
MoBio
A Mobile Application for Collecting Data from Sensors
Petr Je
ˇ
zek and Roman Mou
ˇ
cek
New Technologies for the Information Society, Faculty of Applied Sciences, University of West Bohemia,
Univerzitn
´
ı 8, 306 14 Plze
ˇ
n, Czech Republic
Keywords:
MoBio, Sensor, Android, Mobile Application, Brain Data, Health Data, Domain Terminology, Data Transfer,
EEGBase.
Abstract:
There are a lot of sensors for monitoring human health and/or fitness level on the market. They facilitate
collection of data from the human body and advanced devices even facilitate data transfer to remote servers
where the collected data are further processed. While health data, obtained e.g. from accelerometers or chest
straps, are collected rather frequently, brain electrophysiology data, obtained from surface electrodes, are
still collected relatively rarely. However, integration and correlation of brain signals with other sensory data
would be very interesting for next research of physical and mental health. Although capturing brain signals
in real environment still faces technological difficulties, current development of common infrastructure seems
to be useful. Then this article deals with various architectures and data formats used for storage and transfer
of sensory data and their possible integration with existing neuroinformatics approaches. As a solution we
introduced a terminology describing data from a limited collection of sensors. The terminology is implemented
in the odML format and integrated in a proof-of-concept Android application. Data transfer, storage and
visualisation as well as integration with a remote neuroinformatics resource are presented.
1 INTRODUCTION
There are a lot of factors affecting human health.
Some of them such as genetics, environmental influ-
ence or internal state of an individual cannot be eas-
ily measured. On the other hand, there are the fac-
tors such as blood pressure, glucose level or heart
rate that can be measured relatively easily and non-
invasively using cheap sensors. For a long time elec-
trophysiological measurements have been conducted
in laboratories equipped by common desktop comput-
ers and non-transferable measuring devices. Fortu-
nately, this situation has been rapidly changing. One
of the reasons is increasing popularity of smart de-
vices such as smart-phones or tablets. According to
eMarketer (eMarketer, 2015) two billion people will
own a smart-phone in 2016. Simultaneously, a lot
of relatively cheap sensors for measuring potentials
from the human body are available on the market.
The data produced by these sensors are usually trans-
ferred wirelessly, consequently they can be read and
processed by smart devices. This technical progress
enables a particular shift of treatment from hospitals
to home environments and facilitates collecting data
during outdoor activities. The obtained data can be
used in two fundamental intersecting ways.
At first, assistive technology approach serves to
stimulate, maintain, and improve functional capabili-
ties of people with special needs including disabled
people or aging population. Getting independence
and self-sufficiency increases the quality of life in
general.
The second approach is focused on sportsmen or
actively living people. People are being monitored
when they are performing specific activities (e.g. run-
ning or long distance walking). The data are used for
a long term monitoring of their fitness level.
So far we have not talked about a significant
source of electrophysiology data, the human brain.
Although there are technological improvements for
capturing surface brain data in real environment, this
kind of data collection is still not widespread. How-
ever, integration and correlation of brain signals with
other sensory data would be very interesting for next
research of physical and mental health.
Our research group operates a completely
equipped laboratory (Moucek et al., 2014) for electro-
physiological measurements. We are focused mainly
on experimental work using the methods and tech-
niques of electroencephalography (EEG) and event-
Ježek, P. and Mou
ˇ
cek, R.
MoBio - A Mobile Application for Collecting Data from Sensors.
In Proceedings of the International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AWE 2016), pages 115-121
ISBN: 978-989-758-180-9
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
115

related potentials (ERP). Except of the permanent lab-
oratory we also operate a mobile laboratory equipped
by a set of laptops and portable measuring devices
for performing experiments in the external environ-
ment. With advancing efforts to extend the laboratory
to collect diverse collections of data (e.g. blood pres-
sure, glucose level, heart rate) we are extending our
infrastructure (hardware devices and software tools)
to support measurements of heterogeneous data from
various kinds of sensors.
In this paper we deal with various architectures
and data formats used for storage and transfer of sen-
sory data, and their possible integration with existing
neuroinformatics approaches. Since data from exist-
ing sensors are usually stored in proprietary formats
and transferred to closed databases, it is difficult to
use them in experimental laboratories. As a solu-
tion we introduce a terminology describing data from
a limited collection of sensors. Then a prototype of a
mobile client for collecting data from these sensors is
presented. This client provides integration with a lim-
ited set of devices and enables data to be transferred
and visualized in a remote storage.
2 STATE OF THE ART
According to (Lowe and Laighin, 2012) applications
using sensors can be divided into three categories.
The first category, Smart Phone Applications, use ei-
ther GPS or on-board kinematic sensors as the tech-
nologies of choice for monitoring exercise. The sec-
ond category comprises of any system that uses a cen-
tral controller and an external sensor. The last cate-
gory comes from image processing domain. It uses a
combination of a computer screen and a camera. The
camera monitors the exact movement and position of
the entire body during exercise. The screen is used for
the interaction with the user.
The first category represents applications such as
Endomondo or Runkeeper. The second category is
represented by e.g. Nike+, miCoach, Garmin Heart
Belt or Fora Active tonometer. A typical representa-
tive of the third category is Microsoft Kinect. While
Microsoft Kinect is designed for the indoor use, all
other devices are designed mostly for the outdoor use.
Available tools and sensors are usually designed
for one specific activity. They are able to record only
limited variety of data. In addition, due to their pro-
prietary structure they do not provide suitable inter-
faces for integration with other systems. Issues with
deloying commercial biosensors for a body sensor
network are described in (Seeger et al., 2011).
As well as other communities neuroinformatics
community identified problems with a long-term de-
scription, storage and management of experimental
data/metadata (Teeters et al., 2008). To facilitate solu-
tion of these difficulties International Neuroinformat-
ics Coordinating Facility (INCF) (INCF, 2013) covers
activities for development of infrastructures and data
standards in neuroinformatics community. Ontology
for Experimental Neurophysiology (OEN) (Le Franc
et al., 2014a) uses semantic web approach to describe
terminology used in biomedical science and neuro-
science. Our research groups developed a system for
long term storage and management of EEG/ERP ex-
perimental data and metadata, EEGBase (Jezek and
Moucek, 2012) that was recently extended by a sys-
tem of templates suitable for storing various data and
metadata structures. A mobile EEGBase client (Jezek
and Moucek, 2013) is a supplementary Android tool
that enables collecting experiments out of the labo-
ratory and provides an on-line synchronization with
EEGBase.
3 SENSOR ARCHITECTURES
AND DATA TRANSFER
Sensors use different ways to manipulate data. Some
sensors have their own displays or internal memories
for storing measured data that can be later transferred
to a long term storage. These devices usually provide
a complete history (of course, limited by the capacity
of their internal memories) of stored measurements.
On the other hand, devices such as heart rate meters
do not have any internal memory. Data can be trans-
ferred only at the time of measurement. According
to the way the data are distributed, we defined three
levels of sensor architecture.
• one-layer - The sensor is a stand-alone unit having
a display, memory, and a set of controls. Such a
sensor is completely self-controlled and does not
establish any connection to a remote system.
• two-layers - The sensor captures data that are fi-
nally sent to a remote system (the sensor may/may
not have an internal memory). This remote sys-
tem can be a common computer/laptop or a smart
phone/watch etc.
• three-layers - A cloud service is added to the two-
layer architecture. The captured data are sent to
a remote server where they are stored, managed,
and analyzed.
A lot of sensors belonging to the one-layer archi-
tecture, including various glucometers, tonometers,
and step counters, store captured data in a proprietary
ICT4AWE 2016 - 2nd International Conference on Information and Communication Technologies for Ageing Well and e-Health
116
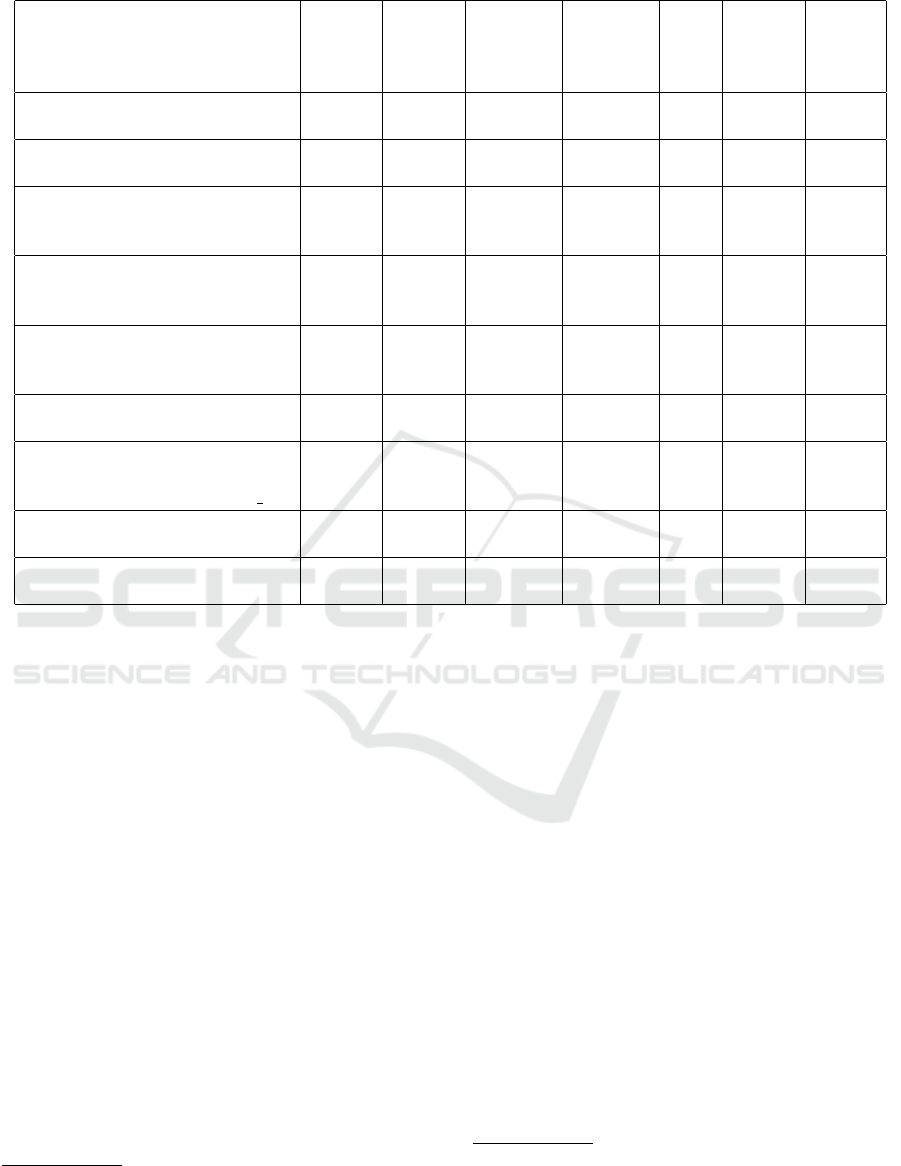
Table 1: Tested e-health and fitness systems.
Intern.
sensors
Extern.
sensors
Conti-
nuous
measure-
ments
Single
measure-
ments
Stats
Cloud
Service
Design
Endomondo
www.endomondo.com
2 3 • • • +
Runstatic
www.runtastic.com
2 2 • • • +
Pedometer
www.runtastic.com
/en/apps/pedometer
1 • • • -
Push-ups
www.runtastic.com/
en/apps/pushups
1 • • • +
Heart rate
www.runtastic.com
/en/apps/heartrate
1 • • • +
Sport Tracker
www.sports-tracker.com
1 2 • • • +
Health Tracker
play.google.com/store/apps/
details?id=com.benoved.phr lite
• -
Madbarz
madbarz.com
• • +
eVito
www.evito.cz
1 5 • • • • +
storage. These devices usually do not require/provide
any connection to other systems. Representatives of
two-layer architecture include sensors such as heart
rate belts or step counters. They often do not have
their own display and usually use Bluetooth or ANT
(resp ANT+)
1
technologies for data transfer. Data are
usually displayed on a screen of a smart device. Be-
cause of the limited performance of mobile devices
only a basic data processing is provided. Advanced
data processing is performed on a remote server that
forms part of the three-layer architecture. There are
few exceptions such as BlibCare tonometer equipped
with WiFi. Then the captured data can be transferred
directly by using e.g. a home WiFi network.
4 TESTED SYSTEMS
E-health and fitness systems that use the three-layer
sensor architecture described above are most suit-
able for long term storage and management of col-
lected data. When providing a remote data transfer
1
ANT is the protocol while ANT+ is a set of mutually
agreed definitions what the information sent over ANT rep-
resents.
to a server, these devices are also suitable to be in-
tegrated with neuroinformatics infrastructures. The
biggest obstacle encountered is usually a data format
of transferred data. When devices implement Blue-
tooth or ANT+ technologies, they can use a lot of de-
fined profiles including Blood Pressure Profile (BLP),
Heart Rate Profile (HRP), Health Thermometer Pro-
file (HTP)
2
for Bluetooth, or Weight Scale and Heart
rate
3
for ANT+.
Table 1 summarizes and provides evaluation of the
e-health and fitness systems we tested. Only the sys-
tems providing at least a cloud service as a storage or
statistical results as an analytic result were selected.
For all systems we considered the number of sup-
ported internal (e.g. a GPS sensor on a smart phone)
and external sensors. This number is very limited as
can be seen in Table 1. Two systems do not support
any sensor; data are inserted manually. The last col-
umn Design is a subjective evaluation of the user in-
terface. Although all tested systems use the ANT or
Bluetooth technology, they implement a proprietary
transfer format instead of using any available profile.
2
https://developer.bluetooth.org/TechnologyOverview/
Pages/Profiles.aspx
3
https://www.thisisant.com/developer/ant-plus/
device-profiles
MoBio - A Mobile Application for Collecting Data from Sensors
117
5 MOBILE APPLICATION
PROTOTYPE
5.1 System Scope
Having difficulties with closed-source dedicated de-
vices we present a prototype of a mobile application
that solves the issues described above. It aggregates
data from various sensors into a flexible data storage
on the server. The data can be used by human readers
or processed by automatic readers. The application
can be easily used outdoors and in areas without In-
ternet connectivity. When the client gets on-line, the
stored data are synchronized with a remote storage.
However, the main core of this solution is a proposal
and implementation of suitable terminology for sen-
sory data.
5.2 Format Selection
Since the aim of the application is to support large
collections of sensors, it must store data in a flexible
data format. Existing data formats are based on differ-
ent levels of abstraction and include low-level binary
formats, highly abstract implementation-independent
data formats, and formats based on ontologies. We
required a format that provided a sufficient level of
abstraction to be system independent, but also easy-
to-use format having no specific demands on users.
Having been involved in neuroinformatics commu-
nity and having been persuaded that the similar so-
lution could be used for other sensory data, we pre-
ferred open-source formats supported and accepted in
this domain.
The working group of the INCF Task Force on
Electrophysiology
4
introduced two approaches to-
wards defining a standard, which may eventually
be merged (Teeters et al., 2013). The first one
uses the Hierarchical Data Format (HDF5) (HDF5
Group, 2013) or respectively epHDF, the specialized
HDF5 format for electrophysiology. The NIX for-
mat (Stoewer et al., 2014) provides a data model
for storing experimental data in HDF5, together with
metadata in the odML format (Grewe et al., 2011).
We selected odML (odML is a free form tree-like
structure of sections, properties and values) as a suit-
able format for the presented application because of
its platform-independence, simplicity, and human-
readability. Moreover, it ensures a compatibility with
other systems developed in neuroinformatics commu-
nity, for example (Zehl et al., 2014), (Le Franc et al.,
4
http://www.incf.org/programs/datasharing/
electrophysiology-task-force
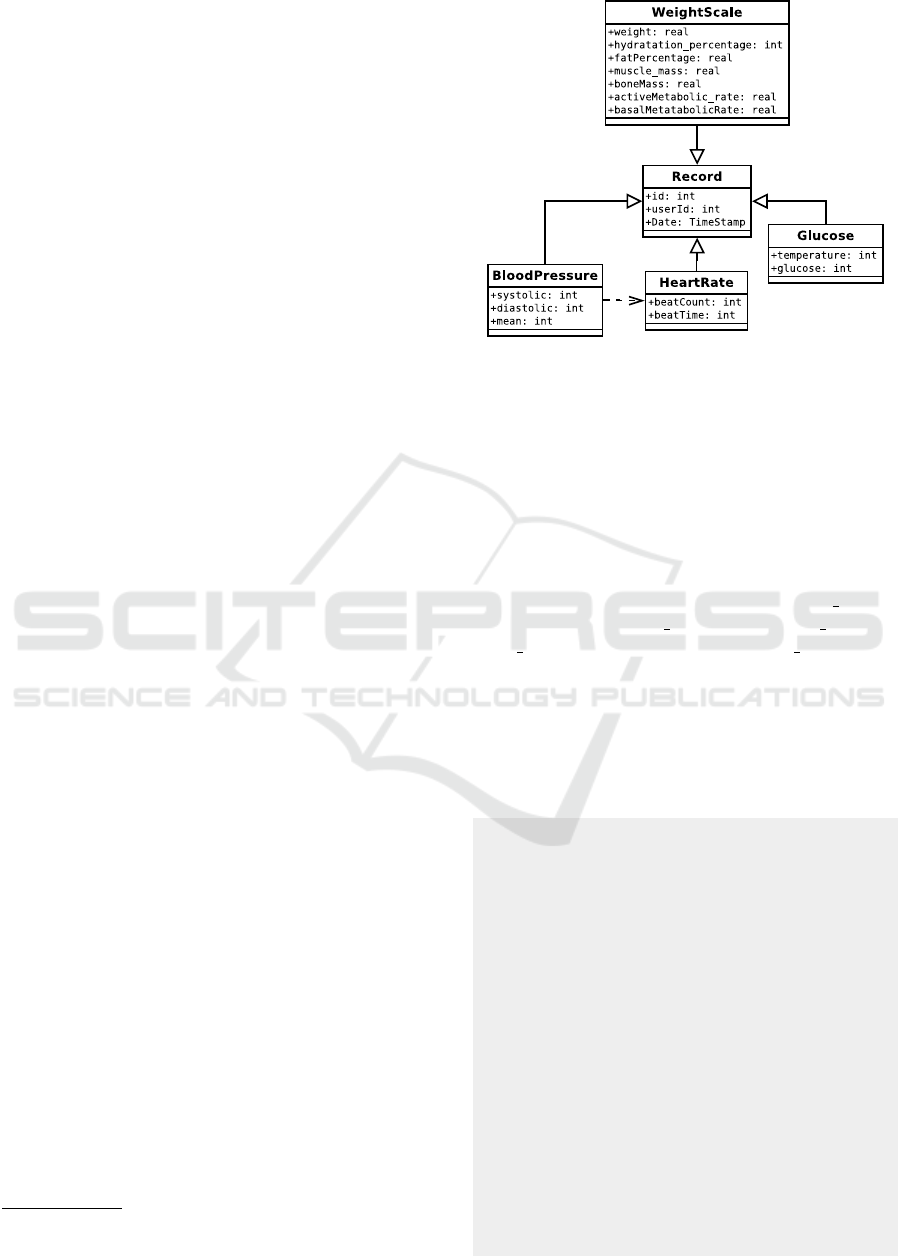
Figure 1: Proposed Terminology.
2014b), and (Davison et al., 2013).
5.3 Terminology
Figure 1 shows a terminology based on the odML for-
mat and proposed for measurements of blood pres-
sure, heart rate, glucose level, and weight scale in-
cluding hydratation, muscle mass or bone mass. List-
ing 1 shows a JSON representation of the blood
pressure data structure, the section blood presure
with properties systolic pressure, diastolic pressure,
mean pressure, and a subsection heart rate are de-
fined. The document is simplified to keep readabil-
ity. A complete document contains a complete set of
properties describing the experiment. Because of the
flexibility of the odML format, this terminology can
be easily extended for other measurements.
Listing 1: Blood pressure example.
{
" me tada t a " :{
" o dM L " :{
" d at e " : " 2 0 1 5 - 0 7 - 0 3 " ,
" x m ln s : g ui " : " ht tp : / / www . g - nod e . or g /
gu im l " ,
" s e c ti o n " : [
{
" n am e " : " b loo d _ p r e ss u r e " ,
" pr o per t y " : [
{
" n am e " : " sy s t ol i c " ,
" v a lu e " :{
" t yp e " : " i nt " ,
" c o n te n t " : 8 0
},
},
{
" n am e " : " dya s t ol i c " ,
" v a lu e " :{
" t yp e " : " i nt " ,
" c o n te n t " : 6 0
ICT4AWE 2016 - 2nd International Conference on Information and Communication Technologies for Ageing Well and e-Health
118
},
},
{
" n am e " : " m ean " ,
" v a lu e " :{
" t yp e " : " i nt " ,
" c o n te n t " : 7 0
},
},
],
" t yp e " : " b loo d _ p r e ss u r e "
}
],
" s e c ti o n " : [
{
" n am e " : " h ea r t _ ra t e " ,
" l in k " : " b loo d _ p r e ss u r e " ,
" pr o per t y " : [
{
" n am e " : " b ea t _ c ou n t " ,
" v a lu e " :{
" t yp e " : " i nt " ,
" c o n te n t " : 5 2
},
},
{
" n am e " : " bea t _ ti m e " ,
" v a lu e " :{
" t yp e " : " i nt " ,
" c o n te n t " : 1
},
},
],
" t yp e " : " h ea r t _ ra t e "
}
],
" v e r si o n " : 1
}
}
5.4 Architecture and Implementation
To support the use of open source technologies we
implemented the MoBio application for the Android
platform that is available on a wide range of devices
including tablets and mobile phones (according to
StatCounter
5
more than 60% of devices are operated
by Android). Moreover, there are other advantages to
use this platform, for example a lot of cheap devices
on the market, large community of developers, and
easy publication of applications.
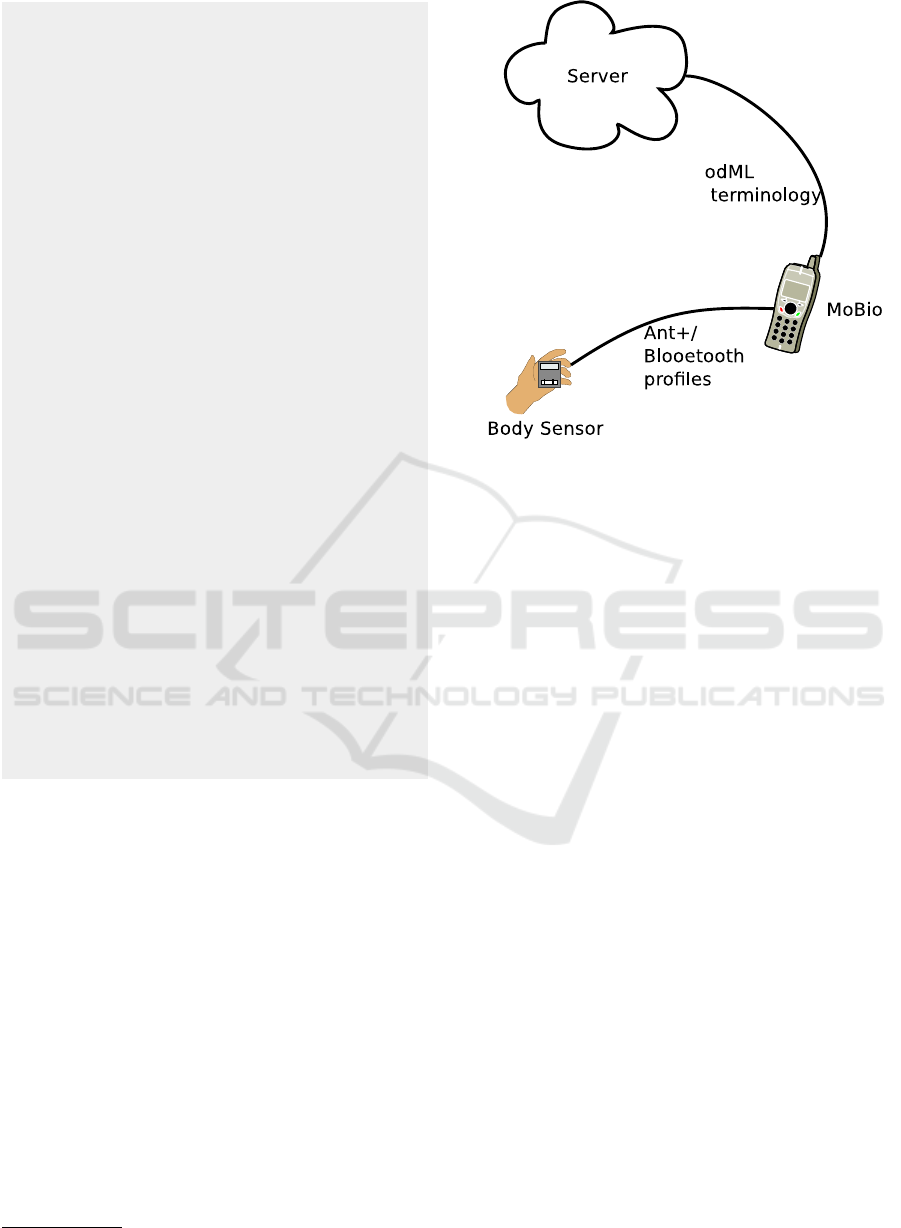
Figure 2 shows the used architecture. The user
collects data using a body sensor. The data are trans-
ferred to MoBio running on a smart phone using
an ANT+ or Bluetooth profile. Then MoBio transfers
data expressed in an odML terminology to the server.
The presented proof of concept implementation
provides basic functionality including management of
5
http://gs.statcounter.com
Figure 2: MoBio System Architecture.
user accounts, support for a limited set of sensors, and
visualization of stored data. Collected data are stored
on a SD card and can be visualized directly on the
mobile phone screen or transferred to the server and
stored there for future processing. The registration
form includes both basic information about the user
such as his/her name or email and advanced infor-
mation such as gender, weight, height, current fitness
level, etc. All these data are required because they can
affect results of measurements.
Supported devices must enable either Bluetooth
or ANT transfer. In the current implementation we
successfully tested a limited set of devices produced
by Garmin and Fora producers. When the device is
paired, the user can transfer data. Figure 3 shows a
basic functionality provided to the logged user. Fig-
ure 3a shows a list with a paired device, the Garmin
Heart Rate Sensor. The current heart rate is shown in
Figure 3b. When the user starts a measurement, data
are continuously stored in the device and the related
heart rate chart is continuously plotted as shown in
Figure 3c.
5.5 Integration with EEGBase
The collected data can be transferred to any storage
system supporting the odML format. EEGBase (con-
trary to the original purpose EEGBase is newly con-
sidered as a storage and management system for any
kind of electrophysiology data), extended with an ad-
vanced system of user templates for storing various
kinds of data and metadata in the odML format, was
selected as a storage. Any template can be defined ac-
MoBio - A Mobile Application for Collecting Data from Sensors
119
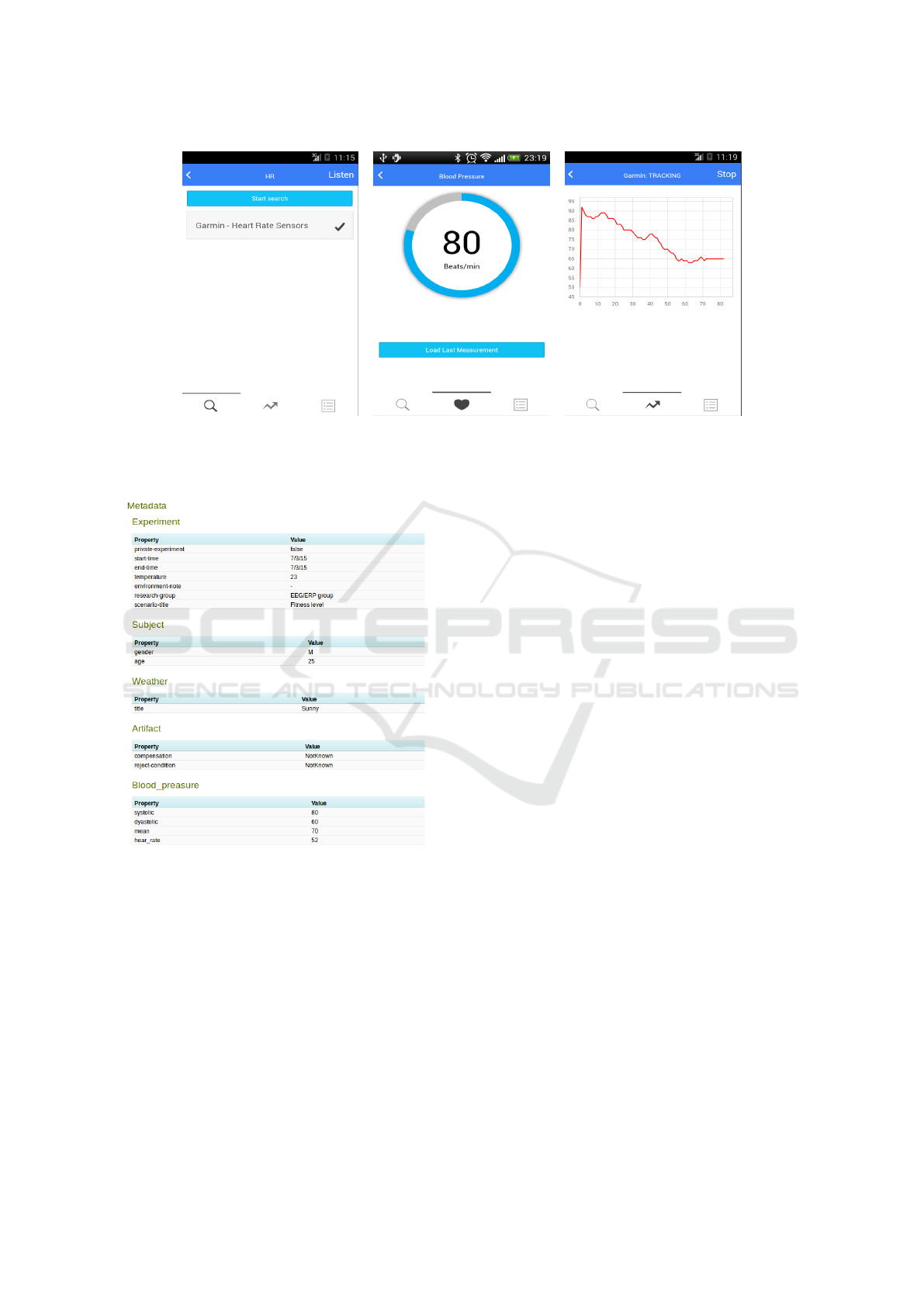
(a) List of available sensors (b) Current heart rate (c) Long term heart rate
Figure 3: Mobio Application Preview.
Figure 4: Data stored in EEGBase.
cording to data coming from a sensor and enriched by
a relevant set of metadata.
Technically EEGBase provides a RESTful API
containing methods for uploading newly collected
data. The user can select an automatic data upload
triggered when the user gets on-line. The user can
also enforce the upload manually. Once the data are
transferred to the server, the odML document is stored
in the ElasticSearch noSQL database and visualized.
Figure 4 shows visualization of the data from List-
ing 1.
6 DISCUSSION
The presented terminology covers a basic set of data
produced by typical sensors. Moreover, the terminol-
ogy is easily extensible since the odML open format
is used. Its JSON serialization is suitable for wireless
transfer due to low memory requirements in compar-
ison with XML. If we suppose a larger network of
sensors for human tracking (e.g. 15 sensors, each
sensor produces 10 signals with sampling frequency
50Hz, the sample size is 1B), then one hour record-
ing produces approximately 27MB of data plus a few
kilobytes for JSON elements. This data size is reason-
able with regard to the current capacity of SD cards.
The data security is ensured by the used protocols be-
cause both Bluetooth and ANT support data transfer
via a secured channel.
7 CONCLUSIONS
In this paper we observed a diverse collection of avail-
able sensors on the market. These sensors vary e.g. in
the used data format and data transfer protocol. We
identified architectures reflecting the ways the data
collected from sensors are managed. Then we se-
lected the three-layer sensor architecture as the most
suitable one for the long-term storage and process-
ing of sensory data. Since sensors producers use their
own systems and proprietary data formats, there are
difficulties with reading, transferring, and processing
data. As a solution we presented a prototype of a
custom terminology describing data collected from
sensors. When creating this terminology we used
ICT4AWE 2016 - 2nd International Conference on Information and Communication Technologies for Ageing Well and e-Health
120

and applied our expertise in neuroinformatics domain.
The terminology is implemented in the Android mo-
bile system MoBio that serves as a proof-of-concept
implementation supporting a limited set of sensors.
Then a complete process from collecting data by the
mobile system, storage of data in a universal format
respecting presented terminology, and final storage of
data in EEGBase is outlined.
Our future work includes extension of the pre-
sented terminology to cover a broader collection of
health sensors. This includes also surface electrodes
capturing electrical signals from the human brain.
Then the extended mobile application will convert
the data transferred using Bluetooth or ANT+ profiles
to an odML document that satisfies this terminology.
The described data can be fully managed in the sys-
tems such as EEGBase. A new version of EEGBase
with a larger support of the presented terminology and
description of the complete terminology itself will be
released. This terminology may well serve to devel-
opers of similar systems when they want to collect
and process data from health sensors.
ACKNOWLEDGEMENTS
This publication was supported by the project
LO1506 of the Czech Ministry of Education, Youth
and Sports
REFERENCES
Davison, A. P., Brizzi, T., Guarino, D., Manette, O. F.,
Monier, C., Sadoc, G., and Fr
´
egnac, Y. (2013).
Helmholtz: a customizable framework for neurophys-
iology data management. Frontiers in Neuroinformat-
ics, (25).
eMarketer (2015). 2 billion consumers worldwide to get
smart(phones) by 2016. http://www.emarketer.com/
Article/2-Billion-Consumers-Worldwide-
Smartphones-by-2016/1011694.
Grewe, J., Wachtler, T., and Benda, J. (2011). A bottom-up
approach to data annotation in neurophysiology. Fron-
tiers in Neuroinformatics, 5(16).
HDF5 Group (2013). Hierarchical data format. http://
www.hdfgroup.org/HDF5/.
INCF (2013). International neuroinformatics coordinating
facility. http://www.incf.org/.
Jezek, P. and Moucek, R. (2012). System for EEG/ERP
Data and Metadata Storage and Management. Neural
Network World, 22(3):277–290.
Jezek, P. and Moucek, R. (2013). Eeg/erp portal for android
platform. Frontiers in Neuroinformatics, (46).
Le Franc, Y., Bandrowski, A., Bruha, P., Pape
ˇ
z, V., Grewe,
J., Moucek, R., Tripathy, S. J., and Wachtler, T.
(2014a). Describing neurophysiology data and meta-
data with oen, the ontology for experimental neuro-
physiology. Frontiers in Neuroinformatics, (44).
Le Franc, Y., Gonzalez, D., Mylyanyk, I., Grewe, J., Jezek,
P., Moucek, R., and Wachtler, T. (2014b). Mo-
bile metadata: bringing neuroinformatics tools to the
bench. Frontiers in Neuroinformatics, (53).
Lowe, S. and Laighin, G. (2012). The age of the virtual
trainer. Procedia Engineering, 34:242 – 247.
Moucek, R., Bruha, P., Jezek, P., Mautner, P., Novotny, J.,
Papez, V., Prokop, T., Rond
´
ık, T.,
ˇ
Stebet
ˇ
ak, J., and
Vareka, L. (2014). Software and hardware infrastruc-
ture for research in electrophysiology. Frontiers in
Neuroinformatics, 8(20).
Seeger, C., Buchmann, A., and Van Laerhoven, K. (2011).
Wireless sensor networks in the wild: Three practical
issues after a middleware deployment. In Proceed-
ings of the 6th International Workshop on Middleware
Tools, Services and Run-time Support for Networked
Embedded Systems, MidSens ’11, pages 1:1–1:6, New
York, NY, USA. ACM.
Stoewer, A., Kellner, C., Benda, J., Wachtler, T., and Grewe,
J. (2014). File format and library for neuroscience
data and metadata. Front. Neuroinform. Conference
Abstract: Neuroinformatics 2014.
Teeters, J., Harris, K., Millman, K., Olshausen, B., and
Sommer, F. (2008). Data sharing for computational
neuroscience. Neuroinformatics, 6(1):47–55.
Teeters, J. L., Benda, J., Davison, A. P., Eglen, S., Ger-
hard, S., Gerkin, R. C., Grewe, J., Harris, K., Jackson,
T., Moucek, R., Pr
¨
opper, R., Sessions, H. L., Smith,
L. S., Sobolev, A., Sommer, F. T., Stoewer, A., and
Wachtler, T. (2013). Considerations for developing
a standard for storing electrophysiology data in hdf5.
Frontiers in Neuroinformatics, (69).
Zehl, L., Denker, M., Stoewer, A., Jaillet, F., Brochier, T.,
Riehle, A., Wachtler, T., and Gr
¨
un, S. (2014). Han-
dling complex metadata in neurophysiological exper-
iments. Frontiers in Neuroinformatics, (29).
MoBio - A Mobile Application for Collecting Data from Sensors
121
