
The data set Ringnorm (RNG) is an artificial
data set that has been used in (Breiman, 1996) and
(Dong et al., 2005). The objects are points in a 20-
dimensional space and belong to one of two classes.
Following the procedure of (Dong et al., 2005), we
generate a Ringnorm data set instance with 10 mil-
lion objects where each object is equally likely to be
in either the positive or the negative class.
For each of the five data sets, we generate five
subsets by randomly sampling 5,000, 10,000, 25,000,
50,000, and 100,000 objects from the full data set.
Table 1: Datasets (after modifications).
Abbr # Objects # Attributes
# Positives
# Negatives
COV 581,012 54 0.574
KDD 4,898,431 122 4.035
RLC 5,749,132 16 0.004
BOW2 8,499,752 234,151 0.037
RNG 10,000,000 20 1.000
5.2 Experimental Design
Sparse computation and sparse-reduced computation
are tested with the two similarity-based machine
learning algorithms KNN, and SNC. In the following,
we use the term classifier to refer to a combination
of a machine learning algorithm and sparse/sparse-
reduced computation. The performance of the clas-
sifiers is evaluated in terms of accuracy (ACC) and
F
1
-scores (F1). The experimental analysis is imple-
mented in MATLAB R2014a and the computations
are performed on a standard workstation with two In-
tel Xeon CPUs (model E5-2687W) with 3.10 GHz
and 128 GB RAM. In Section 5.2.1, we describe the
tuning and testing procedure for a given classifier. In
Section 5.2.2, we discuss the implementation of Ap-
proximate PCA.
5.2.1 Tuning & Testing
We randomly partition each data set into a training set
(60%), a validation set (20%) and a testing set (20%).
The union of the training and the validation sets forms
the tuning set which is used as input for a given classi-
fier. We tune each classifier with and without first nor-
malizing the tuning set. Normalization prevents that
attributes with large values dominate distance or sim-
ilarity computations (Witten and Frank, 2005). For
each of the two resulting similarity matrices the clas-
sifier is applied multiple times with different combi-
nations of tuning parameter values. For KNN, we se-
lected tuning parameter K from the set {1, 2, . . . , 25}.
For SNC, we selected tuning parameter ε from the set
ICPRAM 2016 - International Conference on Pattern Recognition Applications and Methods
228

{1,3, .. ., 15} and the tuning parameter λ from the set
{0,10
−3
,10
−2
,10
−1
,1}.
The testing is performed on the union of the train-
ing and the testing sets, i.e., the same training objects
are used for tuning and testing. Unlike most machine
learning algorithms that first train a model based on
the training data and then apply the trained model to
the testing data, KNN and SNC require the training
data for classifying the testing data. For a given clas-
sifier, and performance measure, we select the combi-
nation of preprocessing option and tuning parameter
values which achieve the best performance with re-
spect to the given performance measure for the vali-
dation set. The performance measure is then reported
for the testing set.
5.2.2 Approximate PCA
The fraction of rows selected for approximate PCA is
one percent for all subsets of the data sets. With this
fraction, approximate PCA requires less than a second
of CPU time for all data sets, and the produced prin-
ciple components are close to the principle compo-
nents returned by exact PCA as determined by man-
ual inspection. For all subsets of data sets other than
BOW2, all attributes (columns) are retained in the
submatrix W . For BOW2, we select the 100 columns
that correspond to the words with the highest absolute
difference between the average relative frequencies
of the word in the positive and the negative training
documents. In that, we deviate from the probabilistic
column selection described in Section 3. For all sub-
sets of BOW2, we compute the similarities in the low-
dimensional space, i.e., with respect to these 100 most
discriminating words. Thereby, we first discretize the
feature vectors by replacing all positive frequencies
with value 1.
5.3 Numerical Results
We compare sparse computation and sparse-reduced
computation in terms of scalability by applying both
methods with the same grid resolution of k = 40 and
the same grid dimensionality of p = 3 to all five data
sets. With these values, the low-dimensional space is
partitioned into 64,000 blocks which is a good starting
point for both methods across data sets. For sparse-
reduced computation, parameter δ =
1
κ
is set to 1.
Tables 2–6 show the results for each of the tested
data sets and both classifiers. The entry “lim” indi-
cates that MATLAB ran out of memory. This happens
when we applied sparse computation to the complete
data sets (except for BOW1). For the data set KDD,
MATLAB already runs out of memory for a sample
size of 100,000.
The main conclusion from these tables is that
sparse-reduced computation scales orders of magni-
tude better than sparse computation while achieving
almost equally high accuracy for all data sets except
COV (to be discussed below). The running time re-
duction is most impressive for KDD and RLC. For
these two data sets both sparse and sparse-reduced
computation perform equally well but the tuning
times obtained with sparse-reduced computation are
up to 1,000 times smaller. Similar results have been
obtained for data sets BOW2 and RNG as reported
in Tables 5, and 6, respectively. For the data set COV,
we observe a reduction in accuracy and F
1
-scores with
sparse-reduced computation. One way to improve ac-
curacy and F
1
-scores is to select a value greater than
1 for κ. Increasing κ results in additional representa-
tives per grid block and thus improves the represen-
tation of the data set. Indeed preliminary work has
shown improvements in COV results with an adjust-
ment of the sparse-reduced approach.
KNN and SNC achieve very similar accuracies
and F
1
-scores for all data sets. KNN has smaller tun-
ing times than SNC because a smaller number tun-
ing parameter combinations is tested. Apart from the
number of tuning parameter combinations, the tun-
ing time is driven by the size and the density of the
similarity matrix. This can be seen from the relation
between the tuning times and the number of repre-
sentatives in Tables 2–6. In sparse computation the
size of the similarity matrix is proportional to the sam-
ple size. On the contrary, in sparse-reduced computa-
tion, the size of the similarity matrix is determined by
the number of representatives. When there are large
groups of highly-similar objects as is the case in the
data sets KDD and RLC, the number of representa-
tives is significantly lower than the number of objects
and an increase in sample size tends to increase the
multiplicity weight of the representatives but not the
number of representatives.
6 CONCLUSIONS
We propose a novel method called sparse-reduced
computation which enables highly scalable data min-
ing by creating a compact representation of a large
data set with minor loss of relevant information. This
is achieved by efficiently projecting the original data
set onto a low-dimensional space in which the con-
cept of grid neighborhoods is applied to discern the
main structure of the data set. The grid structure al-
lows to easily identify highly-similar and identical ob-
jects that are replaced by few representatives to reduce
the size of the data set. A major advantage of sparse-
Sparse-Reduced Computation - Enabling Mining of Massively-large Data Sets
229
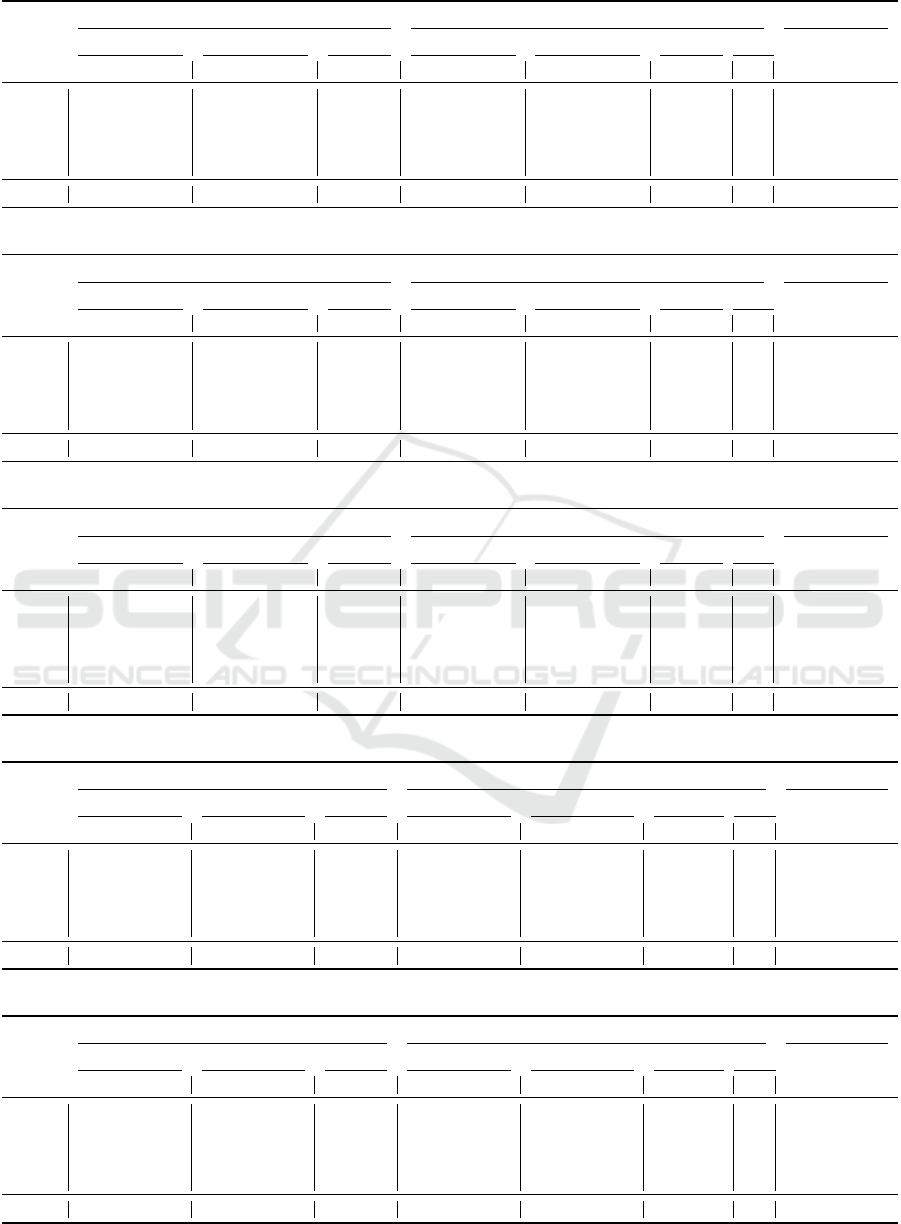
Table 2: Comparison of sparse and sparse-reduced computation for data set COV.
Sparse computation (k = 40, p = 3, r = 0.01) Sparse-reduced computation (k = 40, p = 3, r = 0.01, δ = 1) Speed-up factor
Accuracy [%] F
1
-score [%] Tuning time [s] Accuracy [%] F
1
-score [%] Tuning time [s] # Reps
Sample size KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC
5,000 78.50 80.10 70.26 72.63 2.1 2.6 72.20 72.90 61.13 59.42 2.5 2.6 1,981 0.8 1.0
10,000 81.80 82.15 73.74 72.89 3.6 5.5 73.35 73.55 58.64 60.37 3.7 3.9 3,060 1.0 1.4
25,000 88.04 87.94 83.51 83.23 8.3 19.6 69.78 72.46 57.85 58.74 4.5 4.8 3,504 1.8 4.1
50,000 91.42 91.90 88.22 88.81 24.5 75.7 72.24 73.31 59.12 63.06 6.9 7.3 5,097 3.6 10.4
100,000 94.04 94.20 91.74 91.98 60.1 195.9 72.37 74.06 59.88 61.28 12.3 13.1 5,427 4.9 15.0
581,012 lim lim lim lim lim lim 73.19 73.80 58.93 61.11 17.1 17.6 4,058 - -
Table 3: Comparison of sparse and sparse-reduced computation for data set KDD.
Sparse computation (k = 40, p = 3, r = 0.01) Sparse-reduced computation (k = 40, p = 3, r = 0.01, δ = 1) Speed-up factor
Accuracy [%] F
1
-score [%] Tuning time [s] Accuracy [%] F
1
-score [%] Tuning time [s] # Reps
Sample size KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC
5,000 99.60 99.50 99.75 99.69 3.0 11.8 99.60 99.50 99.75 99.69 0.4 0.4 290 7.5 29.5
10,000 99.80 99.85 99.94 99.91 11.8 53.8 99.85 99.85 99.91 99.91 0.5 0.5 369 23.6 107.6
25,000 99.82 99.80 99.89 99.87 76.1 373.4 99.78 99.78 99.86 99.86 1.0 1.0 634 76.1 373.4
50,000 99.83 99.85 99.89 99.91 303.9 1,520.2 99.71 99.74 99.82 99.84 1.6 1.6 876 189.9 950.1
100,000 lim lim lim lim lim lim 99.82 99.84 99.89 99.90 2.7 2.8 1,085 - -
4,898,431 lim lim lim lim lim lim 99.91 99.91 99.94 99.94 165.0 167.0 4,365 - -
Table 4: Comparison of sparse and sparse-reduced computation for data set RLC.
Sparse computation (k = 40, p = 3, r = 0.01) Sparse-reduced computation (k = 40, p = 3, r = 0.01, δ = 1) Speed-up factor
Accuracy [%] F
1
-score [%] Tuning time [s] Accuracy [%] F
1
-score [%] Tuning time [s] # Reps
Sample size KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC
5,000 100.00 100.00 100.00 100.00 .9 3.2 100.00 100.00 100.00 100.00 0.3 0.4 421 3.0 8.0
10,000 100.00 100.00 100.00 100.00 3.4 14.8 100.00 100.00 100.00 100.00 0.5 0.5 864 6.8 29.6
25,000 100.00 99.98 100.00 96.97 21.6 97.2 100.00 99.98 100.00 96.97 0.8 0.9 903 27.0 108.0
50,000 100.00 100.00 100.00 100.00 94.8 457.1 100.00 100.00 100.00 100.00 1.7 1.8 1,021 55.8 253.9
100,000 100.00 100.00 99.30 100.00 382.0 1,714.7 100.00 100.00 99.30 100.00 2.0 2.1 1,159 191.0 816.5
5,749,132 lim lim lim lim lim lim 99.99 99.99 98.76 98.82 43.8 51.0 1,309 - -
Table 5: Comparison of sparse and sparse-reduced computation for data set BOW2.
Sparse computation (k = 40, p = 3, r = 0.01) Sparse-reduced computation (k = 40, p = 3, r = 0.01, δ = 1) Speed-up factor
Accuracy [%] F
1
-score [%] Tuning time [s] Accuracy [%] F
1
-score [%] Tuning time [s] # Reps
Sample size KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC
5,000 98.70 98.80 81.16 82.86 3.2 3.6 98.70 98.80 81.16 82.86 3.4 3.4 2,418 0.9 1.1
10,000 99.05 98.95 83.76 81.74 6.4 9.0 99.05 98.95 83.76 81.74 5.6 5.7 2,749 1.1 1.6
25,000 98.38 98.22 70.55 67.16 17.8 30.8 98.56 98.46 74.47 72.20 13.6 14.1 8,115 1.3 2.2
50,000 99.46 99.51 91.84 92.61 45.0 147.5 99.54 99.56 93.09 93.43 11.6 12.1 5,137 3.9 12.2
100,000 99.68 99.56 95.13 93.21 126.1 475.7 99.59 99.63 93.72 94.38 20.4 21.1 8,171 6.2 22.5
8,544,543 lim lim lim lim lim lim 99.68 99.67 95.36 95.19 1,181.2 1,196.7 22,287 - -
Table 6: Comparison of sparse and sparse-reduced computation for data set RNG.
Sparse computation (k = 40, p = 3, r = 0.01) Sparse-reduced computation (k = 40, p = 3, r = 0.01, δ = 1) Speed-up factor
Accuracy [%] F
1
-score [%] Tuning time [s] Accuracy [%] F
1
-score [%] Tuning time [s] # Reps
Sample size KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC KNN SNC
5,000 79.40 81.00 78.18 82.36 3.3 3.6 79.40 81.00 81.33 80.78 4.2 4.3 3,206 0.8 0.8
10,000 81.25 81.30 82.29 82.67 6.2 7.1 81.55 81.30 82.52 82.75 7.7 8.0 5,467 0.8 0.9
25,000 79.76 81.46 82.95 83.81 14.3 23.2 81.32 81.42 83.81 83.69 15.1 15.9 8,728 0.9 1.5
50,000 80.34 82.13 83.65 84.11 32.8 87.5 81.43 82.15 83.87 84.19 22.5 23.2 12,925 1.5 3.8
100,000 79.81 82.24 82.89 83.61 88.2 335.0 80.76 82.23 82.63 83.60 33.3 35.3 16,518 2.6 9.5
10,000,000 lim lim lim lim lim lim 82.48 83.76 84.12 85.31 1,960.1 1,990.9 37,511 - -
ICPRAM 2016 - International Conference on Pattern Recognition Applications and Methods
230

reduced computation is that it is applicable to any ma-
chine learning algorithm. A set of computational ex-
periments demonstrates that sparse-reduced compu-
tation achieves significant reductions in running time
with minimal loss in accuracy.
In future research, it is planned to develop a vari-
ant of sparse-reduced computation in which the de-
gree of consolidation of objects to representatives
depends on properties of the region in the low-
dimensional space.
REFERENCES
Andrews, N. O. and Fox, E. A. (2007). Clustering for data
reduction: a divide and conquer approach.
Arora, S., Hazan, E., and Kale, S. (2006). A fast random
sampling algorithm for sparsifying matrices. In Ap-
proximation, Randomization, and Combinatorial Op-
timization. Algorithms and Techniques, pages 272–
279. Springer Berlin.
Asuncion, A. and Newman, D.J. (2007). UCI Machine
Learning Repository.
Baumann, P., Hochbaum, D.S., and Yang, Y.T. (2015). A
comparative study of leading machine learning tech-
niques and two new algorithms. submitted 2015.
Breiman, L. (1996). Bias, variance, and arcing classifiers.
Technical report, Statistics Department, University of
California, Berkeley.
Caruana, R. and Niculescu-Mizil, A. (2006). An empirical
comparison of supervised learning algorithms. In Pro-
ceedings of the 23rd International Conference on Ma-
chine Learning (ICML), pages 161–168, Pittsburgh,
PA.
Chandran, B. G. and Hochbaum, D. S. (2009). A compu-
tational study of the pseudoflow and push-relabel al-
gorithms for the maximum flow problem. Operations
Research, 57(2):358–376.
Chang, F., Guo, C.-Y., Lin, X.-R., and Lu, C.-J. (2010). Tree
decomposition for large-scale SVM problems. Jour-
nal of Machine Learning Research, 11:2935–2972.
Collobert, R., Bengio, S., and Bengio, Y. (2002). A parallel
mixture of svms for very large scale problems. Neural
computation, 14(5):1105–1114.
Dong, J.-X., Krzy
˙
zak, A., and Suen, C. Y. (2005). Fast
SVM training algorithm with decomposition on very
large data sets. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 27(4):603–618.
Drineas, P., Kannan, R., and Mahoney, M.W. (2006). Fast
monte carlo algorithms for matrices II: computing a
low-rank approximation to a matrix. SIAM J. Com-
puting, 36:158–183.
Fix, E. and Hodges, J.L., Jr. (1951). Discriminatory anal-
ysis, nonparametric discrimination, consistency prop-
erties. Randolph Field, Texas, Project 21-49-004, Re-
port No. 4.
Graf, H. P., Cosatto, E., Bottou, L., Dourdanovic, I., and
Vapnik, V. (2004). Parallel support vector machines:
The cascade svm. In Advances in neural information
processing systems, pages 521–528.
Hochbaum, D. and Baumann, P. (2014). Sparse computa-
tion for large-scale data mining. In Lin, J., Pei, J., Hu,
X., Chang, W., Nambiar, R., Aggarwal, C., Cercone,
N., Honavar, V., Huan, J., Mobasher, B., and Pyne, S.,
editors, Proceedings of the 2014 IEEE International
Conference on Big Data, pages 354–363, Washington
DC.
Hochbaum, D.S. (2008). The pseudoflow algorithm: a new
algorithm for the maximum-flow problem. Operations
Research, 56:992–1009.
Hochbaum, D.S. (2010). Polynomial time algorithms for
ratio regions and a variant of normalized cut. IEEE
Trans. Pattern Analysis and Machine Intelligence,
32:889–898.
Hochbaum, D.S., Lu, C., and Bertelli, E. (2013). Evalu-
ating performance of image segmentation criteria and
techniques. EURO Journal on Computational Opti-
mization, 1:155–180.
Hsieh, C.-J., Chang, K.-W., Lin, C.-J., Keerthi, S. S., and
Sundararajan, S. (2008). A dual coordinate descent
method for large-scale linear svm. In Proceedings of
the 25th international conference on Machine learn-
ing, pages 408–415.
Jhurani, C. (2013). Subspace-preserving sparsification of
matrices with minimal perturbation to the near null-
space. Part I: basics. arXiv:1304.7049 [math.NA].
Kawaji, H., Takenaka, Y., and Matsuda, H. (2004). Graph-
based clustering for finding distant relationships in
a large set of protein sequences. Bioinformatics,
20(2):243–252.
Provost, F. and Kolluri, V. (1999). A survey of methods
for scaling up inductive algorithms. Data mining and
knowledge discovery, 3(2):131–169.
Rida, A., Labbi, A., and Pellegrini, C. (1999). Local experts
combination through density decomposition. In Pro-
ceedings of International Workshop on AI and Statis-
tics.
Segata, N. and Blanzieri, E. (2010). Fast and scalable local
kernel machines. The Journal of Machine Learning
Research, 11:1883–1926.
Shalev-Shwartz, S., Singer, Y., Srebro, N., and Cotter, A.
(2011). Pegasos: Primal estimated sub-gradient solver
for svm. Mathematical programming, 127(1):3–30.
Spielman, D.A. and Teng, S.-H. (2011). Spectral sparsifica-
tion of graphs. SIAM J. Computing, 40:981–1025.
Tsang, I. W., Kwok, J. T., and Cheung, P.-M. (2005). Core
vector machines: Fast svm training on very large data
sets. In Journal of Machine Learning Research, pages
363–392.
Witten, I.H.. and Frank, E. (2005). Data mining: practi-
cal machine learning tools and techniques. Morgan
Kaufmann, San Francisco, 2nd ed. edition.
Yu, H., Yang, J., and Han, J. (2003). Classifying large data
sets using svms with hierarchical clusters. In Proceed-
ings of the ninth ACM SIGKDD international confer-
ence on Knowledge discovery and data mining, pages
306–315.
Sparse-Reduced Computation - Enabling Mining of Massively-large Data Sets
231
