
Learning Diagnosis from Electronic Health Records
Ioana Barbantan and Rodica Potolea
Computer Science, Technical University of Cluj-Napoca, G. Baritiu nr. 26-28, Cluj-Napoca, Romania
Keywords: Data Mining, Classification, Value Mapping, Concept Extraction, Semantic Medical Data Alignment.
Abstract: In the attempt to build a complete solution for a medical assistive decision support system we proposed a
complex flow that integrates a sequence of modules which target the different data engineering tasks. This
solution can analyse any type of unstructured medical documents which are processed by applying specific
NLP steps followed by semantic analysis which leads to the medical concepts identification, thus imposing a
structure on the input documents. The data collection, document pre-processing, concept extraction, and
correlation are modules that have been researched by us in our previous works and for which we proposed
original solutions. Using the collected and structured representation of the medical records, informed
decisions regarding the health status of the patients can be made. The current paper focuses on the prediction
module that joins all the components in a logical flow and is completed with the suggested diagnosis
classification for the patient. The accuracy rate of 81.25%, obtained on the medical documents supports the
strength of our proposed strategy.
1 INTRODUCTION
The medical domain continues to capture the interest
of both researchers and industry. There is a
continuous struggle to find the remedy for an
incurable disease that, however, does not harm the
overall health status of the patient. On the other hand,
assistive medical technology is helping patients cope
with their suffering such as hearing loss, hand tremors
(Smith, 2014), or are in need of physical therapy (Lee,
2015).
Understanding how patients will react based on
the medication they are under is an information that
can be explored in the personal records of the
patients. The manual analysis of the medical records
for searching purposes is not feasible due to the heavy
increase of data, especially in unstructured format. On
the other hand, the massive increase of data should
not hinder the exploitation of the Electronic Health
Records (referred to in the following as EHRs). It is
suggested that 70% of the useful medical information
is captured in unstructured text documents (D'Avolio,
2013). The data that can be extracted from these
documents and the inferred knowledge transforms the
medical records into valuable sources of information.
In the attempt to exploit the medical records,
several challenges arise. Typically, the documents are
in unstructured format, are written in a domain
distinctive language, contain domain specific terms,
abbreviations, and acronyms. A number of text pre-
processing steps are required to convert these medical
documents into a format that can be easily exploited
by machines for knowledge inference purposes. The
documents need to be structured and the relevant
concepts must be extracted. Understanding the
context and being able to identify the word meaning
in case of polymorphic words are only a few of the
challenges a knowledge extraction system faces.
While the research benefits are obvious, a knowledge
extraction system enhances the existing medical
knowledge bases and supports the improvement of
the existing health care systems.
The analysis of the medical data in free text
format provides information such as predicting
adverse reactions by analysing the interaction of
drugs (when combined) or identifying co-morbidity
risks, forecasting possible conditions that may occur
based on previous studies and cases. The data can be
used for designing solutions for recommending
investigations for a thorough diagnosis, suggesting
diagnosis or follow-up appointments. The patients
can benefit from upgraded medical care when the
medical records enclosing their medical history,
illnesses, allergies, interventions, and many other
health related characteristics become accessible at
any time by the physicians. On these grounds, the
Electronic Health Records systems have been
introduced to deliver advanced medical services.
344
Barbantan, I. and Potolea, R.
Learning Diagnosis from Electronic Health Records.
DOI: 10.5220/0006069503440351
In Proceedings of the 8th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2016) - Volume 1: KDIR, pages 344-351
ISBN: 978-989-758-203-5
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

The collection of data about patients modelled as
a knowledge base gives more insight into each
patient’s health status and medical needs. The
existence of a knowledge base of former patients
previously investigated and diagnosed, benefits along
all dimensions: health care, costs, diagnosis, and
reduced hospitalization time. Therefore, the ultimate
goal of an medical system is obtaining recommenda-
tions provided by an assistive decision support system
employing such knowledge base. The benefits, to
name a few, are the decrease of patient’s suffering and
a reduced number of medical investigations, qualified
both as costs and time interval between the patient's
hospitalization and start of treatment, thus initiating
the healing process.
The remainder of the paper is organized as
follows into 6 sections. Section 2 describes how the
information extraction and mapping from EHRs is
addressed in literature. Section 3 introduces the
supervised techniques for predicting patient diagnosis
based on the information in their clinical records
while Section 4 introduces the general strategy of
diagnosing patients. Section 5 describes the
experimental setup for classifying the content of an
EHR into a specific diagnosis, while section 6
concludes our work and presents the future
enhancements and directions.
2 BACKGROUND
Along with the EHRs and EHRs systems adoption, a
number of studies were conducted to evaluate their
impact and the users’ satisfaction (Edsall and Adler,
2008). By the year 2011, in the USA, the EHR
systems had been adopted by 54% of the physicians
and 85% of the adopters reported being content and
satisfied while using these systems. However, one
half of the physicians not using an EHR system said
they were planning on purchasing one. These
indicators are provided by (Jamoom, et al., 2012).
The increasing trend on EHRs adoption is introduced
in (Hsiao and Hing, 2014) who report an 18%
adoption by the office-based physicians in 2001. The
adoption rate reached 78% in 2013 on the same
medical cohort.
For tagging the information captured in the EHRs,
a number of annotation tools were made available.
The authors in (Jonquet, et al., 2009) focus their
research on annotating biomedical textual
information via an ontology-based web service. A
collection of over 200 biomedical ontologies and
terminology repositories were integrated. They were
collected from the UMLS ontology repository
(Bodenreider, 2004) and the NCBO bioportal
ontologies (Musen, et al., 2012). The authors propose
a two-step mapping approach. First, a syntactic
concept recognition step is employed using a
dictionary of terms generated from the UMLS
ontology repository and the NCBO bioportal
ontologies. Then, the annotations were augmented
with the knowledge extracted from other ontologies.
A semantic distance was computed to generate new
annotations considering the sibling relations defined
in the ontologies, while an ontology-mapping
component propagated the annotations via the
mappings between the ontologies. One challenge
faced when mapping text to ontology is the ontology
selection, as a consequence of the increasing number
of available ontologies, reported by the authors in
(Jonquet, et al., 2010).
Extracting semantic relations from text is a crucial
step towards natural language understanding, and
towards creating a structured representation of the
content. Although the relation extraction task is a
well-known problem it is still not trivial. Applied to
the healthcare domain it gets even more difficult, due
to the lack of grammar rules and jargon-rich nature of
the text. Some of the approaches dealing with relation
identification between concepts in discharge
summaries are reviewed below.
The task of relation identification is essential in
the automated and semi-automated ontology
development. The authors in (Doing-Harris, et al.,
2015) exploited the synonymy and hierarchical
relations existing between the concepts and based on
the tf/idf frequency, and constructed semantic
vectors. Another use case for the relation
identification between concepts was proposed in
(Henriksson, et al., 2014). The authors examined a
solution for establishing the relations between
synonyms and abbreviations and their corresponding
mappings on concepts from the medical domain. The
generalization of the proposed approach derived from
the use of semantic spaces extracted from two
different corpuses of medical data, namely a corpus
of clinical documents and a corpus of medical journal
articles. The performance measurements of the study
are reported as recall: 0.39 for abbreviations to long
forms, 0.33 for long forms to abbreviations, and 0.47
for synonyms.
The authors in (Albin, et al., 2014) proposed a
method to identify the relations between medical
concepts exploiting the UMLS ontology collection
and implementing the onGrid Web platform. The
platform allowed for efficient transitive queries and
conceptual relation identification. The relations were
assessed between any two sets of biomedical concept
Learning Diagnosis from Electronic Health Records
345

relations and the relations within one set of
biomedical concepts. The proposed solution was
exemplified on the disease-disease relation. The
semantic distance between concepts was computed
based on the semantic type of the concepts as defined
in the UMLS. The relations were defined as weak
when the concepts belong to the abstract types found
closer to the root of the UMLS semantic network. For
imposing an order on the relations, a formula was
introduced to identify the closeness between
concepts, which led to the construction of a relation
matrix.
Supervised Machine Learning techniques and
rule-based methods had been proposed for identifying
the values associated to the medical concepts in
clinical documents. The authors in (Doan, et al.,
2012) proposed an ensemble classifier composed of a
rule-based system, a supervised classifier (SVM) and
a CRF model to recognize medication information
from clinical text. The features extracted to build the
classification model included word features, POS
tags, morphologic features (to capture the affix
information), orthographic features, history features,
and semantic tag features.
Addressing the same task of mapping values to
medical concepts, the authors in (Jiang, et al., 2014)
exploited the information in the RxNorm ontology to
identify the medication concepts. The error analysis
showed that a number of synonyms, abbreviations
and misspelled words contributed to reducing the
value of the recall.
Enabling efficient search across medical
information while submitting primitive or abstract
queries has been investigated by the authors in (Boaz
and Shahar, 2003). They proposed the IDAN project
that allowed for employing temporal ontologies for
querying medical specific information contained in
databases.
3 METHODOLOGY FOR MINING
DATA FROM EHRS
In our previous study (Bărbănțan and Potolea, 2015)
we proposed a strategy for implementing a medical
Assistive Decision Support System. The proposed
system can handle as input any type of unstructured
medical documents from EHRs to radiology reports
or medical prescriptions. The documents need to be
pre-processed using specific Natural Language
Processing tasks, followed by the semantic analysis.
Once the semantic information is associated with the
input data, the extraction of relevant concepts can be
completed. The concept identification and the
specific semantic categories enable the definition for
a structure for the input documents. In the attempt to
provide a structure to the documents the information
must be grouped into sections, such as symptoms,
diagnosis, mediation, follow-up appointments,
investigations, and medical history. The obtained
structured information wss filtered and classified
such that custom decisions about the health status of
the patients can be made. The final objective triggers
the type of solution, that may consist in a search in the
documents for specific concepts, or may represent an
evaluation of the <concept, value> pairs, while the
prediction solution collects all the knowledge and
uses it to evaluate the health status of a new patient.
To evaluate and predict the health status of a
patient we need to understand the methodology of
diagnosis making and we need to be able to extract
from a patient’s EHR the relevant data that can be
used in determining the diagnosis. Figure 1 shows
how the data is extracted and analysed when
assessing the health status of a patient based on his
medical record. In a Data Mining (referred to in the
following as DM) approach, disease understanding is
translated into training models for each diagnoses
while patient evaluation is represented by how similar
the health status of an investigated patient is to the
previously patterns generated in the training phase,
thus to each disease’ specific features.
Figure 1: Methodology for extracting knowledge and
diagnosis prediction.
3.1 Information Extraction from Raw
Data
Collecting representative and valuable data to solve
any particular DM problem is still a challenge. To
identify the relevant features for representing a
disease, several data properties must be satisfied. The
features must have a good coverage upon the
instances and situations such that the false negative
rate is minimized. The features must not be redundant
and should be represented in the most efficient data
formats, whether as numbers, strings or dates. When
KDIR 2016 - 8th International Conference on Knowledge Discovery and Information Retrieval
346

all these conditions are satisfied, a DM algorithm can
be expected to achieve the best performances.
The topic we are currently addressing in this
paper is the prediction of the health status of a patient,
namely if he/she suffers from a specific. To tackle this
challenge, the medical concepts need to be identified
and associated with the corresponding values as
examined for each patient, thus identifying the pair
<concept, value>. The values can either be explicitly
stated in the text as numbers or strings, or they can be
inferred based on the context.
3.2 Information Exploration
Once the medical concepts have been identified along
with their associated values, the data can be exploited
to tackle various tasks using DM strategies. Such
tasks include identifying the relation between the
extracted concepts or predicting the patient’s
diagnosis. The solution for extracting knowledge
from EHRs follows the general DM process as stated
also in (Alag, 2009). The first step in developing the
learning approach is understanding the purpose of the
solution and defining a strategy of achieving it in the
setup imposed by the content. Identifying the
relations between concepts becomes a problem of
finding patterns in data.
3.2.1 Relation Identification
Concept relation identification represents a step
towards establishing the structure of documents,
leading to a conceptual map for representing the
documents. The concept relation identification
process involves an initial step where the relevant
concepts are identified. Identifying the relations
assists in predicting future behaviours or trends and
recognizing patterns in data. Nevertheless,
identifying the relations between the concepts can be
exploited as a learning tool. They are useful for
identifying co-morbidities and help understanding
and learning medical conditions and inferring new
relations between them.
In our previous work (Porumb, et al., 2015),
(Bărbănțan, et al., 2016) we proposed the ReMED
strategy for relation identification between medical
concepts. We tackled this task by constructing a
knowledge base with examples of the different
relation types and generating a model via a SVM
classifier. The created training dataset was assessed
in several feature setups. The relation identification
proposed solution was feature engineered. For each
pair of concepts, a number of features was extracted.
The feature vector was built starting from the bag of
words representation of the input data and
progressively enhanced with features grouped into
the following categories: context, lexical, syntactic,
and grammatical. The features were typically
Boolean, but for a few, the mapped value was integer
or real. To identify the final ReMED model, a best
feature setup was determined employing a trade-off
between precision and recall.
3.2.2 Diagnosis Identification
To automatize the disease identification logical path
followed by a physician, the steps followed need to
be translated in machine understandable rules. All the
knowledge of a physician has to be fed as training
data to an algorithm to learn patterns for classifying
each disease. The health status of the patient has to be
transformed and mapped to be consistent with the
structure of the training data such that the patient can
be evaluated. Employing a Machine Learning
strategy for diagnosis identification, the physician’s
knowledge needs to be transformed into examples
from which the algorithm to learn patterns.
The Clinical Problem Solving course introduces a
general description into the physician’s reasoning
during the diagnosis process (Lucey, MD, 2015).
Following the general flow of understanding what
causes a disease and how to identify it, we defined an
automatic method for making informed decisions
about patients’ health status. The diagnosis making
process follows the general pattern illustrated in
Figure 2 and described in the following.
Figure 2: Understanding what causes a disease.
When assessing the health status of a patient, the
physician performs the following examinations. He
knows how things normally work and classifies the
investigated condition as normal functioning, if
applicable. When things do not follow the normal
path, the patient’s health is labelled as abnormal
functioning which leads to the investigation of the
syndromes and diseases that cause the abnormal
functioning. The physician continues the investiga-
tion by capturing the isolated clinical signs and
symptoms that result from the abnormal functioning,
grouped into syndromes. The ultimate goal is
determining what types of diseases cause the
syndromes and proposing a treatment plan and an
attempt at predicting the evolution of the health status
of the patient.
Normal
functioning
Abnormal
functioning
Syndromes Diseases
Learning Diagnosis from Electronic Health Records
347

4 FROM PATIENT
APPOINTMENT TO
DIAGNOSIS
The flow in section 3.2.2 introduces the flow of
diagnosis identification in a general setup. The
evaluation must consider actual concept values, as a
pair <concept, value>. For example, the existence of
albumin in an EHR is not relevant unless associated
with a laboratory result. To be aligned with the DM
terminology, we will refer to in the following the
medical concepts as features. Because each feature
has to satisfy particular conditions such as normal and
abnormal ranges, data type, or fall in specific
categories, these conditions must be converted into
machine-readable rules. To do this we assign to the
evaluated features their corresponding data types and
define the ranges, either categorical or numerical. The
actual mapping is performed based on the content of
the medical document, and after the actual medical
concepts were identified from the text.
4.1 Medical Concepts Identification
In our previous work, we introduced the MedCIN
solution (Bărbănțan, et al., 2014), (Bărbănțan, et al.,
2015), (Szenasi, et al., 2015), for assigning categories
(disease, symptom, medication, procedure) to the
medical concepts, we have defined several termino-
logy lists, by querying the SNOMED-CT ontology, as
follows: we extracted the instances which are related
to the categories and identified the correspondences
between the concepts and the terminology lists. To
identify the content, the categories, we explored the
semantic class classification as defined in the
ontology. A class includes all the concepts assigned
to a given semantic type. From each semantic class
we extracted all the instances along with the
following information: id, preferred label and all the
alternative labels. The alternate labels as defined in
SNOMED-CT are actually the synonyms of the
concepts, representing other possible terminologies
for a selected concept. Each instance is pre-processed
following the same steps which were employed for
the input documents: POS identification, stop words
removal and lemmatization.
4.2 Assigning Values to Medical
Concepts
Once the medical concepts have been identified, their
values need to be mapped based on the data definition
constraints. Each medical concept has a specific type,
which can fall in one of the specific DM data types:
continuous, Boolean or nominal. To identify each
category, the following strategies were established.
While the solution for quantifying nominal data is the
definition of lists of terms and the solution for
assessing continuous data is the numeric patterns,
when it comes to quantifying the existence of
mentioned medical conditions deep semantic analysis
needs to be involved. When evaluating a medical
concept that is described within a category, post-
processing steps may be involved when the actual
category is not specified in the text.
5 SUPERVISED APPROACH FOR
DIAGNOSING PATIENTS
FROM EHRS
This section describes the instantiation of the
proposed methodology on a specific disease. The
training and testing setups are detailed along with the
evaluations performed. In the attempt to implement
and evaluate the proposed methodology a dataset
containing data about patients that suffer from
Chronic Kidney Disease (CKD) was referenced.
5.1 The CKD Dataset
The dataset used for training the model is the CKD
dataset (Lichman, 2013) from UCI. The dataset is
designed to be used for predicting Chronic Kidney
Disease in patients and the data was collected from an
Indian hospital on a period of 2 months from 400
patients (250 that suffered from CKD and 150 that did
not suffer from CKD). To assess the health status of
the patients, 24 attributes were measured, such as age,
blood components statistics, and the presence of
underlying conditions such as anaemia, hypertension
or coronary artery disease.
The CKD model was created by training the data
using the implementation of the Bayes classifier from
Weka (Hall, et al., 2009). The evaluation accuracy on
the training data on a separate test set using 10-fold
cross-validation, was 98.75% as only the condition of
5 patients was not correctly predicted.
5.2 Feature Vector Mappings
The data structure and representation from the
training data imposes the way the test set is built. For
each type of attribute (nominal and continuous)
specific rules have been defined such that the values
from the text can be mapped to the features. Figure 3
KDIR 2016 - 8th International Conference on Knowledge Discovery and Information Retrieval
348
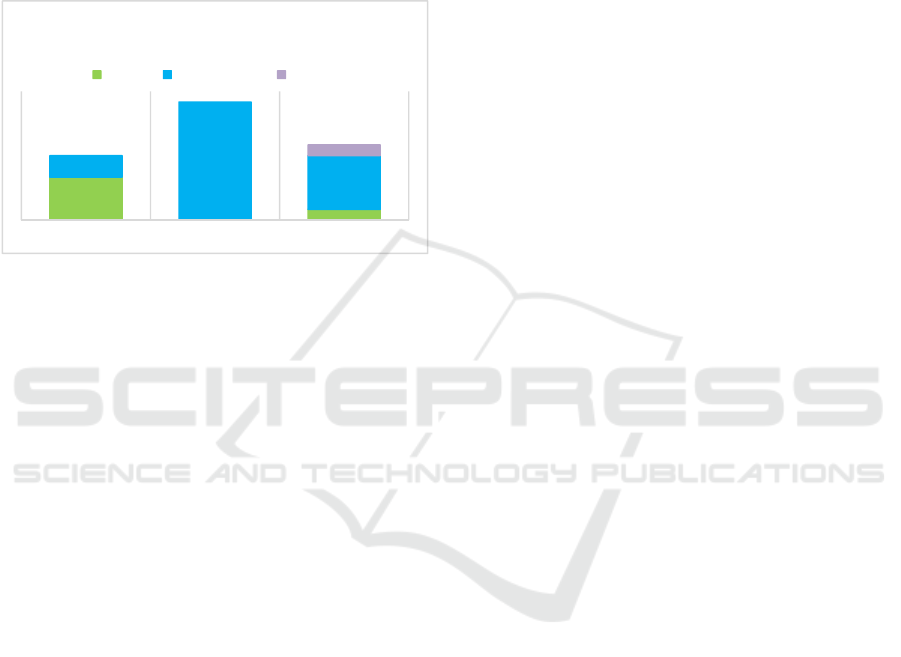
shows that the distribution of continuous and nominal
data types in the training dataset is similar. Each EHR
instance was converted with the proposed knowledge
extraction technique into an instance.
For evaluating whether patients suffer from CKD
based on the content in their EHRs, the document
which is received in an unstructured format is passed
through a number of transformations that allow for
the identification of the features that describe the
health status and assess their corresponding values.
Figure 3: Feature count per data type in the training data.
While the relevant medical concepts are identified
using the strategy described in section 4.1, extracting
their values is presented in the following.
5.2.1 Continuous Value Extraction
Laboratory tests, patient age, blood pressure, weight
or height are evaluated as continuous values. Once
these concepts were identified, an analysis followed
that evaluated the relation of the concept with the
continuous values identified in the text to identify
whether a number is associated to the concept. To
map the results to the correct attribute, equivalence
classes were defined for each attribute. For example,
the age attribute was identified in documents as “y/o”,
“ year old” or “age”, while the pedal edema attribute
was found as “lower extremity edema” or “peripheral
edema”. The condition is imposed by the training data
as the data type associated to age is continuous. Not
mentioning the actual numeric age of the patient
assigns the unknown value to the feature age “?”, just
like in the case of an ambiguous mention of age such
as “elderly” or “young”.
5.2.2 Nominal Value Extraction
Assessing a tumour’s stage or the degree of spread of
a rash imposes a comparison with similar evaluations.
There are cases when a feature can be evaluated only
in a limited number of ways, thus each feature
instance falls under a category. In the particular case
of medical data, the nominal features can be either
strings, such as appetite <good, bad>, or numeric
values, such as Specific Gravity defined by the
following ranges <1.005, 1.010, 1.015, 1.020, 1.025>.
To map the values from the documents to the
categories defined in the training data, a dictionary of
nominal strings has been used and in the case of
numeric ranges, the extracted values were mapped
accordingly.
Syndromes or diseases are usually evaluated
using Boolean values. For these concepts their
presence or ruling out is relevant, that is why, for the
Boolean value extraction a negation analysis is
required. The negation was identified and assessed
with the NegEx (Chapman, et al., 2001) strategy for
syntactic negation identification, and our PreNex
strategy for morphologic negations, introduced in our
previous work (Bărbănțan and Potolea, 2014). In the
particular case, the underlying conditions of
hypertension, diabetes mellitus or coronary artery
disease are cues for the CKD.
5.3 CKD Evaluation
For identifying the patterns in evaluating whether a
patient suffers from CKD, we applied a supervised
learning algorithm on the CKD dataset. The training
consisted of the Bayes classifier evaluated in a 10
folds cross-validation setup. For classifying the medi-
cal instances, the information from a medical record,
was converted into a feature vector which contained
the values extracted from the medical record.
Due to the limited amount of publicly available
annotated medical data, our experiments were limited
in regard to the number of documents. To evaluate the
concept extraction and value extraction, the medical
documents have been processed.
The EHR samples for evaluation were collected
from the MTSamples dataset (Helpline, 2010) and
consisted of 32 EHRs: 16 describing patients
suffering from CKD and 16 not suffering from CKD.
The evaluation of the EHRs consisted in the
classification of 1211 sentences which lead to the
filtering, classification and assessment of 15828
words. The condition for selection these particular
samples related to the investigations presented in the
EHRs, namely all patients have in common the
features used for determining the presence of the
CKD. The results of the classification showed an
accuracy of 81.25%, for the 32 new EHR instances.
5.4 Discussion
There are some issues that need to be discussed
concerning the content of the HER sampled involve
4
1
2
11
5
1
BOOLEAN CONTINUOUS NOMINAL
DISTRIBUTION OF DATA TYPES IN THE
TRAINING DATA
Disease Laboratory test Symptom
Learning Diagnosis from Electronic Health Records
349

in the evaluation and the information contained in the
knowledge base. While the knowledge base is
composed of features that describe the CKD medical
condition, these features were not always present in
the EHR of a patient suffering from CKD. For this
reason, three of the instances belonging to the CKD
class were empty, thus their classification was
incorrect due to the missing information requested by
the knowledge base. This finding deepens our
understanding regarding the way the investigations
are conducted in the medical domain. It is a known
fact that the strategies for determining the cause of
patients’ suffering are different across institutions,
and thus, across medical doctors. These findings
support the premise that the collection and linkage of
as many sources of data as possible, even if their
structure and purpose may seem dissimilar, leads to
more accurate solutions.
6 CONCLUSIONS
The present research explores strategies for handling
the transformation of the unstructured data into
structured format via a knowledge transformation
flow. The output of the transformation enables the
classification of the input unstructured text,
represented by EHRs, into a specific diagnosis. While
the current solution covers a single diagnosis, we
propose extending the training dataset with further
diagnoses, thus extending the feature vector with
features that are present when assessing the presence
of specific diseases. The current status of the medical
Assistive Decision Support System covers complete
solutions for automatically structuring medical
documents and extracting relevant medical concepts
via the PreNex and MedCIN strategies, while the
prediction module is argued in favour, being
validated with an actual use case.
The proposed strategy represents the final step in
our proposed medical Assistive Decision Support
System, introduced in our previous research
(Bărbănțan and Potolea, 2015). Starting from raw
medical data, the proposed solution infers the
appropriate suggestion to each specific task (further
investigations, diagnosis or medication). The solution
enables the transformation of the medical documents
– which are usually stored in unstructured format –
into a structured format by exploring and applying a
taxonomy based mapping technique. This technique
involves the extraction of the relevant terms from the
text assisted by a domain specific terminology and a
context based classification. A number of pre-
processing steps are involved in normalizing both the
input text (unstructured data) and the terminology
sources (structured data), which proved to carry a
significant role. The filtering step which allows for
the discrimination between medical and non-medical
concepts proves to be an efficient method. In the
selection of the terminology sources (WordNet and
SNOMED-CT) their ability to cover the biomedical
domain and also to obtain accurate information was
considered.
ACKNOWLEDGEMENTS
This work has been partially supported by Brained
City - Information Technology based Innovative
Development of Cluj-Napoca Fully Integrated Urban
Ecosystem.
REFERENCES
Alag, S., 2009. Collective Intelligence in Action.
Greenwich: Manning Publications Co.
Albin, A. et al., 2014. Enabling Online Studies of
Conceptual Relationships Between Medical Terms:
Developing an Efficient Web Platform. JMIR Med
Inform, 2(2:e23).
Bărbănțan, I., Lemnaru, C., Potolea, R., 2014. Disease
Identification in Electronic Health Records. An
ontology based approach. Rome, Italy, SCITEPRESS,
pp. 261-268.
Bărbănțan, I., Lemnaru, C., Potolea, R., 2015. Concepts
Identification in Medical Documents. York, University
of Sheffield.
Bărbănțan, I., Porumb, M., Lemnaru, C., Potolea, R., 2016.
Feature Engineered Relation Extraction – Medical
Documents Setting. International Journal of Web
Information Systems (IJWIS), 12(3), pp. 336-358.
Bărbănțan, I., Potolea, R., 2014. Exploiting Word Meaning
for Negation Identification in Electronic Health
Records. Cluj-Napoca, IEEE Computer Society, pp.
283-289.
Bărbănțan, I., Potolea, R., 2015. Knowledge Extraction and
Prediction from Medical Documents. Ohrid, ICT ACT.
Boaz, D., Shahar, Y., 2003. Idan: A distributed temporal-
abstraction mediator for medical databases. Protaras,
Cyprus, Proceedings of the 9th Conference on Artificial
Intelligence in Medicine—Europe (AIME).
Bodenreider, O., 2004. The Unified Medical Language
System (UMLS): integrating biomedical terminology.
Nucleic Acids Res. 32(Database issue), pp. 264-270.
Chapman, W., Bridewell, W., Hanbury, P., Cooper, G. F.,
2001. A simple algorithm for identifying negated
findings and diseases in discharge summaries. Journal
of Biomedical Informatics, 34(5), pp. 301-310.
D'Avolio, L., 2013. 6 Questions to Guide Natural Language
Processing Strategy. Information Week, 18 February.
KDIR 2016 - 8th International Conference on Knowledge Discovery and Information Retrieval
350

Doan, S. et al., 2012. Recognition of medication informa-
tion from discharge summaries using ensembles of
classifiers. s.l., BMC Med Inform Dec Mak, p. 36.
Doing-Harris, K., Livnat, Y., Meystre, S., 2015. Automated
concept and relationship extraction for the semi-
automated ontology management (SEAM) system.
Journal of Biomedical Semantics.
Edsall, R. L., Adler, K. G., 2008. User Satisfaction With
EHRs: Report of a Survey of 422 Family Physicians.
Family Practice Management, 15(2), pp. 25-32.
Hall, M. et al., 2009. The WEKA Data Mining Software: An
Update. s.l.:SIGKDD Explorations.
Helpline, M., 2010. Transcribed Medical Transcription
Sample Reports and Examples. [Online] Available at:
http://www.mtsamples.com [Accessed 4 January
2016].
Henriksson, A. et al., 2014. Synonym extraction and
abbreviation expansion with ensembles of semantic
spaces. J Biomed Semantics.
Hsiao, C., Hing, E., 2014. Use and characteristics of
electronic health record systems among office-based
physician practices: United States, 2001-2013. NCHS
Data Brief, January, Volume 143, pp. 1-8.
Jamoom, E. et al., 2012. Physician adoption of electronic
health record systems: United States, 2011. NCHS Data
Brief, July, Issue 98, pp. 1-8.
Jiang, M. et al., 2014. Extracting and standardizing
medication information in clinical text – the MedEx-
UIMA system. s.l., s.n.
Jonquet, C., Musen, M. A., Shah, N. H., 2010. Building a
biomedical ontology recommender web service.
Journal of Biomedical Semantics (Suppl 1).
Jonquet, C., Nigam, H., Shah, H., Musen, A. M., 2009. The
Open Biomedical Annotator. AMIA Summit on
Translational Bioinformatics, pp. 56-60.
Lee, M., 2015. New stroke therapy uses motion sensor
video game to help rehabilitation, New York: Metro.
Lichman, M., 2013. UCI Machine Learning Data
Repository, s.l.: s.n.
Lucey, MD, C. R., 2015. Clinical Problem Solving -
Coursera. San Francisco: University of California.
Musen, M. et al., 2012. The National Center for Biomedical
Ontology. J Am Med Inform Assoc., 19(2), pp. 190-5.
Porumb, M., Bărbănțan, I., Lemnaru, C., Potolea, R., 2015.
REMed – Automatic Relation Extraction from Medical
Documents. Brussels, s.n.
Smith, C., 2014. Tracking Hand Tremors with Leap
Motion, San Francisco, CA: Leap Motion.
Szenasi, G., Lemnaru, C., Bărbă
nțan, I., 2015. Concept
extraction from medical documents. A contextual
approach. Cluj-Napoca, IEEE.
Learning Diagnosis from Electronic Health Records
351
