
Parameter Identification of Canalyzing Boolean Functions with Ternary
Vectors for Gene Networks
Annika Eichler
1
and Gerwald Lichtenberg
2
1
Automatic Control Laboratory, ETH Zurich, Physikstrasse, Zurich, Switzerland
2
Faculty Life Sciences, Hamburg University of Applied Sciences, Ulmenliet, Hamburg, Germany
Keywords:
Parameter Identification, Networks, Gene Dynamics, Systems Biology, Boolean Functions, Ternary Logic.
Abstract:
In gene dynamics modeling, parameters of Boolean networks are identified from continuous data under vari-
ous assumptions expressed by logical constraints. These constraints may restrict the dynamics of the network
to the subclass of canalyzing functions, which are known to be appropriate for genetic networks. This pa-
per introduces a high performance algorithm, which solves the parameter identification problem by so called
Zhegalkin identification and exploits the restriction to canalyzing functions resulting in reduced calculation
time. The canalyzing constraint is formulated in terms of orthogonal ternary vector lists - which are intrinsi-
cally used in a Branch-and-Cut algorithm obeying this constraint. The algorithm is applied to mRNA micro
array data from mice under different contaminant conditions and good correspondence to a known apoptotic
pathway can be shown.
1 INTRODUCTION
A current field of research in systems biology is gene
dynamics modeling, since understanding the dynam-
ics of the genetic model could help the therapeutic
process (Lin and Khatri, 2013). Canalyzing Boolean
functions have shown to be appropriate to model ge-
netic networks, due to their common characteristics,
as periodicity, global complexity and self organiza-
tion (Kauffman, 1993). In genetic networks canal-
ization is the ability of a genotype to produce the
same phenotype regardless of environmental variabil-
ity (Jarrah et al., 2007). Thus, due to their stabilizing
effect on the discrete dynamical behavior, they turned
out to describe the highly ordered dynamics of gene
networks better than other Boolean models (Kauff-
man et al., 2003).
A successful approach to identify parameters of
Boolean functions from contiuous-valued signals like
microarray data uses Zhegalkin polynomials to rep-
resent these functions, see Lichtenberg et al. (2005);
Faisal et al. (2010); Veliz-Cuba et al. (2010); Breindl
et al. (2013). The Zhegalkin identification problem
is a Mixed Integer Quadratic Program (MIQP) which
can in principle be solved with standard tools like
CPLEX or Xpress, where Branch-and-Cut algorithms
are used. One major problem of Boolean identifica-
tion is the exponential growth of the cardinality of
the solution set with the number of interacting genes.
Thus, those methods are applicable up to a model or-
der of n = 10, where already very large runtimes of
hours or days occur, Faisal (2008).
Furthermore, a clustering problem has to be
solved to determine groups of genes of unknown
cardinality—denoted connectivity degree—which af-
fect each other. Combining the clustering and the
Zhegalkin identification problem leads to a problem
of discrete optimization with even higher complex-
ity. First approximations for the solution of this
combined problem have been found by a preprocess-
ing step based on the Pearson Correlation Coeffi-
cient in Faisal (2008). Next, exploiting efficient rep-
resentations of Zhegalkin polynomials as orthogonal
ternary vector lists (OTVLs), (Bochmann and Stein-
bach, 1991), and adapting tensor decomposition tech-
niques from Kolda and Bader (2009) allows integra-
tion of both steps reported in Lichtenberg and Eichler
(2011). Moreover, the solution set of the identifica-
tion algorithm can be reduced by fixing the maximum
number of rows of the OTVL representing the solu-
tion. This leads to highly efficient computation with
controllable degree of accuracy, because optimality of
the solution is guaranteed by a Branch-and-Cut algo-
rithm used for the reduced solution set.
In this paper, the latter method is restricted to the
subclass of canalyzing functions due to their interest-
110
Eichler, A. and Lichtenberg, G.
Parameter Identification of Canalyzing Boolean Functions with Ternary Vectors for Gene Networks.
DOI: 10.5220/0005978701100118
In Proceedings of the 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications (SIMULTECH 2016), pages 110-118
ISBN: 978-989-758-199-1
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
ing properties. This introduces additional constraints
for the optimization problem, as already reported in
Faisal et al. (2006) and Breindl et al. (2013), but
the reduced solution set is not efficiently exploited
therein. This work shows how to incorporate those
constraints in the identification algorithm by express-
ing canalizing functions as OTVLs. The proposed
algorithm for the identification of canalyzing func-
tions is by orders of magnitude more efficient since
the search space is considerably reduced as obvious
from Table 1. The adapted identification is applied
to gene expression data from mRNA extracted from
mouse liver cells.
This work is organized as follows. Section 2 intro-
duces fundamentals of Boolean functions, Zhegalkin
polynomials and OTVLs. In Section 3 the Branch-
and-Cut Boolean identification algorithm from Licht-
enberg and Eichler (2011) is described. Section 4
presents how to express canalyzing functions as
OTVLs and adapt the identification therefore. The re-
sults on an application to real data are shown in Sec-
tion 5. Finally conclusion are drawn in Section 6.
2 FUNDAMENTALS
The set B={0,1} denotes the set of logicals, U=[0,1]
the unit interval. Negation of Booleans is denoted by
¬z= ¯z, for real ones ¯x= 1−x holds. With ⊗ the Kro-
necker product is denoted.
2.1 Boolean Functions and Zhegalkin
Polynomials
A Boolean function b : B
n
→ B can be represented
by its truth vector b = (b
1
, ...,b
2
n
)
′
∈ B
2
n
, i.e. the last
column of the truth table as shown in Table 2.
Example 1. Consider the Boolean function
b(y
1
, y
2
) = ¬(y
1
∧y
2
), (1)
which is given by the truth table
y
2
y
1
b(y
1
, y
2
)
0 0 1
0 1 1
1 0 1
1 1 0
(2)
with its truth vector. b =
1 1 1 0
′
.
Definition 1. A Zhegalkin polynomial p(y) = l(y)
′
b
is a multilinear polynomial with b ∈ B
2
n
being a truth
vector and l(y) the so called literal vector, given by
Lichtenberg and Eichler (2011) as
l(y) =
¯y
n
y
n
⊗···⊗
¯y
1
y
1
∈ U
2
n
. (3)
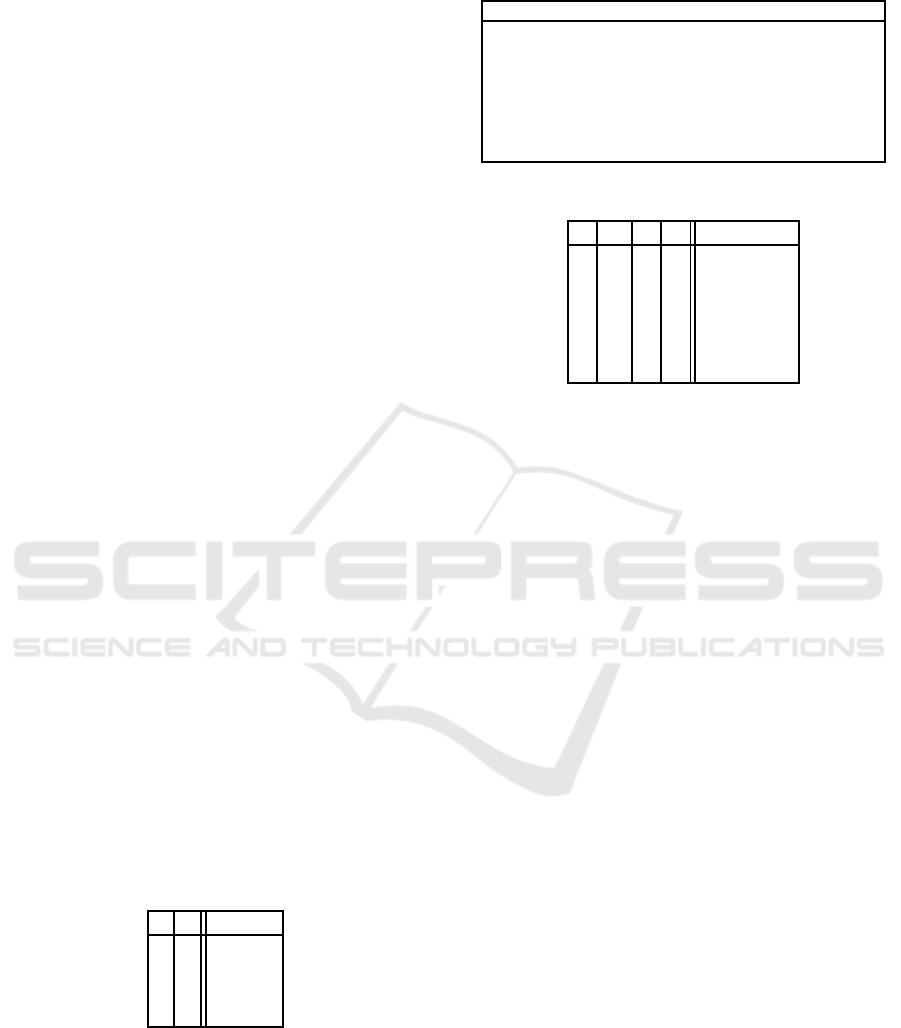
Table 1: Number of all Boolean functions and the canaly-
zing ones.
n Boolean functions CFs
1 4 4
2 16 14
3 256 120
4 65536 3514
5 4.2950·10
9
1292276
6 1.8447·10
19
1.0307 ·10
11
Table 2: Truth table.
y
n
··· y
2
y
1
b(y
1
, ..., y
n
)
0 ··· 0 0 b
1
0 ··· 0 1 b
2
0 ··· 1 0 b
3
0 ··· 1 1 b
4
.
.
.
.
.
.
.
.
.
.
.
.
1 ··· 1 1 b
2
n
Proposition 1 (Zhegalkin (1928)). A Zhegalkin poly-
nomial evaluated at Boolean values y ∈ B
n
gives the
same (Boolean) result as the Boolean function repre-
sented by the truth vector b.
Thus the Zhegalkinpolynomialscan be seen as the
bridge between the Boolean and the real set U. Since
if y ∈ U then p(y) ∈ U as well, if however y ∈ B then
p(y) ∈ B.
Example 1. (continued) To illustrate this for the
Boolean function (1) the corresponding Zhegalkin
polynomial is calculated as
l
′
(y)b =
(1−y
1
)(1−y
2
)
y
1
(1−y
2
)
(1−y
1
)y
2
y
1
y
2
′
1
1
1
0
= 1−y
1
y
2
. (4)
It can be easily seen that if y
1
, y
2
∈ B, then the Zhe-
galkin polynomial leads to the same solution as the
Boolean function (1), as declared in Proposition 1.
2.2 Ternary Vector Lists
Ternary Vector Lists (TVLs) are a common concept
in Boolean algebra, because of its outstanding advan-
tages for large scale problems, Bochmann and Stein-
bach (1991). A TVL of a Boolean function represents
all elements of the Boolean space B
2
n
where the func-
tion is 1 by ternary vectors (TVs). A TV t has the
structure
t ∈ T
n
= {0, 1, −}
n
. (5)
A zero element ’0’ in the TV describes that the corre-
sponding variable appears negated, a one element ’1’
Parameter Identification of Canalyzing Boolean Functions with Ternary Vectors for Gene Networks
111

that it appears not negated. The latter ’−’ is the don’t
care symbol, that can stand for either ’1’ or ’0’.
A TVL with k lines is of the form
T =
t
1
.
.
.
t
k
.
Taking all lines of the truth table with ones always
leads to a valid TVL of a Boolean function. TVLs
with smaller number of lines might be possible by us-
ing ’−’.
Example 1. (continued) With the truth table in (2)
valid TVLs for the Boolean function (1) of the run-
ning example are
T
1
=
"
0 0
0 1
1 0
#
, T
2
=
0 1
− 0
, (6)
T
3
=
0 −
− 0
, T
4
=
0 −
1 0
. (7)
This can easily be checked by replacing ’− ’ with both
’0’ and ’1’.
This example shows that TVLs are not unique,
i.e there exist different TVLs for the same Boolean
function. Another important property is orthogonal-
ity (Bochmann and Steinbach, 1991).
Definition 2. A TVL T is orthogonal, if each binary
vector appears only once in T. This is the case, if
for any pair of lines of T in at least one column a
(0,1)-combination appears. Two TVLs T
A
and T
B
are
orthogonal if T
A
and T
B
have no binary vectors in
common. This is the case if for any pair of lines of
T
A
and T
B
in at least one column a (0,1)-combination
appears.
A binary vector (BV) is a vector with only ’0’s and
’1’s. It can represent only one line of the truth table,
while a ternary vector (TV) due to ’−’ can represent
multiple BVs.
Example 1. (continued) For the TVLs of the exam-
ple it is obvious that all TVL representations are or-
thogonal except of T
3
with no (0,1)-combination in
any column. Here the binary vector
0 0
appears
in both lines.
In the following an orthogonal TVL is denoted
as OTVL. In Bochmann and Steinbach (1991) oper-
ations for OTVLs are described. Important for this
work are the complement and the difference opera-
tors, which are visualized in Table 3 for 3 variables.
The complement CPL(T) =
¯
T of a given OTVL T is
defined as the OTVL of all binary vectors that are not
in T. The difference DIF(T
A
, T
B
) of the OTVLs T
A
and T
B
results in an OTVL of all BVs, that are in T
A
but not in T
B
. If the result is the empty OTVL, T
A
is
totally included in T
B
.
Table 3: Graphical representation of operands for TVLs,
Bochmann and Steinbach (1991).
T
A
=
0 0 −
1− 1
CPL(T
A
) =
0 1 −
1− 0
T
B
=
1−−
DIF(T
A
,T
B
) =
00−
Lemma 1. An OTVL T is orthogonal to its comple-
ment
¯
T.
Proof. With Definition 2 two TVLs are orthogonal, if
they do not have any BVs in common. The comple-
ment of an OTVL T contains all BVs, that are not in
T and is thus orthogonal to T.
Proposition 2. For an OTVL T with k lines the num-
ber of ones in the correspondoing truth vector b is
N
1
= b
′
1 =
∑
k
i=1
2
N
i
−
where N
i
−
is the number of ’−’s
in the i-th line of T.
Proof. The number ones in b is equivalent to the
number of BVs in T. A TV with no ’−’s represents a
single BV and since a ’− ’ stands for either 1 or 0, a
TV with N
−
times the ’−’ symbol, includes 2
N
−
BVs.
Due to orthogonality no BV appears more than once
in T, so that the number of BVs in each line can sim-
ply be added.
2.3 OTVLs and Zhegalkin Polynomials
Since OTVLs and Zhegalkin polynomials are two dif-
ferent representations of Boolean functions, it is pos-
sible to find the corresponding mapping between both
representations.
Proposition 3. Given is an OTVL T of n variables,
that is representing a Boolean function f, then the
corresponding Zhegalkin polynomial, determined by
p
T
, is calculated as
p
T
(y) =
k
∑
j=1
n
∏
i=1
T(t
ji
,y
i
) (8)
with T(t
ji
, y
i
) =
¯y
i
, if t
ji
= 0,
y
i
, if t
ji
= 1,
1, if t
ji
= −.
Proof. Assume T is an OTVL, i.e. without ’−’s, then
∏
n
i=1
T(t
ji
, y
i
) corresponds to the l-th row of the literal
vector. Since t
j
is only a line of T when b
l
= 1 due
to the construction of an OTVL, (8) is equal to l(y)
′
b,
what finishes the proof for OTVLs without ’−’s. If T
is an OTVL with a ’−’ in the k-th column, than this
is equal to a TVL with the same row and a ’1’ in the
k-th column and additionally the same row and a ’0’
in the k-th column. For the row with the ’1’, if it is the
SIMULTECH 2016 - 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
112

n-th row, it is
∏
n
i=1
T(t
ni
, y
i
) = y
k
∏
n
i=1,i6=k
T(t
ni
, y
i
),
and for that with the ’0’, if it is the m-th row, it is
∏
n
i=1
T(t
mi
, y
i
) = ¯y
k
∏
n
i=1,i6=k
T(t
mi
, y
i
). Thus the sum
is (y
k
+ ¯y
k
)
∏
n
i=1,i6=k
T(t
mi
, y
i
) =
∏
n
i=1,i6=k
T(t
mi
, y
i
),
since
∏
n
i=1,i6=k
T(t
mi
, y
i
) =
∏
n
i=1,i6=k
T(t
ni
, y
i
). What
finishes the proof for all OTVLs.
Example 1. (continued) Let’s consider
T
2
=
0 1
− 0
of the running example. Evaluating (8) for T
2
leads to
p(y) = ¯y
1
y
2
+ 1¯y
2
= (1 −y
1
)y
2
+ (1 −y
2
) = 1−y
1
y
2
as derived with the literal form (4) before.
3 ZHEGALKIN IDENTIFICATION
BY BRANCH-AND-CUT
ALGORITHM
Finding the best Boolean model for continuous nor-
malized data is known as Zhegalkin identification
problem, see Faisal et al. (2005), that has been
shown to be well suited for Boolean identification of
gene networks (Faisal, 2008; Veliz-Cuba et al., 2010;
Breindl et al., 2013). In Lichtenberg and Eichler
(2011) the Zhegalkin identification problem is solved
with the help of OTVLs by a Branch-and-Cut algo-
rithm.
In contrast to the first references, the efficient al-
gorithm in Lichtenberg and Eichler (2011) allows to
include this clustering problem in the identification.
A cluster is denoted as the set of genes, which af-
fects the dynamics of a gene of interest, since a gene
is never affected by all others genes, but only a subset,
the cluster. The size of the cluster, called connectivity
degree, and the cluster itself are unknown and have to
be determined in the clustering problem.
Before the main contribution, how OTVLs of ca-
nalyzing functions are structured and how to restrict
the identification to canalyzing functions, the Zhe-
galkin identification algorithm from Lichtenberg and
Eichler (2011) is shortly introduced here.
3.1 Minimization Problem
A Zhegalkin function of n signals can be modeled by
n truth vectors or the respective OTVLs. The state
space model for signal l is then given as
y
l
(t + 1) = l(y(t))
′
b
l
= p
T
l
(y(t)), ∀l = 1, . . . , n
(9)
with p
T
l
(y) as defined in (8). The prediction er-
ror between y
l
(t + 1) predicted with the OTVL T
l
as
model as in (9) and the measurement value ˜y
l
(t + 1)
of signal l at any time t = 0, ··· , T −1 is defined as
d
l
(t + 1) = y
l
(t + 1) − ˜y(t + 1). The task of the Zhe-
galkin identification problem is to find the optimal
OTVL T
⋆
l
and the corresponding Zhegalkin polyno-
mial that solves the minimization problem
min
T
l
J
l
with J
l
=
s
T−1
∑
t=0
d
l
(t)
2
(10)
with J
l
, the 2-norm of the prediction error, being the
error function. It is clear that this minimization prob-
lem has to be solved for all signals l = 1, . . . , n. There-
fore this index is omitted in the following.
One major problem of Boolean and thus Zhe-
galkin identification is the high cardinality of the
search space. There exist 2
(2
n
)
different Boolean
functions of n variables. This fast growth in the num-
ber of variables n is exemplarily shown in Table 1. To
deal with this problem, the algorithm presented here
finds the best approximation T
+
with fixed maximal
number of rows, instead of searching for the optimal
solution. This row restriction significantly reduces the
search space by preserving the basic properties as it is
approved in Section 5 by the numerical example.
3.2 Branch-and-Cut Algorithm
The Zhegalkin identification with rank restriction
from Lichtenberg and Eichler (2011) is a Branch-
and-Cut algorithm, where the nodes represent pos-
sible OTVLs. The algorithm is initialized with the
empty OTVL. The children in the next level are all
3
n
OTVLs with one line. The following levels are
built respectively by adding one TV, that is orthogo-
nal to the parent node, to the OTVL of the parent node
while descending in the search tree. This is equiva-
lent to elongate the OTVL of the parent node by one
line. The algorithm can be summarized in the follow-
ing steps
(1) Initialization
(2) Repeat: Define branching node, branch node, cut
nodes
(3) End: According to stop criteria
The implemented Branch-and-Cut algorithm uses a
best first strategy, therefore the branching node is al-
ways the leaf (node without children) with smallest
error function and with less than the maximal permit-
ted row number. When branching the branching node,
for each TV, that is orthogonal to the OTVL of the
branching node, a leaf where this TV is added to it is
Parameter Identification of Canalyzing Boolean Functions with Ternary Vectors for Gene Networks
113

generated. For each new node the prediction error is
calculated, and when it is clear, that this new branch
can not decrease the current global best solution J
+
,
the node is cut, i.e. deleted from the search tree. The
cutting condition hereby is
cut node j if ∃t ∈ {1, ..., T} : d
j
(t) >
p
ˆ
J
+
. (11)
Here d
j
(t) is the prediction error of node j at time t
and
ˆ
J
+
is the cost of the current best solution. The
cutting condition (11) can be explained by the fact
that y(t) ∈ U and thus non-negative. Therefore the
Zhegalkin polynomial of every TV is non-negative as
well. Thus if the modeled value for one time exceeds
the measured one by more than the current error, the
error can not get smaller if a further TV is added.
For more explanations see Lichtenberg and Eichler
(2011).
Several stopping criteria exist, like a desired lower
threshold of the cost or a maximum number of itera-
tions, can be set manually. If the algorithm stops be-
cause no node is branchable anymore, i.e. every leaf
has reached the maximal permitted row number, then
the optimal T
+
in the restricted search space is found
with minimal cost J
+
.
3.2.1 Including the Clustering Problem
In general the Branch-and-Cut algorithms runs for
each possible cluster, set of genes the considered gene
may depend on, separately. However, if the initial
lower bound
ˆ
J
+
for each new cluster is set to the low-
est optimal bound J
+
of all previously identified clus-
ters, advantage of this information can be taken: if
a cluster with a very good solution has been found,
the cutting condition (11) of the following clusters is
tightened from the beginning on, i.e. a lot of nodes are
cut, leading to reduced calculation effort.
4 CANALYZING FUNCTIONS
Canalyzing functions are a subclass of Boolean func-
tions with the property, that their result is fixed, if one
specific input takes a specific value, no matter what
values the other inputs take.
Definition 3 (Lichtenberg et al. (2005)). A Boolean
function f is canalyzing if there exists an i ∈ {1, ..., n}
and a fixed s, v ∈ {0, 1} such that for all y ∈ B
n
we
have f(y
1
, ..., y
i
, ..., y
n
) = v if y
i
= s.
The variable y
i
is termed as canalyzing variable, s
as canalyzing value and v as canalyzed value. If no i
can be found, so that the condition above is fulfilled,
the function is classified as non-canalyzing. For a ca-
nalyzing Boolean function the following holds
Lemma 2. Given an Boolean function f for n vari-
ables that is canalyzing in y
i
with canalyzing value s
and canalyzed value v, then its complement
¯
f is ca-
nalyzing in y
i
with s and ¯v.
Proof. The complement of the Boolean function f is
defined as
¯
f = 1− f. Thus if f(y
1
, ..., y
i
= s, ..., y
n
) =
v the complement
¯
f evaluated for y
i
= s is
¯
f(y
1
, ..., y
i
= s, ..., y
n
) = 1−v = ¯v.
4.1 OTVLs of Canalyzing Functions
Whereas expressing canalyzing functions as Zhe-
galkin polynomials has been considered in Faisal
(2008); Faisal et al. (2010), this work is focused on
expressing canalyzing in form of OTVLs to be able to
restrict the Branch-and-Cut algorithm of Section 3 to
only canalyzing functions.
If a Boolean function is canalyzing, for the respec-
tive OTVL one of the two following Lemmas holds,
depending on the canalyzed value.
Lemma 3. Given an OTVL T for n variables and with
k lines, then T is canalyzing in variable y
c
with cana-
lyzing value s and canalyzed value v = 0 if and only
if t
jc
= ¯s for all j = 1, . . . , k.
Proof. The corresponding Zhegalkin polynomial is
calculated by (8). Since t
jc
= ¯s for all j = 1, . . . , k,
(8) can be written as
p
T
(y) = T( ¯s, y
c
)
k
∑
j=1
n
∏
i=1,i6=c
T(t
ji
, y
i
). (12)
If y
c
= s, i.e. the canalyzing value is taken, then
T( ¯s, y
c
) = T( ¯s,s) = 0, thus p(y) with y
c
= s is equal
to v = 0.
Lemma 4. Given an OTVL T for n variables and with
k lines, then T is canalyzing in variable y
c
with cana-
lyzing value s and canalyzed value v = 1, if and only
if T includes a TV t
c
defined as t
c
= [t
c
1
, . . . , t
c
n
] with
t
c
c
= s and t
c
i
= − for all i ∈ {1, . . . , n}\c.
Remark 1. To be included in T, the TV t
c
must not
be a line of T, but all BVs in t
c
must appear in T,
i.e. DIF(t
c
,T) = {}. The empty TVL corresponds to a
Boolean vector with only zeros.
Proof. If T is canalyzing with v = 1 its complement
¯
T
is canalyzing with v = 0, see Lemma 2. According to
Lemma 1 the complement
¯
T is orthogonal to all TVs
in T. Thus there has to be a (0,1)-combination for any
pair of rows out of T and
¯
T. As proposed T has to
include t
c
, where are only ’−’s in row j except of in
the c-th column. To be orthogonal to t
c
in every line
SIMULTECH 2016 - 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
114
[
−−0
]
[
1−−
]
[
1−−
]
[
−0−
]
[
0−−
]
h
1−−
0 1 0
i
h
1−−
0− 0
i
h
1−−
0 1 −
i
h
−0−
1 0 0
i
h
0−−
1 0 0
i
h
0−−
1− 1
i
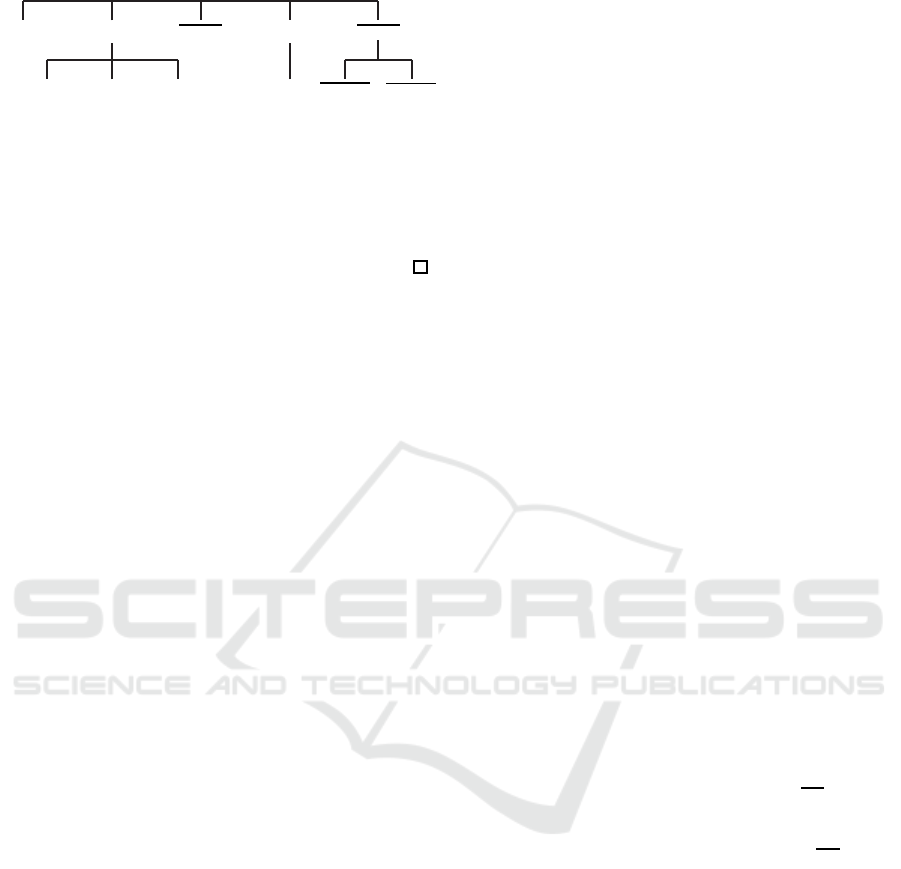
Figure 1: Search tree for Boolean identification restricted to
canalyzing functions with n = 3 and row number restricted
to two.
of the complemented
¯
T in the cth-column there has
to be the element ¯s. Thus
¯
T is canalyzing with v = 0
according to Lemma 3.
Example 1. (continued) The Boolean function (1)
from the running example is canalyzing with canaylz-
ing variable y
1
as well as y
2
, both with canalyzing
value ’0’ and canalyzed value of ’1’: if y
1
or y
2
, re-
spectively, takes the value ’0’, than the result of the
Boolean function is ’1’, independently of the other
variable. This is also obvious from the OTVL rep-
resentations in (6), which fall in the class of OTVLs
described in Lemma 4.
4.2 Zhegalkin Identification with
Canalyzing Constraints
In Faisal et al. (2005); Faisal (2008); Faisal et al.
(2010); Breindl et al. (2013) it is shown how to ex-
press canalyzing functions as Zhegalkin polynomi-
als and integrate those constraints in the Zhegalkin
identification. Here it is shown how to restrict the
Branch-and-Cut algorithm in Section 3 to canaliz-
ing constraints. In addition to its good biological
properties another worthwhile advantage of canaly-
zing functions is their reduced number compared to
all Boolean functions, see Table 1. There the num-
ber of canalyzing Boolean functions for n variables is
compared all existing Boolean functions. A signifi-
cant decrease of the number of canalyzing functions
compared to all Boolean ones is obvious. The adap-
tion introduced here of the identification algorithm
takes advantage of that and can considerably reduces
the calculation time thereby.
To restrict the Branch-and-Cut algorithm from
Lichtenberg and Eichler (2011) to canalyzing func-
tions, only few adaptions are necessary. First instead
of initializing the search tree with the empty OTVL as
before, it is to initialize with the 2n TVs of n variables,
which are canalyzing with v = 1.
Example 1. For 3 variables, due to Lemma 4 all TVs,
which are canalyzing with v = 1 are given as
1−−
,
−1−
,
−−1
,
0−−
,
−0−
,
−−0
,
where the canalyzing variable of the two TVs in the
first columns is the first variable with the canalyzing
value 1 and 0, e.g. for the second and third variable.
Due to Lemma 4 any orthogonalTVs can be added
to these root-nodes, without loosing the canalyzing
property. Furthermore each existing canalyzing func-
tion with v = 1 (with respect to the maximum line
constraint) is in the search space, because by initial-
ization all existing combinations of canalyzing vari-
able and value are covered, and can thus be identified.
To cover also the canalyzing functions with v = 0
as additional roots those 2n TVs, which are canaly-
zing with v = 1, are taken again, but subtracted from
the TV only consisting of ’−’s, describing the whole
Boolean space. Note that the subtraction operation
for Zhegalkin polynomials is equivalent to the Differ-
ence operation for the corresponding OTVLs. Sub-
tracting a TV of the whole Boolean space is equiva-
lent to building the complement, thus due to Lemma
2 the resulting OTVL is canalyzing with v = 0. If one
of these root-nodes with v = 0 should be branched,
then instead of adding all orthogonal TVs, all orthog-
onal TVs are subtracted. Hereby the canalyzing prop-
erty with v = 0 is preserved. Note that for checking
if a TV is orthogonal, it is more efficient to check if
it is orthogonal to all TV’s that are substracted, then
from the difference itself. To distinguish between the
OTVLs canalyzing with v = 1 and v = 0, v is added as
further variable to each node. In the branching step,
if for the branching node we have v = 1, orthogonal
TVs have to be added, otherwise subtracted. For the
cutting step, the cutting condition also depends on v
as follows
cut node j
with v = 0 if ∃t ∈ {1, ..., T} :
d
j
(t)>
√
J
+
,
with v = 1 if ∃t ∈ {1, ..., T} :
d
j
(t)> −
√
J
+
.
5 APPLICATION OF THE
CONSTRAINED
IDENTIFICATION
ALGORITHM
The presented identification algorithm is applied to
gene expression data also used in Faisal et al. (2010).
The considered gene expression data are measure-
ments of mRNA extracted from mouse liver cells us-
ing microarray technology (GeneChip Human Exon
1.0 ST Array). The measurements were repeated four
times (T = 3) after 2, 4, 12 and 24 hours. In total the
expression levels of 21799 genes could be detected.
Parameter Identification of Canalyzing Boolean Functions with Ternary Vectors for Gene Networks
115
Two different mRNA samples were tested, one treated
with the contaminant Benzo(a)pyrene (BaP) with a
concentration of 5µM and one with a lower one of
50nM, called T5µM and T50nM in the following.
This contaminant BaP is found in cigarette smoke
and automobile exhaust and is connected to deadly
diseases such as cancer. Geneticists assume that the
contamination of cells with BaP with the high con-
centration of 5µM leads to the cellular process apop-
tosis, programmed cell death, but not the contamina-
tion with the low concentration. Therefore the present
gene data is analyzed with regard to apoptosis.
The apoptotic pathway for mice can be found
in the KEGG database, Kanehisa and Goto (2000),
hosted by Kanehisa Laboratory. From all detected
genes, 78 are, due to the database, known to be in-
volved in the apoptotic pathways. These are extracted
and considered in the following. The database gives
for each gene a set of genes where it may depend on.
This knowledge is taken into account for a first identi-
fication, where these sets are taken as possible clusters
for the identification of the respective gene. Thereby
possible solutions of clusters are a priori reduced.
The identification with canalyzing constraints as pre-
sented in Section 4.2 and without constraints as given
in Lichtenberg and Eichler (2011) is applied. The
maximum number of rows of the resulting OTVLs
is restricted to two. For the identification for each
gene a model for connectivity degree two up to the set
size given in the database is identified with canalyzing
constraints. For the identification without constraints
the maximal connectivity degree for each gene is re-
stricted to 5, although for some genes the database
give a possible larger cluster, since already for 5 the
average calculation time for one possible cluster is
with 71 s more than a minute. And if a gene may
depend on 11 genes, according to the database, with a
connectivity degree of 5 this results in
11
5
= 462 pos-
sible clusters, and thus in more than 546 minutes for
only one gene. In comparison with canalyzing con-
straints, one cluster takes 0.022s for a connectivity
degree of 5. For a connectivitydegree of 11, the maxi-
mum one found in the database, the identification with
canalyzing constraints takes 28.66×10
3
s.
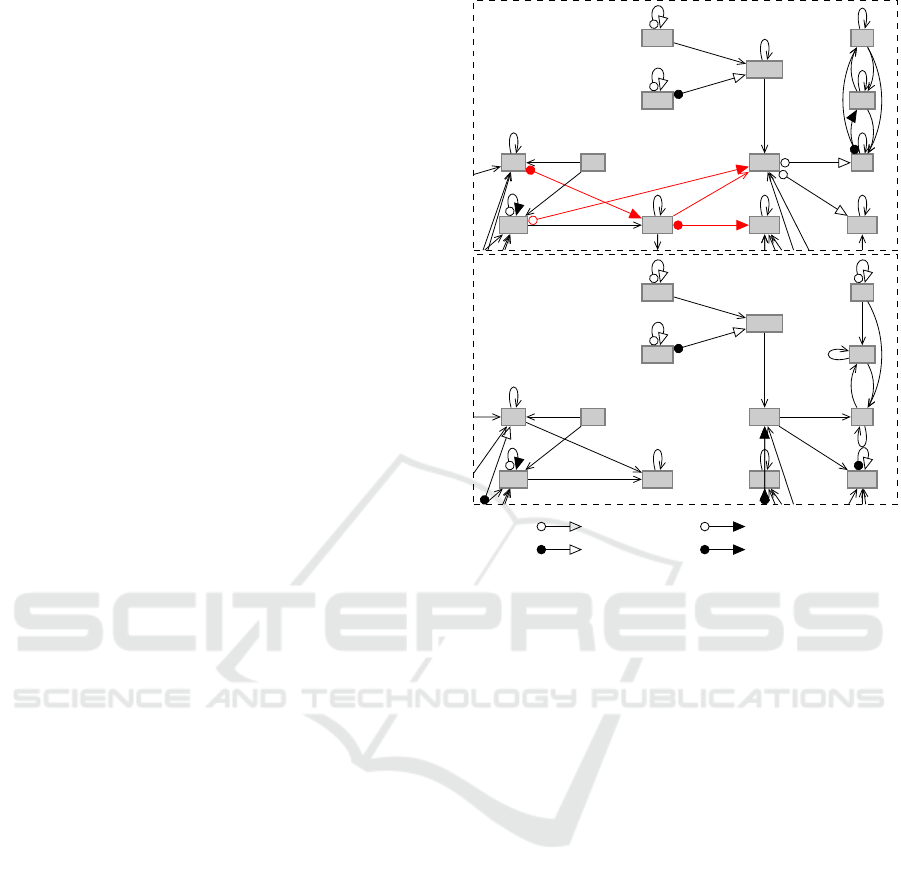
A cutout of the identified network is shown in Fig-
ure 2 for both concentrations. In general the apoptotic
pathways consists of the extrinsic pathway and the in-
trinsic one. Here the extrinsic one is shown in de-
tail. The expectation, that the concentration of T5µM
leads to apoptosis, while that of T50nM does not, is
affirmed here. According to the database the extrinsic
pathway is triggered by engagements at the death lig-
ands, which activate caspase-8. That induces a signal-
ing cascade, resulting in an activation of caspase-3,
T5µM
Fadd
Tradd
Cflar
Capn1
Capn2
Casp8
Casp12
Casp3
Casp7
CAD
Dffb
Dffa
Casp6
T50nM
Fadd
Tradd
Cflar
Capn1
Capn2
Casp8
Casp12
Casp3
Casp7
CAD
Dffb
Dffa
Casp6
s = 0, v = 0 s = 0, v = 1
s = 1, v = 0 s = 1, v = 1
Figure 2: Identified extrinsic pathway for T5µM and
T50nM with given clustering constraints, (canalyzing func-
tions in red canalyzing functions, with no constraints in
black, that with minimum error is shown).
what leads to cell death. This can be seen for T5µM,
where caspase-8 is activated leading to and activation
of caspase-3. In Figure 2 the connections of impor-
tance here are marked in red. For T50nM there is
no connection between caspase-8 and caspase-3 de-
tected. The arcs with circled tail and triangular head,
denote the canalyzing genes, thus the major influenc-
ing one. If the tail is colored, its canalyzing value
is one, if the head is colored, the canalyzing value is
one, and zero otherwise. Thus, for the interconnec-
tion from caspase-8 to caspase-7 in the network of
T5µM this, e.g. means that an activation of caspase-
8 always activates caspase-7, irrespectively of other
genes, whereas a deactivation of Tradd always acti-
vates caspase-3.
An a posteriori analysis of the models identified
by the identification, where no canalyzing constraints
were imposed, shows that a significant ratio of identi-
fied models are canalyzing functions. These ratios of
canalyzing functions compared to all identified func-
tions for a certain connectivity degree are shown in
Figure 3. For comparison the overall ratios of canaly-
zing function in all Boolean functions, as calculated
from Table 1, are given. It is obvious that expect for
the connectivity degree of two the identified models
SIMULTECH 2016 - 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
116
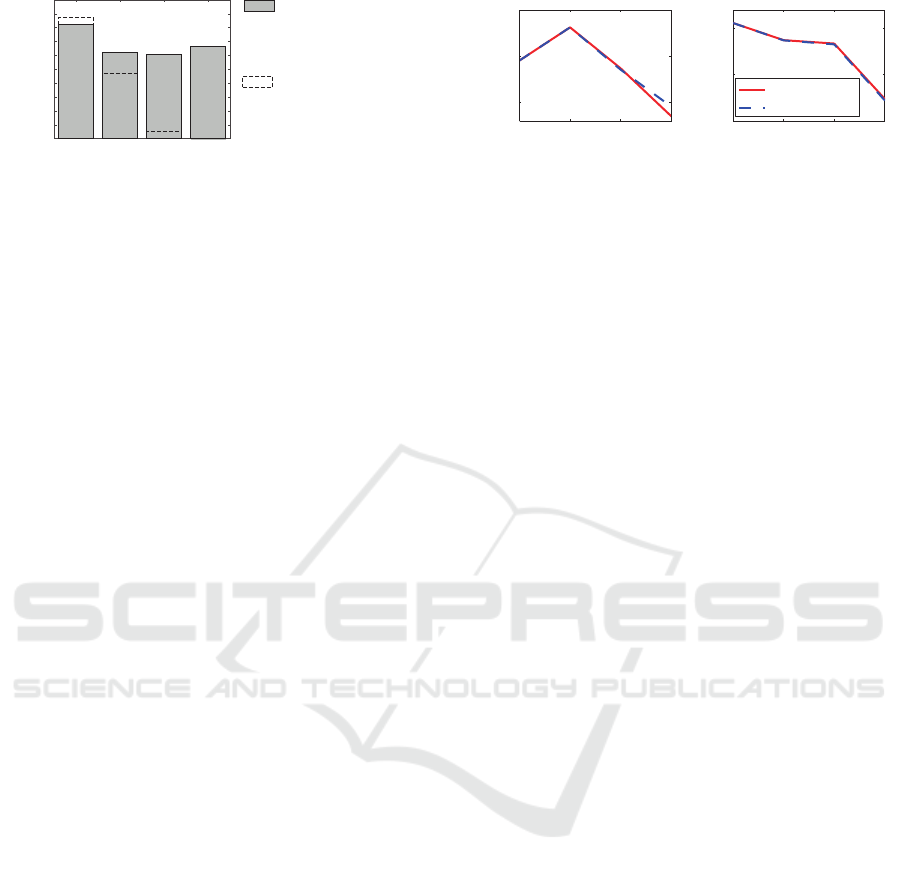
0
0.2
0.4
0.6
0.8
1
2 3 4 5
replacements
connectivity degree
ratio of canalyzing func-
tions resulting from the
identification without ca-
nalyzing constraints
ratio of canalyzing func-
tion in all Boolean func-
tions
ratio of canalyzing
functions
Figure 3: Ratio of canalyzing functions.
show a significant higher ratio of canalyzing functions
than there exists in total. This confirms the choice to
restrict the identification to canalyzing functions, not
only due to the restricted search space and thus re-
duced calculation time, but also because genetic mod-
els obviously tend to be canalyzing, as also reported
by biologists.
The identification is repeated, without consider-
ing the dependency sets given by the database, but
testing all possible clusters with connectivity degree
from two to four with maximum number of rows of
the OTVL restricted to two. Note that thus for one
gene,
78
4
+
78
3
+
78
2
= 1505504 different clusters
have to be checked. Here only identification with ca-
nalyzing constraints is performed, since without con-
straints, this is not tractable anymore. For T5µM in
average an error of 6.87×10
−5
is achieved, where
the root mean square error is taken as error measure.
Biologists talk about good approximations if an er-
ror < 10
−3
is achieved. This is not reached for only
two out of the 78 genes. Remark that for the identi-
fication the maximum number of lines of the identi-
fied OTVLs was restricted to two, which is necessary
to reduce the solution space and make the problem
tractable. This seems to be very small. Nevertheless
the very good fit of the identified models confirms that
this might be enough. For T50mM the average error
is with 1.43×10
−4
slightly larger. This also let sus-
pect, that the high concentration rather lead to apop-
tosis than the low one. Here only the genes involved
in apoptosis are considered, but if other processes are
executed, other genes may be involved.
To analyze the continuous gene expression level
dynamics, the measurements and the prediction using
the identified model are compared. The prediction of
gene l, initialized with the measured values
˜
y(0), is
determined by
y
l
(t + 1) = p
T
l
(y(t)) with y(0) =
˜
y(0).
The dynamic of two genes for T5µM is shown in
Figure 4. Here with caspase-3 and caspase-8 , two
genes right in the center of the extrinsic pathway are
depicted. The prediction fits very well, what is not
astonishing since errors of 1×10
−7
and 5.6×10
−8
0 . 7 5
0 . 8
0 . 8 5
0 1 2 3
discrete timesteps
gene expression data
caspase-3
0 1 2 3
0 . 5
0 . 5 5
0 . 6
discrete timesteps
gene expression data
caspase-8
measurement
prediction
Figure 4: Measured vs. predicted gene expression level dy-
namics for T5µM.
are achieved. For the sample T50nM the error of the
identified model is with 1. 2 ×10
−4
and 3.2×10
−4
al-
most 10
3
-times worse. This also supports the conclu-
sion, that other processes then apoptosis with other
genes involved occur for that sample.
6 CONCLUSIONS
The paper presents how to express canalyzing func-
tions in terms of OTVLs. Based on that, it is shown
how to restrict the solution space of the Boolean iden-
tification algorithm in Lichtenberg and Eichler (2011)
to canalyzing functions by simple adaptions mainly
in the initialization step. Thereby the restriction to a
maximum number of lines, that as a core of the algo-
rithm leads efficiently to a suboptimal solution, does
not need to be given up. The advantage of the restric-
tion to canalyzing function is twofold, first from the
biological point of view, since canalyzing functions
are known to describe gene networks better than other
functions, and second from the computational point of
view. By the adaption of the Zhegalkin identification
algorithm presented in this paper, the search space
is enormously reduced by the canalyzing constraints,
what leads to managable computation times even for
larger data. The presented algorithm has been applied
to experimental gene data. By the canalyzing con-
straints the problem of identification and clustering
of 78 genes got tractable and has shown very good
fits. Further assumptions of the biologists regarding
the network structure of specific processes could be
approved by the algorithm presented here.
ACKNOWLEDGEMENTS
The authors acknowledge Saskia Trump (UFZ
Leipzig) for her support and experimental data.
Parameter Identification of Canalyzing Boolean Functions with Ternary Vectors for Gene Networks
117

REFERENCES
Bochmann, D. and Steinbach, B. (1991). Logikentwurf mit
XBOOLE. Verlag Technik.
Breindl, C., Chaves, M., and Allg¨ower, F. (2013). A linear
reformulation of boolean optimization problems and
structure identification of gene regulation networks. In
Proc. 52nd IEEE Conf. Decision Control, pages 733–
738.
Faisal, S. (2008). Discrete-Time Modelling of Gene
Networks by Zhegalkin Polynomials. Ingenieurwis-
senschaften. Dr. Hut Verlag.
Faisal, S., Lichtenberg, G., Trump, S., and Attinger, S.
(2010). Structural properties of continuous represen-
tations of boolean functions for gene network mod-
elling. Automatica, 46(12):2047–2052.
Faisal, S., Lichtenberg, G., and Werner, H. (2005). A poly-
nomial approach to structural gene dynamics mod-
elling. In Proc. 16th IFAC World Congr., pages 2119–
2119.
Faisal, S., Lichtenberg, G., and Werner, H. (2006). Canali-
zing Zhegalkin polynomials as models for gene ex-
pression time series data. In Proc. 1st Int. Cong. Eng.
Intell. Syst.
Jarrah, A. S., Raposa, B., and Laubenbacher, R. (2007).
Nested canalyzing, unate cascade, and polyno-
mial functions. PhysicaD: Nonlinear Phenomena,
233(2):167–174.
Kanehisa, M. and Goto, S. (2000). Kegg: Kyoto encyclo-
pedia of genes and genomes. Nucleic Acids Research,
28(1):27–30.
Kauffman, S. (1993). The Origins of Order, Self Organi-
zation and Selection in Evolution. Oxford University
Press.
Kauffman, S. A., Petersen, C., Samuelsson, B., and Troein,
C. (2003). Random boolean network models and the
yeast transcriptional network. PNAS, 100(25):14796–
14799.
Kolda, T. and Bader, B. (2009). Tensor decompositions and
applications. SIAM Review, 51(3):455–500.
Lichtenberg, G. and Eichler, A. (2011). Multilinear alge-
braic boolean modelling with tensor decompositions
techniques. In Proc. 18th IFAC World Congr., pages
5603–5608.
Lichtenberg, G., Faisal, S., and Werner, H. (2005). Ein
Ansatz zur dynamischen Modellierung der Genex-
pression mit Shegalkin-Polynomen. Automatisierung-
stechnik, 53:589–596.
Lin, P. and Khatri, S. (2013). Logic Synthesis for Genetic
Diseases: Modeling Disease Behavior Using Boolean
Networks. Springer.
Veliz-Cuba, A., Jarrah, A. S., and Laubenbacher., R. (2010).
Polynomial algebra of discrete models in systems bi-
ology. Bioinformatics, 26(13):1637–1643.
Zhegalkin, I. (1928). Arithmetics of symbolic logic. Mat.
Sb., 35(3-4):311–377.
SIMULTECH 2016 - 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
118
