Stitching Grid-wise Atomic Force Microscope Images
Mathias Vestergaard
1
, Stefan Bengtson
1
, Malte Pedersen
1
, Christian Rankl
2
and Thomas B. Moeslund
1
1
Institute of Media Technology, Aalborg University, Aalborg, Denmark
2
AFM R&D, Keysight Technologies, San Fransisco, U.S.A.
Keywords:
Atomic Force Microscopy, RANSAC, Template Matching, Image Stitching.
Abstract:
Atomic Force Microscopes (AFM) are able to capture images with a resolution in the nano metre scale. Due to
this high resolution, the covered area per image is relatively small, which can be problematic when surveying
a sample. A system able to stitch AFM images has been developed to solve this problem. The images exhibit
tilt, offset and scanner bow, which are counteracted by subtracting a polynomial from each line. To be able to
stitch the images properly template selection is done by analyzing texture and using a voting scheme. Grids
of 3x3 images have been successfully leveled and stitched.
1 INTRODUCTION
Atomic Force Microscopes (AFM) are able to create
topography images with very high resolution by trac-
ing the surface of a sample, line-by-line with a tiny
probe. The resolution of the AFMs is in nanometer
scale while conventional optical microscopes, in com-
parison, are able to achieve a maximum resolution
of some hundred nanometers. The limited resolution
of optical microscopes is imposed by the way light
diffracts (Eaton and West, 2010). The high resolution
of the AFM images allows for studies of molecules
(Last et al., 2010) and living cells (Johnson, 2011).
However, the improved resolution comes at the
cost of increased capturing time, as tracing the sur-
face of a sample, line-by-line, is a slow process which
can take several minutes. Another drawback is the
decrease in field of view, i.e. the area of the sample
contained in a single image. It is therefore not uncom-
mon for the size of the sample to exceed the viewable
area of the AFM and to counteract this, most AFMs
feature a motorized stage which allows them to auto-
matically capture the sample piece-by-piece.
Figure 1: Topography images of bacteria.
The result of this method is a series of images which
provides a fragmented view of the sample as seen in
Figure 1. The three images are overlapping in the hor-
izontal direction, but it is difficult and tedious to work
with this fragmented kind of view. A method to pro-
cess and stitch the images into a single image without
visible seams is therefore wanted.
Because of the high resolution of the AFM even
the smallest flaws will be apparent in the images.
These flaws complicate the stitching process and re-
quire steps to remove potential irregularities. This is
necessary as the final image, consisting of several iso-
lated images, should have a uniform background.
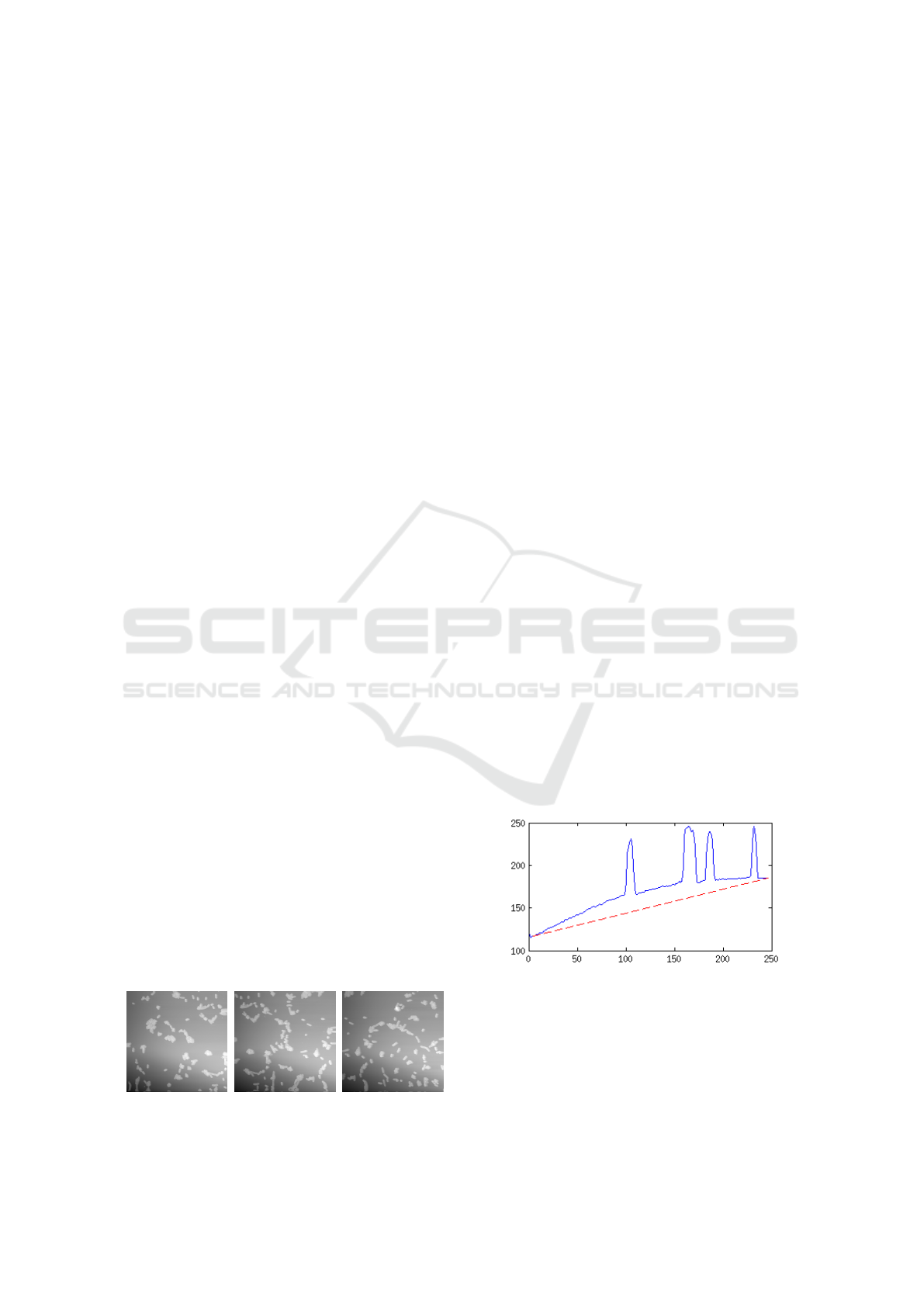
Figure 2: A line from one of the bacteria images. The red
line is added to visualize the tilt and scanner bow.
It is difficult to place the sample with enough preci-
sion to ensure a horizontally leveled surface and this
results in a visible tilt, where the images are darker on
one side. An example can be seen in Figure 2, which
illustrates a horizontal line taken from one of the bac-
teria images. The tilt can be seen as the red dashed
112
Vestergaard, M., Bengtson, S., Pedersen, M., Rankl, C. and Moeslund, T.
Stitching Grid-wise Atomic Force Microscope Images.
DOI: 10.5220/0005716501100117
In Proceedings of the 11th Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2016) - Volume 3: VISAPP, pages 112-119
ISBN: 978-989-758-175-5
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
line, which is more or less the same within a specific
sample, but may vary from one sample to another.
Most AFMs have only one moving part which is a
piezoelectric block that can be placed either beneath
or above the sample. This block is controlled by ap-
plying current to the sides, which results in a move-
ment pattern similar to that of a pendulum, see Fig-
ure 3.
Scanner
tube
Probe
Figure 3: Illustration of the bow is created when a piezo-
electric scanner tube is used to move the probe.
This is especially clear in images where the piezoelec-
tric block is forced to approach its maximum range. A
phenomenon occurs where an even surface appears to
bow and the intensity near the edge of the image is
lower than at the center. This bow can be observed in
Figure 2.
Beside scanner bow and tilt another problem can
occur where some of the objects move or change
shape between the capturing of two adjacent images.
This can happen if the sample consists of living or-
ganisms where movement creates a difficulty in find-
ing the right blending. The same object can end up
having two different positions, relative to the rest of
the image, if it has moved. In worst case scenarios
objects are totally missing as seen in Figure 4.
Figure 4: Two subimages showing the same area of a sam-
ple. The subimages are taken from the overlapping part of
two images.
2 APPROACH
The proposed solution contains four modules as illus-
trated in Figure 5. The boxes symbolize the modules
which are: Preprocessing, Feature Extraction, Recog-
nition and Post Processing. Between the modules
are arrows which represent the way of the workflow.
These modules will be described in the following sec-
tions.
Preprocessing
Image Leveling
Feature Extraction
Find
Template Positions
Processed Image
Recognition
Template Matching
Template Positions
Post Processing
Translation
and
Stitching
Position of
best match
Figure 5: The overall design of the proposed solution.
2.1 Image Leveling
The objective of the leveling process is to remove
scanner bow, tilt and offset, while preserving the ob-
jects in the images. Traditional image processing
techniques are possible solutions, such as the rolling
ball algorithm (Lundtoft, 2014).
However, this approach does not account for the
characteristics of the artifacts. Namely how they can
be described as 2nd order polynomials acquired in a
line-by-line pattern.
A common method to level AFM images is therefore
to fit and then subtract a polynomial line-by-line
for the entire image (Eaton and West, 2010). When
fitting the polynomial it is important to exclude the
objects to avoid overcompensation which could result
in new artifacts (Figure 6).
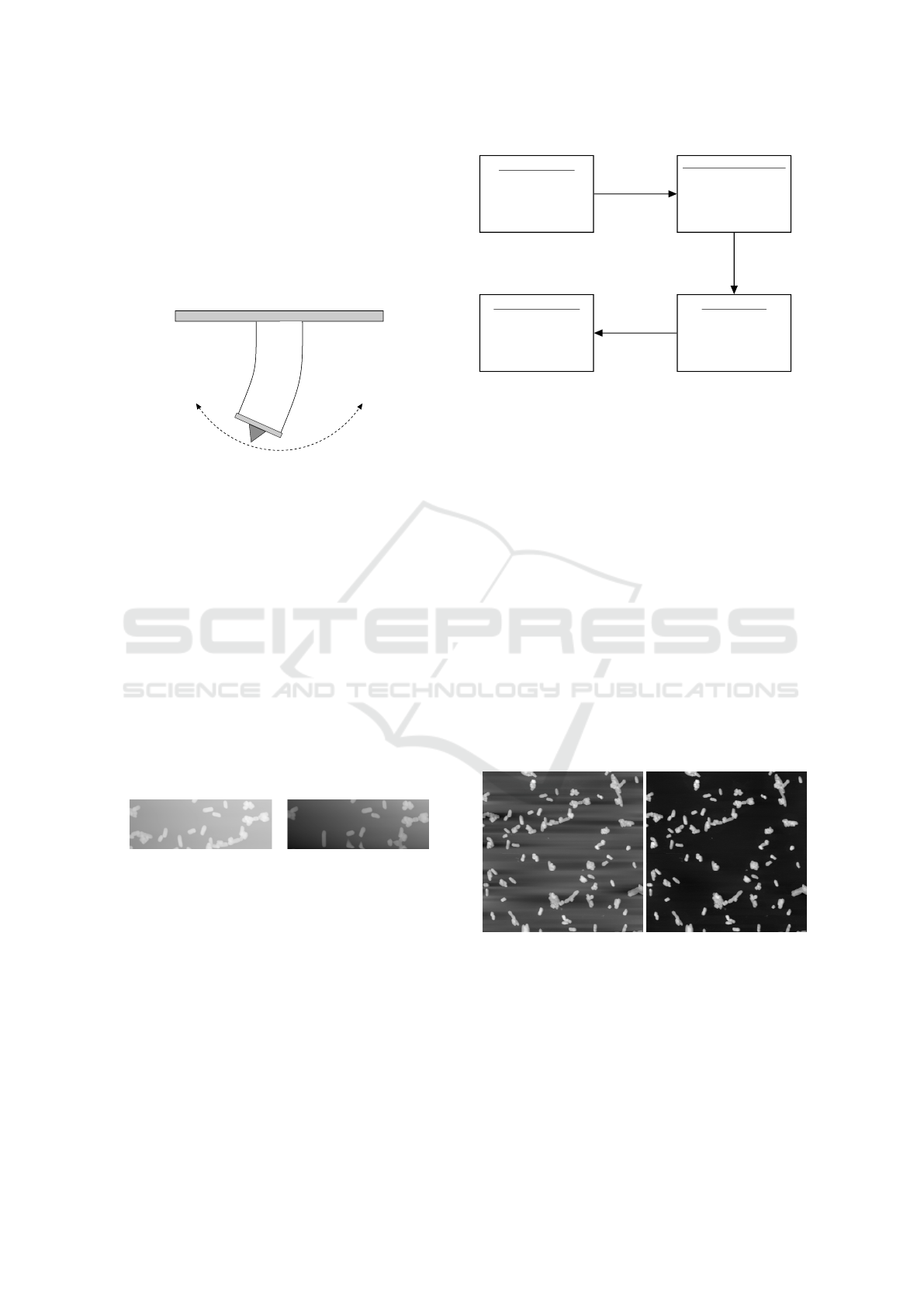
(a) Objects not excluded. (b) Objects excluded.
Figure 6: The same image leveled with and without object
exclusion.
Some approaches rely on manual selection of objects
to exclude (Eaton and West, 2010), which is very
labour intensive and not useful when dealing with a
lot of images. Another approach is to use K-means
clustering to identify objects in every line (Pan, 2014)
(Tsaftaris et al., 2008).
Stitching Grid-wise Atomic Force Microscope Images
113

2.1.1 Leveling Algorithm
The developed leveling algorithm expands upon the
idea of line-by-line leveling by introducing a new
way of automatically excluding objects through the
use of RANSAC (Random Sample Consensus) (Fis-
chler and Bolles, 1980). RANSAC is suitable as it
is able to estimate parameters of a given model (in
this case a 1st or 2nd order polynomial) from a data
set (a line from the image) contaminated with outliers
(objects). RANSAC basically works by repeatedly
making guesses at a solution based on n randomly se-
lected initial points, where n is the minimum number
of points required to determine the parameters of the
model. Each solution is then evaluated and compared
against the current best guess, yielding the best guess
as the solution in the end.
RANSAC is therefore non-deterministic and its
success depends on the number of iterations k which
RANSAC is allowed to execute. k can be determined
theoretically based on two assumptions:
The solution is considered decent if:
• All the n initial points are inliers, i.e. part of the
polynomial to be removed.
• The n initial points are not closely grouped to-
gether, which is evident be contemplating e.g. 2nd
order polynomials.
For the sake of simplicity, the number of data points
is divided into n evenly sized intervals and the n
initial points are considered to be appropriately
spaced if each resides in an interval of its own.
Hence, the probability of never reaching a de-
cent solution out of k iterations is given by:
P(bad) = (1 − P(in)
n
× P(spaced))
k
(1)
where P(in) is the probability of a point being an
inlier and P(spaced) is the probability of the n
points being appropriately spaced, i.e. P(spaced) =
(1/n)
n
. The probability of reaching a decent solution
P(decent) out of k iterations must then be given by:
1 − P(decent) = (1 − P(in)
n
× (1/n)
n
)
k
(2)
from where k can be isolated:
k =
log(1 − P(decent))
log(1 − P(in)
n
× (1/n)
n
)
(3)
Considering a scenario with 50% outliers, i.e. P(in) =
0.50, and it would take:
k =
log(1 − 0.999)
log(1 − 0.50
3
× (1/3)
3
)
≈ 1488 (4)
iterations to find a decent solution with a probability
of P(decent) = 99.9% when using n = 3 initial
points, which is sufficient for a 2nd order polynomial.
Another solution could be to divide each line
into n evenly sized intervals and then randomly
choose one point from each interval as the n initial
point. This would reduce the number of iterations k
to:
k =
log(1 − P(decent))
log(1 − P(in)
n
)
(5)
as it is no longer necessary to check the spacing be-
tween the points, i.e. take P(spaced) into account.
However, this method was found to be inconsis-
tent during tests on real data, as it would repeat-
edly fail at images with un-evenly distributed outliers,
where the majority of outliers would reside in one of
the fixed intervals. The idea of using fixed intervals
was therefore discarded.
2.1.2 Performance Considerations
A runtime test of a generic RANSAC algorithm is
performed to determine whether the k = 1488 from
Equation 4 is realistic. The algorithm is implemented
in C++ and is performed on a laptop with a P8600
@ 2.4GHz CPU. On average (100 repetitions) it
took 0.88 seconds to execute the algorithm. Hence,
it would take 3 minutes and 45 seconds to apply
RANSAC on each horizontal line in a relatively small
image of 256 × 256 pixels, which is not acceptable.
In order to reduce runtime, RANSAC is only
used to exclude objects in the first line of the image.
In the following lines, the exclusion of objects is
achieved by ignoring every point x in the line l(x)
which is not within a fixed threshold t from the
polynomial p(x)
latest
found in the previous line. I.e.
only points which fulfils
t
2
> (p(x)
latest
− l(x))
2
(6)
are used when fitting the polynomial to be subtracted
in every line, apart from the first line where RANSAC
is used.
2.1.3 Final Leveling Algorithm
The final leveling algorithm is as follows:
1. Fit a polynomial p(x)
latest
to the first line l
1
in the
AFM image using RANSAC.
2. Find inliers in the next line based on p(x)
latest
and
a fixed threshold t.
3. Fit a polynomial to the found inliers and subtract
the found polynomial from the current line.
VISAPP 2016 - International Conference on Computer Vision Theory and Applications
114

4. Overwrite p(x)
latest
with the polynomial just
found.
5. Repeat step 2-4 until all lines have been processed
in the AFM image.
The developed leveling algorithm does well when the
percentage of background is noticeably larger than the
object percentage (Figure 7).
The leveled images will however start to exhibit
an offset in height when the object percentage
approaches or exceeds 50% (Figure 8). This issue
originates in the initial step of the algorithm, where
RANSAC might mistakenly fit a polynomial to the
top of the objects instead of the background.
The above issue can be detected by comparing
the mean of the current line µ
line
with the mean of
the fitted polynomial µ
poly
. The polynomial is then
considered to be fitted correctly if:
µ
line
> µ
poly
(7)
(a) Original image. (b) Leveled image.
Figure 7: Leveling of image with a high background per-
centage.
(a) First image. (b) Second image.
Figure 8: Two adjacent images of SiO
2
stripes after lev-
eling. The difference in intensity is caused by the height
offset between the images.
The correct polynomial can be found by fitting a poly-
nomial to the outliers, instead of the inliers. This san-
ity check and correction are easily executed at the end
of the developed leveling algorithm.
The average processing time of the developed level-
ing algorithm, including the above sanity check, is
found to be roughly 1 second (see Table 1). This is
deemed acceptable considering the time it takes to
capture each AFM image (several minutes).
The test was performed on a laptop with a P8600
@ 2.4GHz CPU with datasets containing 9 images
each. The bacteria and SiO
2
stripes datasets contains
images with a resolution of 256 x 256 pixels, while
the bacteria HQ dataset contains images of 512 x 512
pixels. Hence the noticeable difference in runtime.
Table 1: Runtime test of the developed leveling algorithm.
Number of iterations is fixed at k = 1488.
Runtime (seconds)
Dataset min avg max
Bacteria 0.84 0.92 0.95
Bacteria HQ 1.32 1.43 1.49
SiO
2
stripes 0.77 0.81 0.83
2.2 Estimating Translation
A prerequisite for stitching images is the knowledge
of how they are related, in this case translation, due to
the nature of how the images are acquired using the
motorized stage in the AFM. As only translation is
present, it is possible to express the relation between
two adjacent AFM images as:
x
1
y
1
+
x
shi f t
y
shi f t
=
x
2
y
2
(8)
where [x
1
y
1
]
T
is the position of an arbitrary object in
image 1, [x
2
y
2
]
T
is the position of the corresponding
object in image 2 and [x
shi f t
y
shi f t
]
T
is the translation.
If the position for an object pair (i.e. [x
1
y
1
]
T
and
[x
2
y
2
]
T
) is known for a pair of images, it is possible
to calculate [x
shi f t
y
shi f t
]
T
, i.e. the translation between
these images.
SIFT (Scale-invariant Feature Transform) (Lowe,
1999) is often used to identify object pairs, when
stitching images with varying scale or rotation, e.g.
creating panorama images (Brown and Lowe, 2007).
However, complications such as scale and rotation
are not an issue in the AFM images, which is why a
simpler approach is chosen in the form of template
matching.
2.2.1 Template Extraction
Some AFM images contain only a small amount of
objects and the probability of choosing a region con-
sisting of background is therefore relatively large if
no precautions are taken. Templates taken from such
regions are difficult to recognize and a solution for
finding usable template positions is therefore needed
Stitching Grid-wise Atomic Force Microscope Images
115

to ensure that the matching-process has the best con-
ditions.
Good templates are defined as templates contain-
ing a high level of texture. In this work we define
texture as a region with strong vertical and horizontal
edges measured by a Sobel filter. The gradient mag-
nitude found using this filter does not help determine
whether a template is unique, but gives an indication
of the proportion of the edges contained within. A
high level of texture is nevertheless found to be as-
sociated with uniqueness and further filtering is not
considered necessary.
2.2.2 Template Matching
Normalized Cross Correlation (NCC) (Briechle and
Hanebeck, 2001) is used to match the templates, as
the AFM images are only exposed to translation. The
templates will be extracted from one image and cor-
related with another (the reference image). NCC is
chosen over basic correlation to increase the proba-
bility of a successful match even if the leveling does
not succeed.
The NCC is given by
cos(v) =
A
p
• T
||A
p
|| ||T ||
(9)
where v is the angle between the template T and the
image patch A
p
of the reference image A.
The angle v is an expression for the match rate
between A
p
and T , so when cos(v) approaches 1 the
similarity increases. The interval of the match rate is
[0;1] as the respective images only consist of positive
values.
2.2.3 Region of Interest
The overlapping region is selected as the region of in-
terest during the extraction and matching of templates
between the two images. Any template matches out-
side this region must obviously be wrong and should
be discarded anyway.
However, it is possible to limit the ROI even fur-
ther when looking for a template match in image 2.
This is due to the grid-like pattern in which the AFM
acquires images, resulting in the following assump-
tions:
1. The translation between two adjacent images only
changes drastically in one direction. I.e. transla-
tion in either the horizontal or vertical direction is
always negligible or not present at all.
2. The translation is roughly the same between all
adjacent images.
From assumption 1 the ROI can be limited to either
a horizontal or vertical slice (Figure 9) when only the
orientation of the two images are known (i.e. image 1
is to the left of image 2).
Image 1 Image 2
Figure 9: The highlighted area is the overlapping region,
the blue square is the template and the blue area is the ROI
in image 2.
If a rough translation is known as well (from a pre-
vious image pair) it is possible to estimate where the
template match is likely to be found on image 2 due
to assumption 2 (Figure 10).
Image 1 Image 2
Figure 10: The ROI in image 2 (blue area) is narrowed down
even further than in the previous figure.
The translation is therefore estimated as follows:
1. Limit the ROI on both image 1 and 2 to the over-
lapping region.
2. If a rough translation is known, jump to step 5.
3. Calculate the initial overlap from meta data from
the image files and adjust ROI accordingly on im-
age 1.
4. Use assumption 1 to limit the ROI on image 2.
Skip the next step.
5. Use assumption 2 to limit the ROI on image 2.
6. Extract a template T from image 1.
7. Find a match for T in image 2.
8. Calculate translation based on the position of T
and the found match.
2.2.4 Multiple Templates
Using a single template match to calculate the trans-
lation should be sufficient in theory. However, it is
VISAPP 2016 - International Conference on Computer Vision Theory and Applications
116

possible that a match is not correct due to some of the
following reasons:
• Living Samples - If the sample is alive and mov-
ing around, it it possible that a template might not
be present in both images.
• Noise - Random noise in the images could mess
up the template matching and yield a false-
positive match.
• ROI Selection - The ROI selection might not
be spot on and the same template might not be
present in the ROIs for both images.
To avoid the impact of these issues, a voting-scheme
is utilized to decide which translation to accept.
Translation candidates are repeatedly calculated and
then allowed to cast a vote. This process stops when
a single translation candidate reaches a set number of
votes w and wins, or until no more templates can be
extracted from image 1. In the latter case, the candi-
date with the most votes wins.
Each vote cast is weighed:
v
weighted
= (m
rate
)
2
(10)
where v
weighted
is the weighted vote and m
rate
is the
similarity measure returned by the NCC in Equa-
tion 9. m
rate
is squared in order to punish translations
with a low match rate even further.
2.2.5 Stitching
The last step of the proposed solution is the actual
stitching of the images according to the found trans-
lations. The images are stitched together two images
at a time, first horizontally into rows and then verti-
cally (Figure 11).
1 2 3
4 5 6
7 8 9
1 2 3
4 5 6
7 8 9
1 2 3
4 5 6
7 8 9
Figure 11: Stitching order.
The stitching order is similar to the order in which
the images are captured by the AFM, i.e. row-by-row.
This is an intentional choice, as it reduces the impact
of movement from living objects. The impact is re-
duced as the elapsed time between the capturing of
e.g. image 1 and 2 is less than the time between im-
age 1 and 4, thereby leaving less time for the objects
to move.
Two blending methods have been proposed:
P
mean
=
P
1
+ P
2
2
(11)
P
min
= min(P
1
,P
2
) (12)
where P
1
is the respective pixel from the overlapping
part of image 1 and P
2
the pixel from image 2.
P
mean
is used in cases where the trails of move-
ment (ghosting artifacts) should be visible whereas
P
min
is used where these trails are unwanted. A down-
side of the P
min
method is that parts of the objects can
disappear as the minimum value of the two pixels is
used. An example of the two blending methods can be
seen in Figure 12, where a slightly wrong translation
is used to highlight how the methods behave.
mean
(a) Mean blending.
min
(b) Min blending.
Figure 12: The two blending methods used on the same
overlapping region.
3 RESULTS
A demonstration of how the proposed solution works
is illustrated in the bacteria images in Figure 13. The
result can be seen in Figure 15 and Figure 16 where
the mean and min blending methods have been used,
respectively.
The difference between the two blending methods
is not as clear as in the example in Figure 12 although
some lines, where the images are stitched, can be seen
when the mean blending method is used. These lines
occur as a result of the missing or moving objects as
explained earlier.
The solution is also tested on a dataset exhibit-
ing a high percentage of objects (Figure 14), which
is known to cause trouble for the leveling algorithm.
The purpose is to test the translation estimation and
stitching capabilities when the intensity is not consis-
tent due to the shortcomings of the leveling.
The result of the leveling and stitching process can
be seen in Figure 17. It is clear that the leveling al-
gorithm struggles, which is evident due to the many
artifacts. Most notable are the sharp seams caused by
different intensities and sudden fluctuations in inten-
sity between lines. However, the translation estima-
tion and stitching does appear successful, as the ob-
jects agree with the objects found in the original 3x3
image grid.
A runtime test of the entire system is performed
on a laptop with a P8600 @ 2.4GHz using grids of
Stitching Grid-wise Atomic Force Microscope Images
117
3x3 images. Four different image grids were used,
one with a resolution of 512x512 pixels and three of
256x256 pixels. The high resolution grid took 43 sec-
onds to process, while the three lower resolution grids
took 13 seconds on average.
Figure 13: Topography images of bacteria - 3x3 grid.
Figure 14: Topography images of CHO cells - 3x3 grid.
4 DISCUSSION
The leveling algorithm starts to fail when the objects
occupy a percentage above 90% of each line (Fig-
ure 18). This high percentage increases the risk of
finding an inferior solution in the initial step where
RANSAC is used.
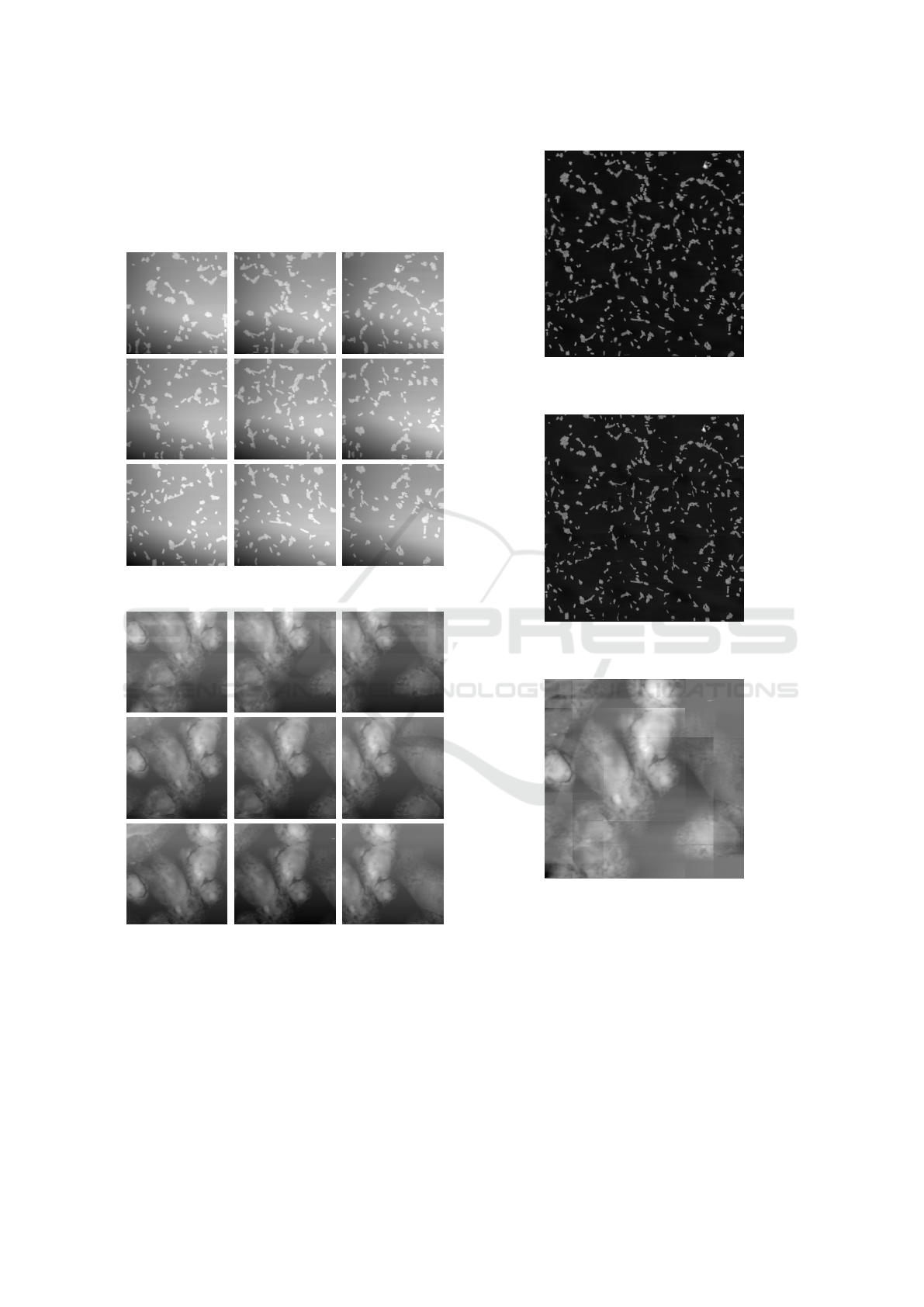
Figure 15: Stitched image of bacteria using the mean blend-
ing method.
Figure 16: Stitched image of bacteria using the min blend-
ing method.
Figure 17: Stitched image of CHO cells using the min
blending method.
A possible solution could be to apply RANSAC,
with fewer iterations, at several random lines in the
image, in order to find a line with a decent amount of
background. Further tests on diverse datasets could
be used to identify the actual limit of the leveling al-
gorithm. Another issue is the impact of the points,
which are mistakenly classified as inliers (Figure 19).
In images with a relatively low object percentage this
is not an issue, as the false inliers will be outweighed
by the true inliers when fitting a polynomial.
Reducing the threshold used when identifying in-
VISAPP 2016 - International Conference on Computer Vision Theory and Applications
118
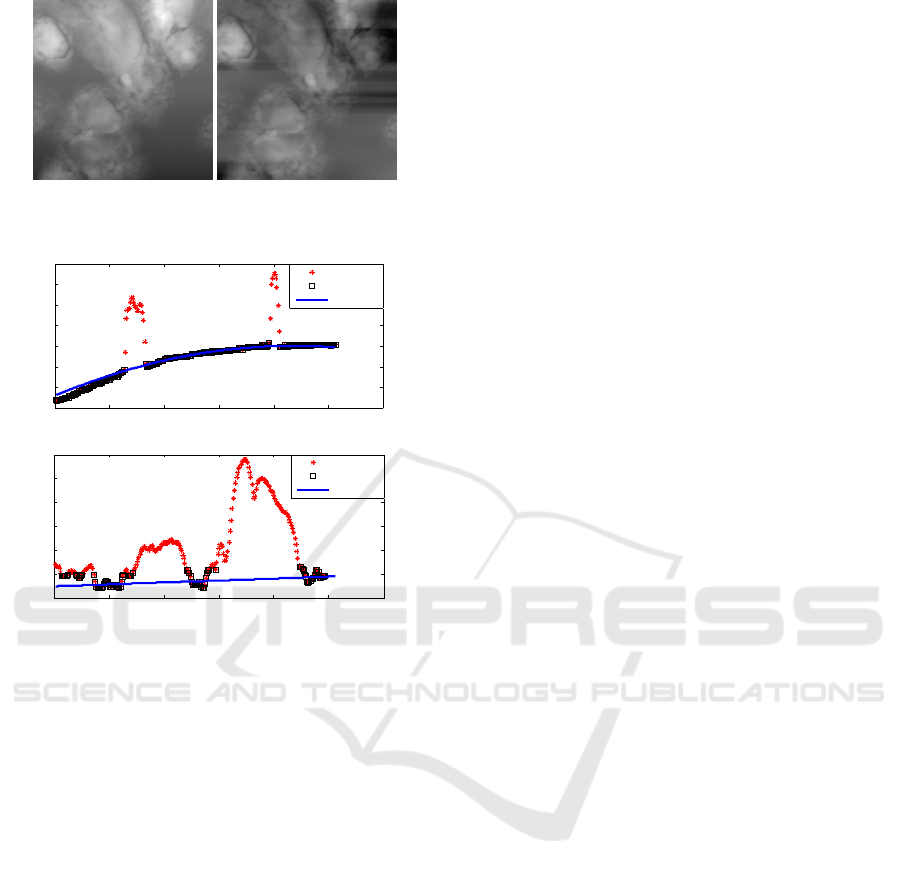
(a) Original image. (b) Leveled image.
Figure 18: Image with a high object percentage.
0 50 100 150 200 250 300
120
140
160
180
200
220
240
260
line
inliers
last poly
(a) Image 1 - easy to level.
0 50 100 150 200 250 300
120
140
160
180
200
220
240
line
inliers
last poly
(b) Image 2 - difficult to level.
Figure 19: Example of inlier detection.
liers could be a possible solution to this problem, but
it has not been tested. Even if the leveling fails to
some degree, the NCC allows for intensity irregular-
ities between the images and is able to find the right
translation. An improvement on the template recog-
nition could be to check the uniqueness of extracted
templates before attempting to match them.
The choice of the NCC and the region-of-interest
selection is based on the assumption that the im-
ages are only related through translation. This sim-
ple model seems to be a good choice as the images
appear to be correctly stitched when disregarding ar-
tifacts not caused by the stitching, like failed leveling
or disappearing objects.
The mean blending method is able to show how ob-
jects have moved between the images and the min
method can be used in cases where the overview is
more important than the detail. However, a blending
method that maintains the original shape of the ob-
jects, without ghosting artifacts, such as de-ghosting
(M. Uyttendaele and Szeliski, 2001) could improve
the output in certain scenarios.
5 CONCLUSION
The proposed solution is able to level and stitch grid-
wise captured AFM images with a few limitations as
discussed above. It takes between 12-43 seconds for
the system to stitch a grid of 3x3 images depending
on the size of the images.
The two blending methods work as intended al-
though there are disadvantages to both of them. The
decision of which method is the best in a specific sit-
uation depends on the user of the system.
REFERENCES
Briechle, K. and Hanebeck, U. D. (2001). Template match-
ing using fast normalized cross correlation. Optical
Pattern Recognition XII, 4387:95–102.
Brown, M. and Lowe, D. G. (2007). Automatic panoramic
image stitching using invariant features. International
Journal of Computer Vision.
Eaton, P. and West, P. (2010). Atomic Force Microscopy.
Oxford University Press.
Fischler, M. A. and Bolles, R. C. (1980). Random sample
consensus: A paradigm for model fitting with appli-
cations to image analysis and automated cartography.
http://www.dtic.mil/dtic/tr/fulltext/u2/a460585.pdf.
Johnson, W. T. (2011). Imaging cells with the agilent
6000ilm afm.
Last, J. A. et al. (2010). The applications of atomic force
microscopy to vision science. Invest Ophthalmol Vis
Sci, 51:6083–6094.
Lowe, D. G. (1999). Object recognition from local scale-
invariant features. The Proceedings of the Seventh
IEEE International Conference on Computer Vision.
Lundtoft, D. H. (2014). Internship report from keysight
technologies. Technical report, Aalborg University.
M. Uyttendaele, A. E. and Szeliski, R. (2001). Eliminat-
ing ghosting and exposure artifacts in image mosaics.
Computer Vision and Pattern Recognition, pages 509–
516.
Pan, X. (2014). Processing and feature analysis of atomic
force microscopy images. Master’s thesis, Missouri
University of Science and Technology.
Tsaftaris, S. A. et al. (2008). Automated line flattening
of atomic force microscopy images. Master’s thesis,
Northwestern University.
Stitching Grid-wise Atomic Force Microscope Images
119
