
Open Implementation of DICOM for Whole-Slide Microscopic Imaging
S
´
ebastien Jodogne
1
,
´
Eric Lenaerts
1
, Lara Marquet
1
, Charlotte Erpicum
2
,
Roland Greimers
2
, Pierre Gillet
3
, Roland Hustinx
1
and Philippe Delvenne
2
1
Department of Medical Physics, University Hospital of Li
`
ege, Avenue de l’H
ˆ
opital, 4000 Li
`
ege, Belgium
2
Department of Pathology, University Hospital of Li
`
ege, Avenue de l’H
ˆ
opital, 4000 Li
`
ege, Belgium
3
University Hospital of Li
`
ege, Avenue de l’H
ˆ
opital, 4000 Li
`
ege, Belgium
http://www.chuliege.be/
Keywords:
Whole-slide Imaging, Telepathology, DICOM.
Abstract:
This paper introduces an open implementation of DICOM for whole-slide microscopic imaging, following
Supplement 145 of the DICOM standard. The software is divided into two parts: (a) a command-line tool to
convert an whole-slide image to the DICOM format, and (b) a zero-footprint Web interface to display such
DICOM images. The software architecture leverages the DICOM server Orthanc. The entire framework is
available as free and open-source software. The existence of this software supports the development of digital
pathology and telepathology in clinical environments, featuring a smooth integration with existing EHR and
PACS solutions.
1 INTRODUCTION
Anatomopathology plays important medical roles by
detecting preneoplastic lesions and by giving a di-
agnosis, prognosis and evaluation of therapeutic re-
sponse based on visual observations of cellular or
tissue samples. Telepathology is an application of
telemedicine that allows the practice of the anato-
mopathology over a long distance with the use of im-
ages in an electronic format rather than viewing glass
slides (Ling and Krishnappa, 2012). Telepathology is
potentially useful for several applications:
• Intraoperative Consultation: Intraoperative con-
sultations are emergencies in anatomopathology
since the purpose of this examination is to guide
immediate surgical management. For such con-
sultations, the results should be given within 30
minutes (Ribback et al., 2014). Telepathology can
be used in this context for obtaining consultation
by a pathologist present in a remote physical lo-
cation.
• Secondary Consultation from Experts: Secondary
consultation refers to situations where a primary
diagnosis has been performed on the primary ma-
terial and further opinion is needed. Telepathol-
ogy is beneficial by accelerating the process and
by reducing the risks of material loss or break
compared to postal services (Farahani and Pan-
tanowitz, 2016).
• Education and Research: Telepathology is highly
promising in medical education and clinical re-
search. Currently, digital imaging starts to replace
traditional glass slides and high quality micro-
scopes to discuss interesting cases during lessons,
symposia and conferences. Telepathology facili-
tates interactions between multiple users and al-
lows the consistency and longevity of imaged ma-
terials (Mar
´
ee et al., 2016).
• Pathology Archiving: The storage of histolog-
ical slides is mandatory for a long period of
time. Since the virtual slide perfectly reproduces
the glass slide without any loss of information,
telepathology could enable the storage of images
in an entirely electronic way. As a consequence,
the currently-used, costly physical archives could
become legacy systems in the future (Webster and
Dunstan, 2014).
Telepathology requires microscopic images to be
put quickly and easily online, from a slide scanner
onto a secured Web server. To this end, at least three
technical difficulties must be overcome.
Firstly, any pathology laboratory requires high
electronic storage capacities to store their whole-slide
images, as the size of the latter may range from hun-
dreds of megabytes to hundreds of gigabytes, de-
pending on the scanning objective and the tissue sec-
Jodogne S., Lenaerts ÃL’., Marquet L., Erpicum C., Greimers R., Gillet P., Hustinx R. and Delvenne P.
Open Implementation of DICOM for Whole-Slide Microscopic Imaging.
DOI: 10.5220/0006155100810087
In Proceedings of the 12th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2017), pages 81-87
ISBN: 978-989-758-227-1
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
81

tion. A typical, uncompressed whole-slide image of
80,000×60,000 pixels acquired in 24-bit color (RGB)
weights over 10GB (DICOM Standards Committee,
Working Group 26, Pathology, 2010). Image com-
pression (JPEG or JPEG 2000) can divide this size by
a factor 10, but this still roughly corresponds to the
size of 3D medical images (e.g. computed tomogra-
phy or magnetic resonance imaging). Storing and in-
dexing such a large amount of data on the long run is
obviously not compatible with the manual administra-
tion of a filesystem: It implies the use of automated,
scalable, enterprise-ready database systems similar to
the PACS of the hospital (Picture Archiving and Com-
munication System).
The second technical difficulty is the lack of inter-
operability between the proprietary ecosystems that
are implemented by the manufacturers of slide scan-
ners. A single hospital might host several scanners
from different manufacturers, making it hard to con-
solidate all the whole-slide images inside a central-
ized tool because of a large variety of file formats. For
the same reason, it is hard to link the EHR (Electronic
Health Record) of a patient to her anatomopathology
images. This lack of interoperability in the clinical
workflow is also an obstacle to the exchange of im-
ages between hospitals, as well as to the use of stan-
dalone post-processing tools that are necessary for
big-data analysis, medical research or education.
A possible solution to this problem consists in
taking advantage of the DICOM standard for medi-
cal imaging (NEMA — National Electrical Manufac-
turers Association, 2016). Indeed, in 2010, the DI-
COM Committee defined a standard way of exchang-
ing whole-slide microscopic imaging that is vendor-
independent and that is fully compatible with exist-
ing PACS systems (DICOM Standards Committee,
Working Group 26, Pathology, 2010). Unfortunately,
there is currently no open implementation of this re-
cent standard, most probably because of the existence
of many patents (Cucoranu et al., 2014): As a con-
sequence, even if any modern PACS can ingest such
DICOM images, few will render them. There is also a
lack of sample files, which strongly calls for an open
infrastructure to share knowledge about this complex
standard, to the benefit of hospitals, manufacturers
and researchers.
The third technical challenge for telepathology
consists in serving the images over Internet. Be-
cause of the size of whole-slide images, a raw transfer
of the files would not be compatible with real-time
constraints or mobile applications. It is also highly
desirable to avoid the installation of some heavy-
weight client software on the computers of an hos-
pital, as this operation is often prevented for secu-
rity reasons: A telepathology viewer should there-
fore be able to run entirely inside a standard Web
browser, as such a browser is almost always installed
by default on any computer. A Web application also
has the advantage of being compatible with mobile
devices. The OpenSlide software can already serve
whole-slide images generated by commonplace scan-
ners onto the Web (Goode et al., 2013). Similarly,
the Cytomine software builds upon the OpenSlide ex-
perience to provide an advanced collaborative anal-
ysis framework for research and education in digital
pathology (Mar
´
ee et al., 2016). Unfortunately, nei-
ther of those platforms currently supports the DICOM
standard that is mandatory for clinical workflow.
The previous discussion calls for the engineer-
ing of a framework built upon the DICOM standard
that would support the development of workflow for
digital pathology and telepathology in clinical envi-
ronments, featuring a smooth integration with exist-
ing hospital information systems (EHR and PACS).
The needed software would be able to convert whole-
slide images encoded using proprietary formats, to
a standard DICOM file in accordance with the “VL
Whole Slide Microscopy Image Information Object
Definition (IOD)”, as specified in the “PS3.3: Infor-
mation Object Definitions” part of the DICOM stan-
dard (NEMA — National Electrical Manufacturers
Association, 2016). It would then be able to for-
ward the converted images to a generic PACS sys-
tem, while serving them directly over Internet, using
a zero-footprint Web client.
This paper introduces such an innovative frame-
work and makes it available as free and open-source
software to support the development of digital pathol-
ogy and telepathology in hospitals.
2 SYSTEM AND METHODS
According to the Introduction section, our software
framework for digital pathology and telepathology is
divided into two separate components:
• A standalone command-line tool that takes as in-
put a non-DICOM whole-slide microscopic im-
age, and that generates a compliant DICOM file.
• A DICOM server that can easily publish such im-
ages on the Web.
This section will fully describe our technical solu-
tion, after an introduction to the DICOM file format
for whole-slide microscopic imaging.
VISAPP 2017 - International Conference on Computer Vision Theory and Applications
82

2.1 DICOM for Visible Light
Whole-Slide Microscopic Images
Because images for digital pathology are very large
(possibly dozens of gigabytes), they most often can-
not entirely fit inside the memory of a standard com-
puter. For this reason, whole-slide images are in prac-
tice divided as a regularly-spaced set of tiles, each tile
being a small patch taken from the full image. A tile is
most often a square whose sides contain between 256
and 1024 pixels. Each individual tile can be accessed
separately, allowing a digital pathology application to
bring a tile into memory only when needed.
Compressing some whole-slide image amounts to
compressing each individual tile either using a loss-
less algorithm (JPEG 2000), or a destructive algo-
rithm (JPEG). Furthermore, to enable the navigation
over the entire whole-slide image, the full-sized im-
age is downscaled several times, leading to a pyramid
of images with decreasing spatial resolutions. Each
level of the pyramid is encoded as a separate tiled im-
age. This process is illustrated in Figure 1. Note that
similar encoding schemes are also commonly used to
serve cartography maps on the Web.
The DICOM standard models the real world as
follows: A given patient benefits during her life from
a set of medical imaging studies. Each study is made
of a set of series. Each series is in turn a set of in-
stances, the latter being a synonym for a DICOM file.
A single DICOM instance can be multi-frame, mean-
ing it can store several independent images (provided
all of its individual frames share the same size). As
a consequence, the whole-slide pyramid represented
in Figure 1 corresponds to one DICOM series, whose
parent study might contain other series acquired dur-
ing the same clinical episode, possibly coming from
other medical imaging modalities. This series is made
of several instances (the DICOM files), each instance
storing the individual tiles of one given pyramid level
as separate frames. A single instance is not allowed to
store tiles from multiple pyramid levels, yet the same
level can be spread over multiple instances, so as to
prevent the appearance of huge DICOM files.
Besides its individual frames, each DICOM in-
stance is associated with clinical data under the form
of a recursive key-value associative array. Such keys
are called the DICOM tags and are indexed with two
16-bit hexadecimal numbers. The DICOM standard
lists which DICOM tags are mandatory, conditional
or allowed for whole-slide images in the so-called
“VL Whole Slide Microscopy Image IOD” (NEMA —
National Electrical Manufacturers Association, 2016,
PS3.3, Section A.32.8).
A full enumeration of this set of DICOM tags is
obviously out of the scope of this paper, and the inter-
ested reader is kindly invited to refer to the DICOM
standard. In the context of this paper, it is sufficient
to know that each single instance must specify the
size of the pyramid level it is related to: The DICOM
tag “Total Pixel Matrix Columns” (0x0048,0x0006)
stores the width of the pyramid level, whereas “To-
tal Pixel Matrix Rows” (0x0048,0x0007) stores its
height. These two tags allow to know to which level
of the pyramid a given DICOM instance belongs.
Similarly, because the tiles of one pyramid level
can be shuffled over several multi-frame DICOM in-
stances, each frame is associated with the (x, y) po-
sition of the corresponding tile in the corresponding
pyramid level: The tag “Column Position In Total Im-
age Pixel Matrix” (0x0048,0x021e) contains the x-
position of one frame, and “Row Position In Total Im-
age Pixel Matrix” (0x0048,0x021f) its y-position.
This information is collected for each frame of the DI-
COM instance inside the tag “Per Frame Functional
Groups Sequence” (0x5200,0x9230). Our frame-
work almost exclusively relies on this set of tags.
2.2 DICOM-izer
The first component of our software framework for
telepathology is the tool that converts a whole-slide
image from a non-DICOM format to DICOM. This
tool will be referred to as the DICOM-izer. It takes
the form of a standalone, cross-platform command-
line tool, so that it can easily be integrated into any
pathology department.
The DICOM-izer features built-in support to read
the most widespread open file format for whole-slide
imaging (i.e. hierarchical TIFF), as well as common-
place image formats (PNG, JPEG and JPEG 2000). It
is also able to decode proprietary file formats for slide
scanners (SVS, BIF, VMS. . . ) through the OpenSlide
toolkit (Goode et al., 2013)
1
. It extracts as much clin-
ical information as possible from the meta-data of the
input image (such as the scanner manufacturer), but
additional information that is not verbatim available
inside the source file must be provided alongside (e.g.
the identifier of the patient, or the optical parameters
of the acquisition). The output of the DICOM-izer is
a set of compliant DICOM files that can be sent to any
DICOM modality, including the PACS of the hospital,
thanks to the standard DICOM C-Store command.
If the input image does not contain the full pyra-
mid but only its finest level, the DICOM-izer can au-
tomatically generate all the upper levels of the pyra-
mid. Similarly, the DICOM-izer can change the com-
1
Note that OpenSlide does not support the generation of
DICOM files by itself.
Open Implementation of DICOM for Whole-Slide Microscopic Imaging
83
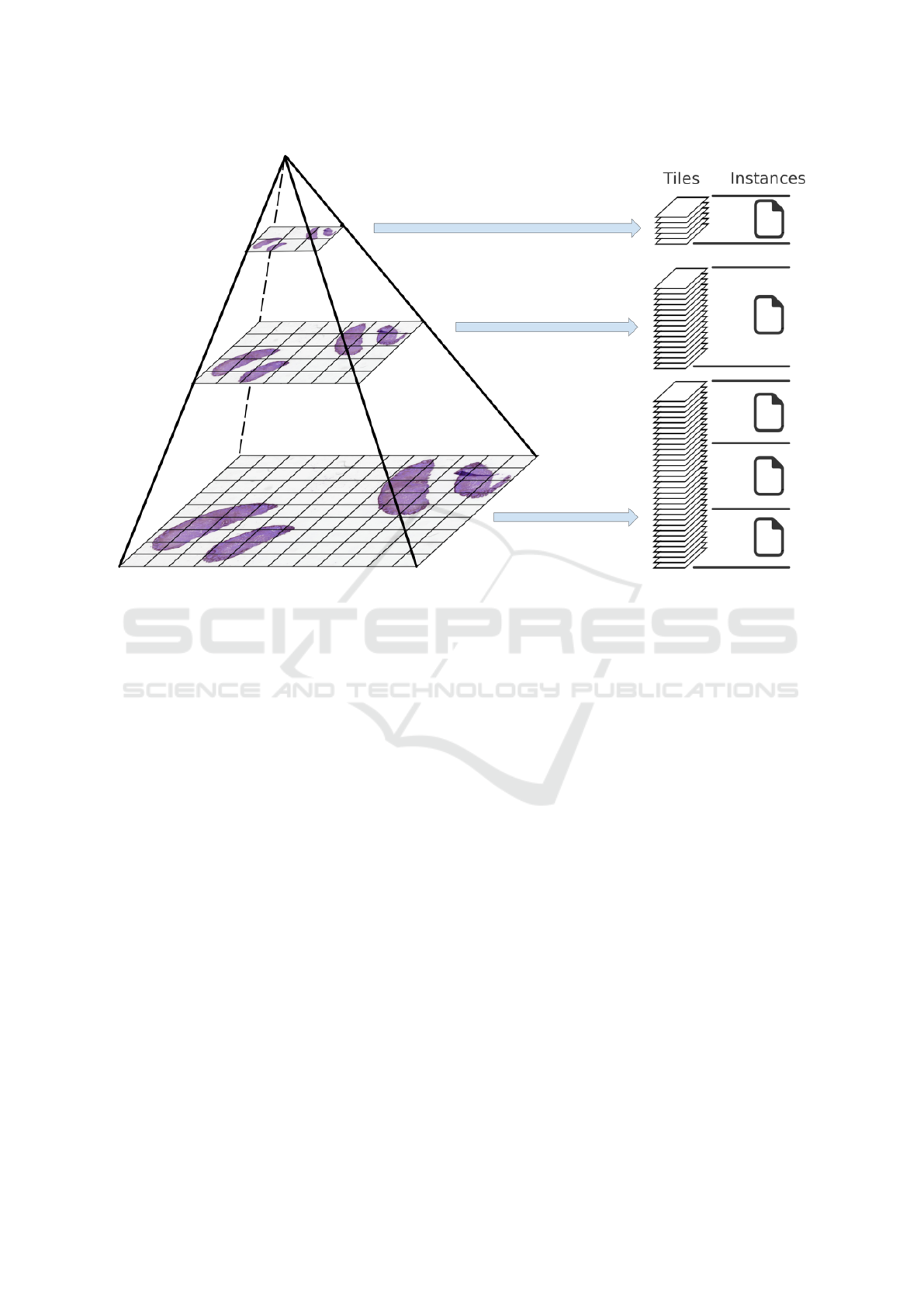
Figure 1: Mapping a multi-resolution pyramid according to the DICOM standard (DICOM Standards Committee, Working
Group 26, Pathology, 2010). Each level of the pyramid is a downscaled version of the whole-slide image, and is decomposed
as a set of tiles. The tiles are encoded as separate frames of multi-frame DICOM instances (files).
pression scheme of the input image to one of the al-
gorithms supported by the DICOM standard (i.e. no
compression, JPEG or JPEG 2000). Unsurprisingly,
decoding a proprietary format, changing the compres-
sion, and/or rebuilding the pyramid are CPU-intensive
re-encoding operations that can last dozens of sec-
onds, even though our DICOM-izer efficiently takes
advantage of multi-threading. However, the baseline
process of simply transcoding a hierarchical TIFF to
DICOM only takes a few seconds, with almost no
CPU usage.
Note that microscopy images are often stained or
multi-spectral. This means that multiple channels
might be needed to store the full microscopy image.
The DICOM standard requests to store the various
spatial and spectral channels in separate DICOM se-
ries. In such situations, the DICOM-izer can be sepa-
rately invoked for each channel, which will generate a
set of DICOM series, all belonging to the same parent
DICOM study.
2.3 Orthanc Plugin for Whole-Slide
Imaging
The second component of our software framework
for telepathology allows laboratories to immediately
publish the DICOM images that are produced by the
DICOM-izer over Internet. As argued in the Introduc-
tion, this component is a necessary companion to the
DICOM-izer, as most PACS do not currently support
the rendering of whole-slide images. Because our
framework is designed to be as open as possible to the
benefit of the worldwide community of pathology lab-
oratories and researchers, this component leverages a
free and open-source DICOM server.
The two most well-known free and open-source
DICOM servers are DCM4CHE (Warnock et al.,
2007, written with Java and JBoss) and Orthanc
(Jodogne et al., 2013, written in C++). Besides its
small footprint, Orthanc has the advantage of propos-
ing a RESTful API that makes it ready for Web ap-
plications (Fielding, 2000), and to propose a plugin
mechanism that can be used by third-party developers
to extend the core REST API without using an addi-
tional Web server. As a consequence, our Web pub-
lishing component is built upon the Orthanc vendor
neutral archive, and takes the form of a C++ plugin.
Our Orthanc plugin is a shared library that is dy-
namically loaded by Orthanc during its startup, and
that is responsible for:
1. Transparently indexing all the tiles of a given DI-
COM series as a whole-slide pyramid.
VISAPP 2017 - International Conference on Computer Vision Theory and Applications
84
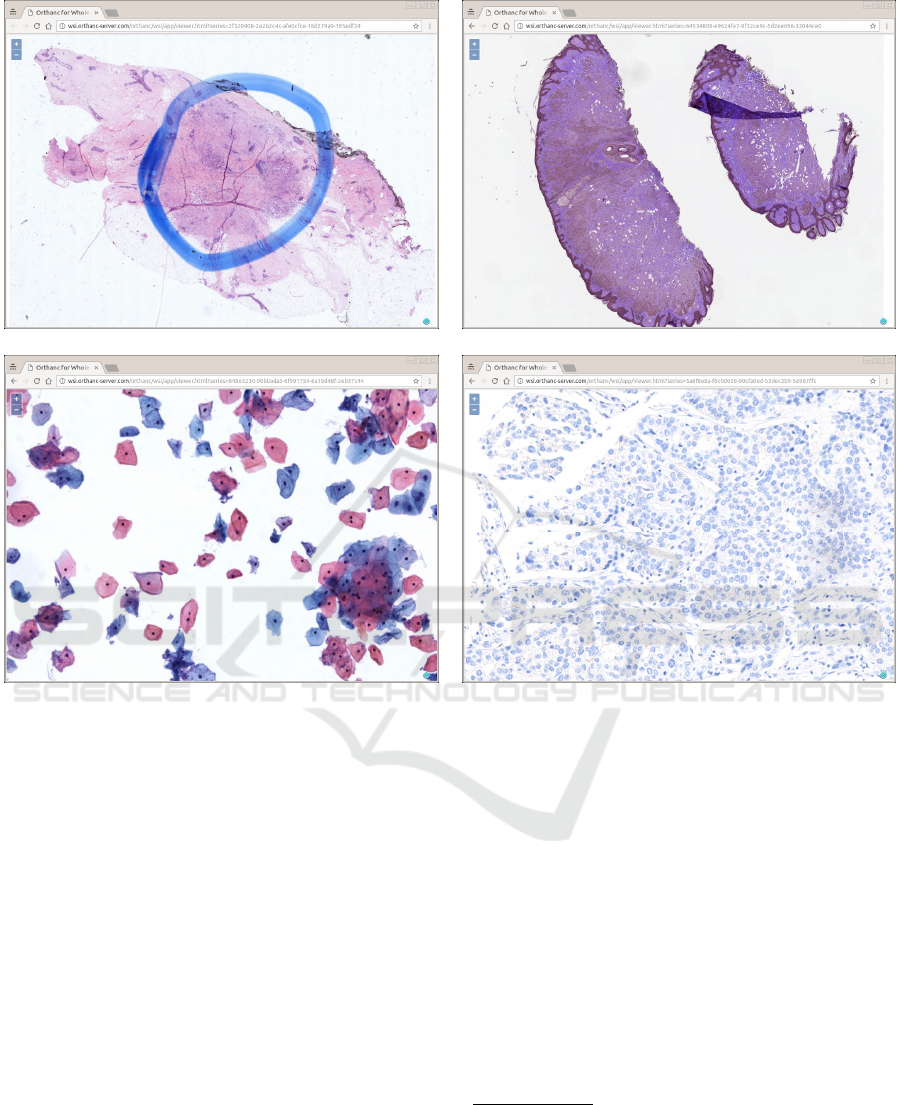
Figure 2: Some screenshots of a Web browser displaying real-world pathology images stored inside Orthanc, at various zoom
levels. The Web application is zero-footprint: It is entirely written in JavaScript, and no heavyweight client must be installed
(all is done by the Web browser). The Web interface is built upon OpenLayers version 3, a free and open-source JavaScript
library for displaying raster tile maps (Open Source Geospatial Foundation, 2010). Note that the Web application can be
served through the HTTPS protocol, meaning that the medical communication can be secured through proper authentication
and encryption.
2. Serving the individual tiles according to their
(x, y, z) location (z corresponds to the level of the
tiles in the pyramid), after dynamically transcod-
ing them to an image format that is compatible
with Web browsers (either PNG for uncompressed
or losslessly-compressed whole-slide images, or
JPEG for destructively-compressed images).
3. Publishing all the HTML and JavaScript static re-
sources that are necessary for the Web viewer.
The indexing process of Step (1) first groups the
instances according to the size of their associated
pyramid level z, then extracts the (x, y) position of
each frame in these instances, only by considering
the DICOM tags that were introduced at the end of
Section 2.1. Figure 2 shows screenshots of a Web
browser accessing our viewer of DICOM whole-slide
images
2
.
The DICOM-izer can be configured to automat-
ically push its output images to any Orthanc server
through its RESTful API. Note also that because
Orthanc is a fully-featured vendor neutral archive, it
can be used to re-transmit (resp. query/retrieve) the
whole-slide images to (resp. from) other DICOM
modalities, including the PACS of the hospital. Fi-
nally, an official plugin is available to make Orthanc
use a PostgreSQL database, making it fully scalable
and enterprise-ready if need be.
2
The demonstration server from which these
screenshots were taken is publicly available online at:
http://wsi.orthanc-server.com/demo/. This demonstration
server illustrates the fact that the Web interface is entirely
zero-footprint: Any modern Web browser will display
it out-of-box, without having to install any additional
software.
Open Implementation of DICOM for Whole-Slide Microscopic Imaging
85
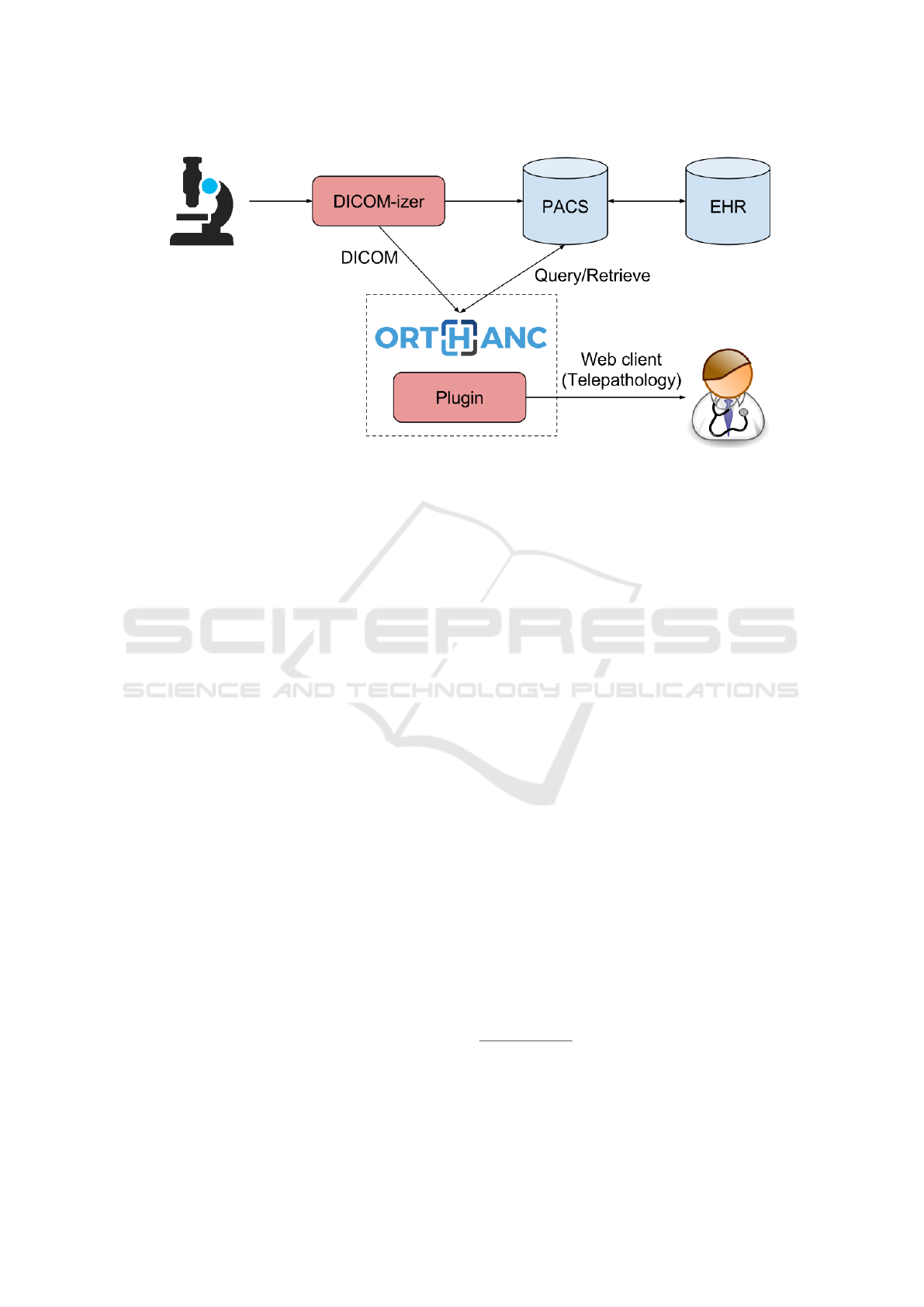
Figure 3: Possible clinical workflow for digital pathology. The innovative contributions introduced by this paper are high-
lighted in red.
3 APPLICATIONS
Figure 3 illustrates a real-world clinical workflow that
becomes possible thanks to our open framework for
digital pathology and telepathology. In this work-
flow, the pathology department sends its images to the
PACS of the hospital, which enables the continuous
integration with the electronic health record (EHR) of
the patient: This would solve the “pathology archiv-
ing” objective explained in the Introduction.
In parallel, the DICOM-izer also sends its out-
put images to an Orthanc server equipped with our
plugin. As soon as the DICOM images are received
by Orthanc, they are immediately made available on-
line for real-time viewing: Opening an image in the
browser takes less than one second, and the user ex-
perience is similar to well-known Web mapping sys-
tems. This allows telepathologists to quickly and eas-
ily review whole-slide images remotely, using any
standard Web browser. This remote, read-only access
is also possible using a smartphone or a tablet.
The CPU power that is required by the Orthanc
server is very low if the images are encoded using
JPEG, which is the most common case: As Web
browsers natively support JPEG, the plugin can serve
the compressed tiles without decoding them. Fur-
thermore, as the telepathology plugin is entirely Web-
based, the network administrators of the hospital can
setup a reverse proxying system and HTTPS encryp-
tion to ensure the proper authentication and confi-
dentiality for the access to the medical information:
This would solve the “intraoperative consultation”
and “secondary consultation” objectives. Also note
that Orthanc can query/retrieve images archived in the
PACS so as to put them back online.
Finally, whole-slide images that are found to be
of interest for research or education, can be exported
as hierarchical TIFF directly from our Web viewer to
a richer Internet application for collaborative analysis
such as Cytomine (Mar
´
ee et al., 2016). This would
solve the “education and research” objective.
4 CONCLUSION
This paper introduces an implementation of the “VL
Whole Slide Microscopy Image IOD”, as specified in
the “PS3.3: Information Object Definitions” part of
the DICOM standard (NEMA — National Electrical
Manufacturers Association, 2016), in order to support
the development of digital pathology and telepathol-
ogy in clinical laboratories and hospitals. The source
code of our framework is provided as free and open-
source software, under the terms of the AGPL li-
cense
3
. It is notably compatible with Microsoft Win-
dows, Apple OS X and GNU/Linux environments.
To the best of our knowledge, this is first pub-
lic, open, reference implementation of DICOM for
whole-slide microscopic imaging. Our framework
consists of a standalone DICOM-izer, that converts
3
The source code is accessible from the official home-
page of Orthanc: http://wsi.orthanc-server.com/. Full tech-
nical documentation of the framework and of the un-
derlying open Web API is part of the Orthanc Book:
http://book.orthanc-server.com/.
VISAPP 2017 - International Conference on Computer Vision Theory and Applications
86

whole-slide images to compliant DICOM files, to-
gether with a dedicated plugin extending the vendor
neutral archive Orthanc. The latter plugin extends Or-
thanc with a lightweight, zero-footprint Web viewer
of whole-slide images. This viewer can be accessed
remotely from any Web browser or mobile device.
It has also been discussed how our framework for
telepathology can be integrated inside a typical hos-
pital workflow to meet real-world challenging objec-
tives such as remote consultation, education, clini-
cal research, or long-term archiving. Future work
will consist in taking advantage of our framework
to support a multi-centric clinical study that will de-
velop and assess new algorithms to quantify relevant
biomarkers on digitized immunostained slides of neo-
plastic lesions.
REFERENCES
Cucoranu, I. C., Parwani, A. V., Vepa, S., Weinstein, R. S.,
and Pantanowitz, L. (2014). Digital pathology: A sys-
tematic evaluation of the patent landscape. Journal of
Pathology Informatics, 5(1):16.
DICOM Standards Committee, Working Group 26, Pathol-
ogy (2010). Supplement 145: Whole slide micro-
scopic image IOD and SOP classes.
Farahani, N. and Pantanowitz, L. (2016). Overview of tele-
pathology. Clinics in Laboratory Medicine, 36(1):101
– 112.
Fielding, R. (2000). Architectural Styles and the Design
of Network-based Software Architectures. PhD thesis,
University of California, Irvine.
Goode, A., Gilbert, B., Harkes, J., Jukic, D., and Satya-
narayanan, M. (2013). OpenSlide: A vendor-neutral
software foundation for digital pathology. Journal of
Pathology Informatics, 4(1):27. Freely available at:
http://openslide.org/.
Jodogne, S., Bernard, C., Devillers, M., Lenaerts, E., and
Coucke, P. (2013). Orthanc – Lightweight, RESTful
DICOM server for healthcare and medical research.
In IEEE 10th International Symposium on Biomedical
Imaging (ISBI), pages 190–193, San Francisco, USA.
Freely available at: http://www.orthanc-server.com/.
Ling, C. and Krishnappa, P. (2012). Telepathology – An up-
date. International Journal of Collaborative Research
on Internal Medicine and Public Health, 4(12).
Mar
´
ee, R., Rollus, L., St
´
evens, B., Hoyoux, R., Louppe,
G., Vandaele, R., Begon, J.-M., Kainz, P., Geurts, P.,
and Wehenkel, L. (2016). Collaborative analysis of
multi-gigapixel imaging data using cytomine. Bioin-
formatics. Freely available at: http://cytomine.be/.
NEMA — National Electrical Manufacturers Association
(2016). NEMA PS3 / ISO 12052, Digital Imaging
and Communications in Medicine (DICOM) standard.
Freely available at: http://medical.nema.org/.
Open Source Geospatial Foundation (2010). Openlay-
ers: Free maps for the web. Freely available at:
http://www.openlayers.org/.
Ribback, S., Flessa, S., Gromoll-Bergmann, K., Evert, M.,
and Dombrowski, F. (2014). Virtual slide telepathol-
ogy with scanner systems for intraoperative frozen-
section consultation. Pathology - Research and Prac-
tice, 210(6):377 – 382.
Warnock, M. J., Toland, C., Evans, D., Wallace, B., and
Nagy, P. (2007). Benefits of using the DCM4CHE DI-
COM archive. Journal of Digital Imaging, 20(Supp.
1):125–129.
Webster, J. D. and Dunstan, R. W. (2014). Whole-slide
imaging and automated image analysis: Considera-
tions and opportunities in the practice of pathology.
Veterinary Pathology, 51(1):211–223.
Open Implementation of DICOM for Whole-Slide Microscopic Imaging
87
