
A Qualitative Framework Dedicated to Toxicology
Benjamin Miraglio
1
, Gilles Bernot
1
, Jean-Paul Comet
1
and Christine Risso-de Faverney
2
1
Universit
´
e C
ˆ
ote d’Azur, CNRS, I3S, Sophia Antipolis, France
2
Universit
´
e C
ˆ
ote d’Azur, CNRS, ECOMERS, Nice, France
Keywords:
Computational Toxicology, Discrete Dynamic Systems, Rule-Based Modelling, Temporal Logic.
Abstract:
Emerging constraints have led the toxicology community to complete the classical paradigm of toxicology
with the study of molecular events underlying the toxicity of a chemical substance. This evolution motivates
the emergence of new modelling approaches for toxicology. In this article, we introduce a qualitative rule-
based formalism dedicated to the domain of toxicology. This new formalism departs from other rule-based
formalisms such as BioChAM because it directly encodes possible alterations of equilibrium, instead of mak-
ing equilibriums emerge from the dynamics of the model. Using a simple example of the energy metabolism,
we show that this formalism is able to describe both the normal evolution of a biological system and its possible
toxic disruptions.
1 INTRODUCTION
Toxicology can be defined as the study of adverse
effects caused by exogenous chemical substances to
biological systems. The classical paradigm assumes
that the more an organism is exposed (in dose and/or
time) to a compound, the greater the compound ef-
fects will be. In these conditions, any chemical sub-
stance can therefore cause harmful effects to an or-
ganism if this organism is exposed during a long
enough time to a high enough dose of chemical.
That concept serves as a basis to the dose-response
relationship, which enables toxicologists to establish
a causality between the exposure to a chemical and its
induced observed effects. It also allows toxicologists
to determine the threshold of toxicity, namely the low-
est exposure (in dose and/or time) where an induced
effect is observed.
Many experiments carried out recently have
pointed out the limitations of this paradigm. Indeed,
toxicity assessment is quite complex and, besides the
dose and time of exposure, a lot of other factors can
affect the results of toxicity tests. In particular, these
factors include temperature, food, light, the route of
exposure and the chemical interactions of the tested
substance with other chemical compounds. Other fac-
tors related to the test subject itself, including age,
sex, genetics, health status, hormonal status or win-
dow of exposure may also greatly influence the vul-
nerability of an organism to a chemical substance.
To answer these limitations, an increasing trend in
toxicology is to focus on the causal sequence of key
events occuring during the toxic response and lead-
ing to an observable effect. These sequences, called
pathways of toxicity, lay the basis of the mechanistic
toxicology and include events from molecular, cellu-
lar and even organ scales.
As mechanistic toxicology allows a better under-
standing of molecular mechanisms leading to adverse
effects, it can cope with many difficulties mentioned
earlier, such as the extrapolation of toxicity findings
obtained from laboratory animals to humans or the
consideration of additional factors in toxicity assess-
ments. Moreover, as distinct pathways of toxicity can
share the same key events, knowledge obtained when
studying one chemical could be reused when assess-
ing other chemicals.
Concurrently, as the potential toxicity of chemical
exposure became an area of great concern to both the
public (Colborn et al., 1996; Kepner, 2004) and the
regulatory authorities (Backhaus et al., 2010), the pro-
duction of chemical compounds is increasingly reg-
ulated worldwide. Manufacturers must now conduct
extensive studies to demonstrate the innocuity of their
products, considerably increasing the cost of develop-
ment of such products.
This context favours the emergence of different
mathematical modelling approaches, and so far, most
of these approaches are quantitative and aim at either
inferring the toxic threshold of a chemical substance
Miraglio B., Bernot G., Comet J. and Risso-de Faverney C.
A Qualitative Framework Dedicated to Toxicology.
DOI: 10.5220/0006168200930103
In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2017), pages 93-103
ISBN: 978-989-758-214-1
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
93

or confirming its specific pathway of toxicity. These
objectives require a lot of toxicological data. This
can be restrictive given the current acquisition cost of
new biological data. There is thus an incentive to de-
velop methods that do not focus on toxic thresholds
but instead, aim at describing pathways of toxicity
in a qualitative manner. Such an approach would fo-
cus on equilibrium shifts and would therefore require
comparatively less toxicological data.
In this article, we present a new qualitative formal-
ism allowing to enumerate all the conceivable path-
ways of toxicity linked to a compound present in a
given biological system. As a lot of these modelled
pathways are biologically improbable, it is then pos-
sible to encode into temporal logic basic toxicolog-
ical knowledge and filter out the less relevant ones.
The remaining modelled pathways can finally serve
as a basis to design more informative experiments and
help toxicologists in their search of new pathways of
toxicity.
The next section of this article is dedicated to the
brief description of related work focusing on formal
frameworks dedicated to model biological systems.
As our formalism is presented alongside examples in-
spired from the energy metabolism, section 3 sketches
an overview of the energy metabolism and its key
components. In section 4, we explain how to use the
new formalism to describe the equilibrium changes
of a system. In section 5, we show how to integrate
toxicological knowledge in the system using temporal
logic. Finally, this formalism is applied to a simplified
model of the energy metabolism in section 6.
2 RELATED WORK
The rule-based formalism presented throughout this
article was originally inspired from the Boolean se-
mantics of BioChAM (Calzone et al., 2006), an en-
vironment able to model biological systems as net-
works of chemical reactions. However, several speci-
ficities of toxicology make this environment not op-
timal when handling toxicological models, motivat-
ing the development of a more domain-oriented for-
malism. For instance, the notion of abnormal con-
centrations required us to adopt multivalued seman-
tics. Furthermore, the presence of modulations of
reactions, crucial in toxicology, is difficult to han-
dle with BioChAM. This motivated us to directly de-
scribe equilibrium changes in a biological system, ab-
stracting any quantitative computing steps.
Pathway logic (Talcott, 2008) is another frame-
work dedicated to the description of biological sys-
tems while highlighting structural aspects of the cell.
Unfortunately, this framework is also not adapted to
deal with specificities of toxicology such as abnormal
concentrations or modulations of reactions.
Finally, Bio-PEPA (Ciocchetta and Hillston,
2009) is a modelling approach where biological sys-
tems are formalised as discrete models. These mod-
els include precise biological information such as ki-
netic laws and stoechiometry. Different analysis can
then be performed on these models, ranging from the
construction of continuous time Markov chains to the
translation of models into ordinary differential equa-
tions. However, the discretisation required for con-
tinous time Markov chains and probabilistic model
checking approaches is based on a precise knowl-
edge of kinetic parameters. Unfortunately, this pre-
cise knowledge is often unavailable, leaving room for
a framework where reaction rates are abstracted. Of
course, such a framework should maintain the possi-
bility for toxicologists to easily express concurrency
between equilibrium changes.
3 A QUICK INTRODUCTION TO
METABOLISM
The metabolism can be described as the set of chem-
ical reactions allowing cells to survive. A major dis-
tinction in metabolism is made between catabolism
and anabolism. Catabolism refers to the set of reac-
tions degrading molecules in smaller parts. Catabolic
reactions are mainly oxidations and tend to produce
energy for the cell. On the contrary, anabolism gath-
ers synthesis reactions producing key molecules for
the cell and tend to require energy.
In addition to the catabolism/anabolism separa-
tion, metabolic reactions can also be clustered de-
pending on the type of molecules they handle. Hence,
carbohydrate metabolism refers to pathways manag-
ing glucose and other carbohydrates and the lipid
metabolism regroups reactions that both break down
and synthesise lipids. Obviously, these different
clusters of reactions are heavily interconnected, and
the most notable intermediary molecules are called
metabolic crossroads.
The formalism developed in this article will be il-
lustrated by a simple model of the energy metabolism
of a mammalian hepatic cell in aerobic conditions.
Here, the energy metabolism can be understood as a
set of reactions including pathways belonging to car-
bohydrate and lipid metabolisms as well as additional
chemical reactions occuring in mitochondria. This set
of reactions is summarised in Figure 1 and frequent
references to this figure will be made during the de-
scription of the reactions. Note that the subcellular lo-
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
94
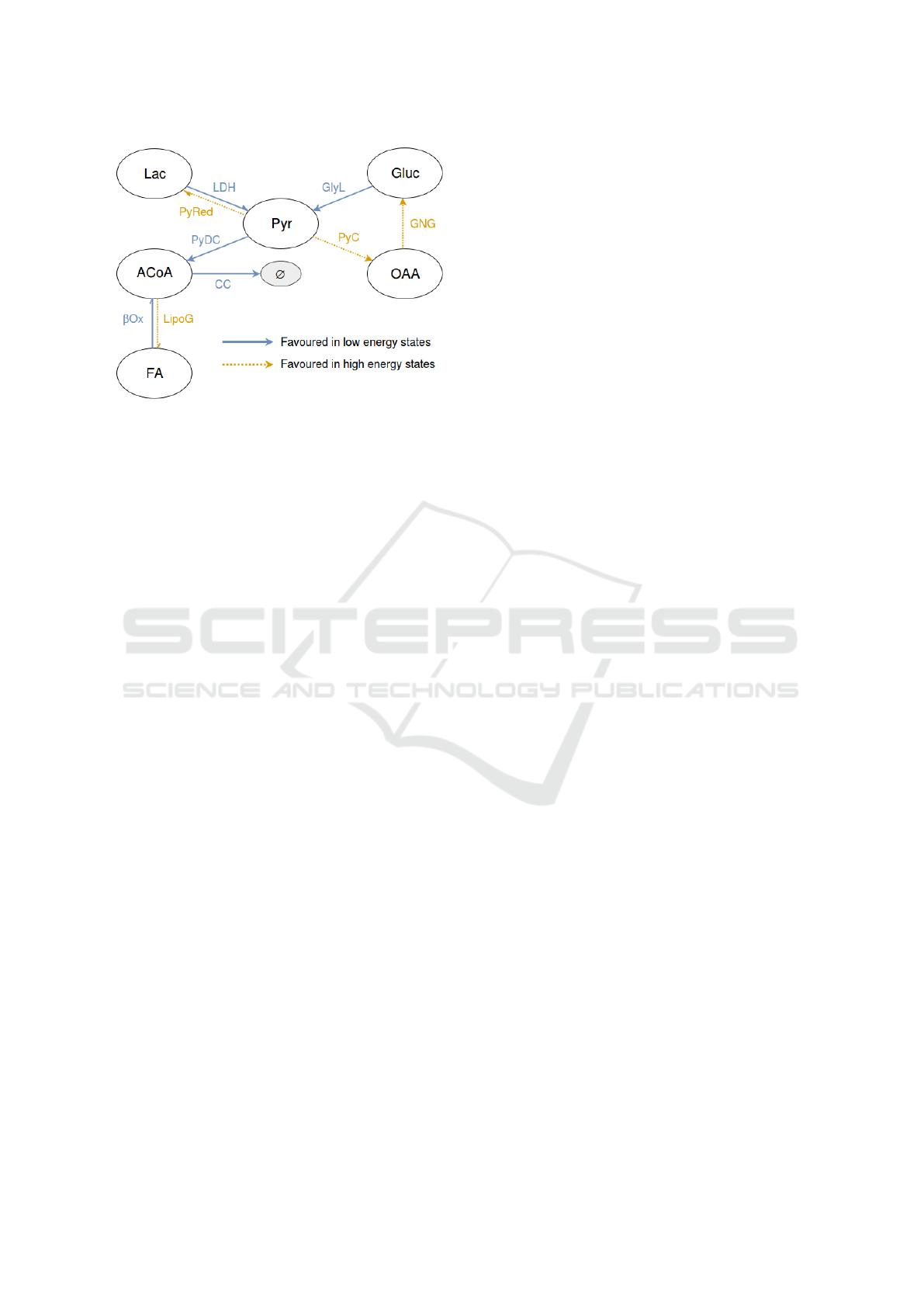
Figure 1: Representation of a simplified energy metabolism
model. Arrow captions correspond to rule identifiers devel-
oped in Section 6.
cation of the reactions (cytosol or mitochondria) is ab-
stracted in this figure. Moreover, reactions favoured
in low energy situations are depicted by using plain
arrows while reactions promoted in high energy cases
are shown with dotted arrows.
Carbohydrate Metabolism. Carbohydrates consti-
tute one of the main energy supply for cells and their
metabolism is essentially located in the cell cytosol.
The main catabolic pathway is glycolysis (GlyL),
which converts glucose (Gluc) and several other car-
bohydrates into pyruvate (Pyr) (Pilkis and Granner,
1992). Pyr then migrates to mitochondria where it
is decarboxylated (PyDC) into acetyl-CoA (ACoA),
an important metabolic crossroads. Note that the de-
carboxylation of Pyr into ACoA is performed by the
pyruvate dehydrogenase complex (PDH) (Pettit et al.,
1975). This enzymatic complex will not be included
in the application model developed in Section 6 and
is not represented in figure 1. However, it is used in
the running example illustrating Section 4.
The anabolic counterpart of glycolysis, the glu-
coneogenesis (GNG), uses energy to create new
molecules of Gluc (Pilkis and Granner, 1992). It
should be noted that Pyr carboxylation (PyC) into ox-
aloacetatic acid (OAA) - the first step of gluconeo-
genesis - directly competes with Pyr decarboxylation
(PyDC) to ACoA. The competition outcome depends
on the quantities of available ACoA: if there is a short-
age, Pyr is decarboxylated to ACoA, otherwise, it is
carboxylated in OAA (Owen et al., 2002). The other
reactions composing gluconeogenesis are mainly re-
versed glycolysis reactions.
Lipid Metabolism. Lipids are another important
source of energy for cells. The β-oxidation (βOx) is
the main catabolic pathway and degrades fatty acids
(FA) into ACoA in mitochondria (Schulz, 1991).
Conversely, lipogenesis (LipoG) occurs in the cytosol
and converts ACoA into FA (Hellerstein et al., 1991).
It is interesting to note that both lipid and car-
bohydrate catabolic pathways result in the produc-
tion of ACoA. Indeed, ACoA is actually an important
metabolic crossroads and can be involved in the pro-
duction of either new molecules - notably lipids - or
energy, through its inclusion in the citrate cycle (CC).
The inclusion of ACoA in the citrate cycle results in
the destruction of ACoA, represented in figure 1 by
the symbol ”∅”.
Citrate Cycle. The citrate cycle (CC), also known
as tricarboxylic acid cycle, is a key component of en-
ergy production (Owen et al., 2002). It starts with
the conjugation of ACoA and OAA to form citrate
(Cit) and is then composed of a sequence of reac-
tions resulting in the regeneration of OAA and the
production of energy (Owen et al., 2002). Hence, the
overall cycle only decreases ACoA levels to produce
energy, therefore, only ACoA is consumed in Figure
1. On top of the production of energy, the citrate cy-
cle also produce reduced compounds. These reduced
compounds are then used to fuel the mitochondrial
respiratory chain (MRC, not represented in Figure 1),
generating even more energy (Chance and Williams,
1956).
Reduction-oxidation Reactions. The reduced
compounds are actually coenzymes such as nicoti-
namide adenine dinucleotide or flavine adenine
dinucleotide that act as electron transporters in the
cell and can have either an oxidised or a reduced
form. For the sake of simplicity, we will summarise
the ratio between reduced and oxidised coenzymes
as the reducing potential of a cell (pRed). A high
(resp. low) reducing potential therefore means high
(resp. low) concentrations in reduced coenzymes
and subsequent low (resp. high) levels of oxidised
coenzymes.
As a matter of fact, the whole metabolism can be
seen as a network of reduction-oxidation (redox) re-
actions and the reducing power of a cell can inform
on the state of the cell metabolism. For instance, both
carbohydrate and lipid catabolic pathways reduce oxi-
dised coenzymes in their reduced form (increasing the
cell reducing potential), while conversely, both an-
abolic pathways oxidise reduced coenzymes in their
oxidised form (decreasing the reducing potential).
A Qualitative Framework Dedicated to Toxicology
95

For the sake of clarity, modifications of the reduc-
ing potential are not directly shown in Figure 1 but
can be deduced from the arrows nature. Indeed, re-
actions favoured in low energy states tend to produce
reducing potential (solid arrows in figure 1) while re-
actions favoured in high energy states tend to decrease
the reducing potential of a cell (dotted arrows in figure
1).
As previously said, mitochondria are able to create
energy from the reduced coenzymes. This process de-
creases the reducing power, regenerating the oxidised
coenzymes supplies of the cell, allowing the continu-
ation of catabolic reactions.
Another important source of coenzymes oxidation
lays in the cytosolic reduction of Pyr into lactate (Lac)
(Brooks, 1998). If mitochondria functioning falters,
the pyruvate reduction (PyRed) is still able to resup-
ply the cell in oxidised coenzymes, preventing the in-
terruption of catabolic pathways. Since pyruvate re-
duction is reversible, its reversed reaction (lactate de-
hydrogenation or LDH), can also generate reducing
potential while converting Lac in Pyr. The newly cre-
ated Pyr can, in turn, either generate a lot of energy
through its decarboxylation in ACoA or participate to
the gluconeogenesis through carboxylation in OAA
(see Carbohydrate metabolism paragraph).
Disruptions of the Cell Reducing Potential. As
the cell reducing potential is central to a lot of
metabolic reactions, disruptions of the redox equilib-
rium can have great impacts on the organism. For
instance, alcohol dehydrogenase is able to convert
ethanol (Eth, not represented in Figure 1) to acetalde-
hyde while greatly increasing the cell reducing poten-
tial. If this increase is strong enough, it can saturate
the mitochondrial respiratory chain and trigger the re-
duction of Pyr into Lac. The subsequent accumula-
tion of Lac in cells can then lead to troubles such as
metabolic acidosis.
Furthermore, as the presence of Eth triggers Pyr
reduction into Lac, the amount of Pyr available to car-
boxylation in OAA is decreased. The subsequent pro-
duction of Gluc from OAA through the gluconeogen-
esis is then strongly impacted. Since gluconeogenesis
is an important pathway to address hypoglycemia, a
fasting organism can thus see its capacity to recover
from hypoglycemia strongly damaged after its expo-
sition to ethanol (Field et al., 1963).
4 DESCRIBING EQUILIBRIUM
CHANGES
A biological system can be described as a set of bio-
logical entities interacting with each other at different
concentrations. In a given organism, each entity has
a concentration regarded as normal in standard condi-
tions. For instance, the normal blood concentration of
glucose is about 1 g/L in an adult human.
Our domain-oriented formalism allow us to rep-
resent the evolution of the concentration of each en-
tity and to depict abnormal concentrations from which
toxicity can arise. Indeed, we introduce four qualita-
tive abstract levels, which are listed here in increasing
order:
• ε reflects a negligible concentration of a given en-
tity, that is to say a concentration too low to trigger
any reaction in the biological system.
• ι conveys an abnormally low concentration, i.e.
a relative lack of this entity that can affect some
mechanisms in the biological system.
• ∆ indicates a normal concentration.
• θ shows an abnormally high concentration,
namely an excess of this entity.
Notation 1. [Concentration levels] We note L the set
{ε, ι,∆, θ} equipped with the total order relation such
that: ε < ι < ∆ < θ. The elements of L are called
concentration levels.
In a given biological system and depending on the
studied issue, not all entities have concentrations re-
garded as abnormally low or high. Therefore, only
the levels ε and ∆ are mandatory for each entity, ι and
θ being optional.
Taking this variation in consideration, the signa-
ture of a biological system allows the definition of the
set of biological entities considered in the system and,
for each entity, its admissible concentration levels.
Definition 1. [Signature] A signature is an applica-
tion E : E → P (L) where E is a finite set and for all
e ∈ E, {ε, ∆} ⊂ E(e). Elements of E are called en-
tities and for each entity e, E (e) is called the set of
admissible levels of e.
For instance, the signature of a ba-
sic energy metabolism model may involve
E = {PDH, Pyr, ACoA, OAA, Cit} can correspond to
the set of five entities where each entity has its own
set of admissible levels. For example, we may have
E(Pyr) = {ε, ι, ∆, θ}.
After defining the system signature, the state of
the system can be defined as the qualitative level
of each entity present in the system. The previous
example model can be at a state η
0
where PDH
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
96

is at the level ∆, noted η
0
(PDH) = ∆ and where
η
0
(Pyr) = θ, η
0
(ACoA) = ι, η
0
(OAA) = ∆ and
η
0
(Cit) = ε. This state can also be written:
η
0
= (∆, θ, ι, ∆, ε) (1)
where the order of variable is (PDH, Pyr, ACoA,
OAA, Cit).
Definition 2. [State] A signature E being given, the
set of states ζ is the set of functions η : E → L such
that for all e ∈ E, η(e) ∈ E (e).
In this formalism, the evolution of the system
is represented by two functions: the incrementa-
tion, noted incr, and the decrementation, noted decr.
These functions apply to one entity at a time and
return the level of this entity just above (resp. be-
low) its current level. Because all entities do not
have the same set of admissible levels, there is
one function defined for each entity. For instance,
if E (OAA) = {ε, ∆, θ} and η
0
(OAA) = ∆, then
incr
OAA
(η
0
(OAA)) = θ and decr
OAA
(η
0
(OAA)) =
ε. It should be noted that the incrementation
(resp. decrementation) function is not defined on
the maximal (resp. minimal) level of the admis-
sible levels. Therefore, in our previous example,
incr
OAA
(η(OAA)) is not defined if η(OAA) = θ.
Besides these functions, the formalism also makes
use of formulas to describe properties about the enti-
ties concentration levels.
Definition 3. [Formula] The set A of atomic formu-
las on a signature E is the set of expressions of the
form a 6 b where a and b can be any element of E ∪L.
The set F of formulas on a signature E is induc-
tively defined by:
• A ⊂ F .
• if ϕ and ψ are elements of F , then ¬ϕ, ϕ ∧ ψ,
ϕ ∨ ψ, ϕ ⇒ ψ are also elements of F .
Definition 4. [Satisfaction relation] A state η and
a formula ϕ ∈ F on a signature E being given, the
satisfaction relation η ϕ is inductively defined by:
• if ϕ is an atom of the form a 6 b, then η ϕ if and
only if η(a) 6 η(b) where η is the extension of η
to E ∪ L by the identity on L.
• if ϕ is of the form ϕ
1
∧ϕ
2
then η (ϕ
1
∧ϕ
2
) if and
only if η ϕ
1
and η ϕ
2
. We proceed similarly
for the other connectives.
Moreover, ”η ϕ” is read ”η satisfies ϕ”.
We may also use the abbreviation a = b as a short-
cut for (a 6 b) ∧ (b 6 a). We proceed similarly for
a < b, a > b and a > b.
Examples of formulas can be ϕ ≡ (Pyr = θ),
stating an excessive presence of Pyr or ψ ≡ (Cit >
ACoA), stating that the qualitative level of Cit is
strictly superior to the one of ACoA. The state η
0
,
previously described in eq. 1, satisfies ϕ but not ψ.
To determine the evolution of the system, a set of
rules is then used. A rule can be interpreted as possi-
ble modifications in the state which can be abstracted
by the following representation:
r : A
1
+ ··· + A
m
⇒ A
m+1
+ ··· + A
n
boost(ϕ) block(ψ)
Beside its identifier r, each rule includes two sets
of entities. The first one, for all i in [1, m], constitutes
the set of “reactants” whose level can be reduced by
the application of the rule. The other one, for all i in
[m+1, n], represents the set of “products” whose level
can be increased by the application of the rule. A rule
also includes two modulating conditions boost(ϕ) and
block(ψ) (ϕ and ψ being formulas) representing re-
spectively a possible positive and negative modula-
tion of the rule. The boost(ϕ) (resp. block(ψ)) mod-
ulation takes only effect if ϕ (resp. ψ) is satisfied and
its effects are further detailed later on. Of course, if
no modulation is known for a given rule, boost and
block regulations are not displayed in the rule repre-
sentation.
Definition 5. [Biological action network] A biologi-
cal action network on a signature E, or E -action net-
work, is a set R of rules of the form:
(1) r : A
1
+ ··· + A
m
⇒ A
m+1
+ ··· + A
n
boost(ϕ) block(ψ)
where:
• r is an identifier such that there are not two rules
in N with the same r.
• ∀i = 1 . . . n, A
i
∈ E.
• {A
1
. . . A
m
} ∩ {A
m+1
. . . A
n
} = ∅.
• ϕ and ψ are elements of F .
For short, we will call such rules E-rules and we
will call state of R a state on the signature of R.
Let us note that a rule representing possible alter-
ations of a biological state, it makes no sense to have
an entity being part of both reactants and products of
a same rule.
Moreover, please notice that a rule can be devoid
of any reactant or product. In the previous definition,
the index m can be equal to zero (the rule does not
need any reactant) or m can be equal to n (the rule
has no product). A rule without reactant can be con-
sidered as the constitutive production of an entity in a
given model and a rule without product can be inter-
preted as the degradation of an entity. In either cases,
the empty solution is depicted using the symbol.
It is worth mentioning that despite the strong re-
semblance between a rule and a chemical reaction,
a rule must not be interpreted as quanta of reactants
converted into quanta of products but as a possible
evolution of the levels of entities present in the rule.
A Qualitative Framework Dedicated to Toxicology
97

As a basic example of rule, the condensation of
acetyl-CoA and oxaloacetate to form Cit can be rep-
resented by the following rule:
r
A
: ACoA + OAA ⇒ Cit
Since neither positive nor negative modulating
conditions are considered here, only reactants and
products are displayed.
In order to be applicable at a given state, a rule
must meet basic criteria inspired from biology. First,
since the level ε is interpreted as a negligible concen-
tration, a rule is applicable only if all its reactants are
present at least at the level ι. In addition, a rule can-
not be applied if the negative modulating condition
block() applies, namely if the corresponding formula
is satisfied.
Definition 6. [Applicable rule] Let us consider a
state η on a signature E. An E -rule r ∈ R of the form
(1) is said applicable at the state η if and only if:
• ∀i = 1 . . . m, η(A
i
) 6= ε.
• η 2 ψ.
For instance, let us consider the conversion of Pyr
into acetyl-CoA by the enzyme name Pyr dehydroge-
nase (PDH). If we assume that E(PDH) = {ε, ι, ∆, θ},
the conversion can be written as:
r
B
: Pyr ⇒ OAA block(PDH < ∆)
This rule is applicable if and only if the level of
Pyr is strictly greater than ε and the level of PDH is
at least ∆, namely if there is Pyr in the system and a
normal concentration of PDH. Note that the catalysis,
namely the necessary presence of an enzyme to the
proper conduct of a reaction, can be expressed using
the block() condition as in the previous example.
Considering rules as possible alterations of the
equilibrium led us to a difficult choice on the way of
handling the emergence of abnormal levels of prod-
ucts.
On one hand, we can consider that every reactant
of a rule has to be in excess to shift the product(s)
equilibrium(s) to excessive levels. This vision is la-
belled optimistic since we suppose the system less
prone to drift towards excessive states. On the other
hand, with a more pessimistic perspective, only one
reactant in excess is enough to propagate the excess
to the product(s).
Both the optimistic and the pessimistic approaches
need an exception system to take into account par-
ticular biological cases. In the optimistic approach,
the exception is a boost() statement that relaxes the
conditions for a product to reach excessive levels. In
the pessimistic approach, the exception would be a
brake() statement selectively preventing excesses to
propagate.
As the optimistic approach is predominant in tox-
icological data, our formalism implements it thanks
to stricter conditions for a product level to increase.
These constraints can be found in the way of comput-
ing the potential next level of products.
Definition 7. [Potential next level] Let R be an E -
action network, let η be a state of R, let r ∈ R. We
note η
r
: E → L the partial function such that η
r
(e)
is defined if and only if r is applicable and if one of
the following conditions is satisfied:
• e ∈ {A
1
. . . A
m
} and in this case η
r
(e) =
decr
e
(η(e)).
• e ∈ {A
m+1
. . . A
n
}, η(e) < max(E(e)), and in this
case:
– if η 2 ϕ and η(e) < min
i∈{1...m}
(η(A
i
)) then
η
r
(e) = incr
e
(η(e)).
– if η ϕ then η
r
(e) = incr
e
(η(e)).
The potential next level of an entity through an
applicable rule refers to the next level of the entity
after the application of that rule. If the entity acts as a
reactant, its potential next level is the one returned by
the decrementation function applied to that entity.
If the entity acts as a product, its potential next
level depends on the boost() statement:
• if the boost() statement is not satisfied, a product
level can increase only if all the reactants levels
are strictly greater (this is due to the optimistic
vision explained previously). In this case, the po-
tential next level of a product is thus the one re-
turned by the incrementation function applied to
the product.
• if the boost() statement is satisfied, the previous
restriction no longer applies. In such cases, the
potential next level of a product is returned by
the incrementation function applied to it, indepen-
dently of the reactant levels.
Let us note that the potential next level is returned
either by the incrementation or decrementation func-
tion. Therefore, when these functions are not defined,
the potential next level of an entity is also not defined.
Keeping the conversion of Pyr as an example, we
can also specify that an excess of Pyr dehydrogenase
can cause trouble in oxaloacetate levels by adding a
boost() condition to the rule r
B
:
r
C
: Pyr ⇒ OAA block(PDH < ∆) boost(PDH > ∆)
Here, assuming that the rule is applicable at the
state η
0
and that η
0
(OAA) = ∆, the potential next
level of oxaloacetate by this rule can be θ only if
η
0
(Pyr) = θ or if η
0
(PDH) > ∆ (so, η
0
(PDH) = θ).
Among all the applicable rules at a given state,
only one is applied at a time. When a rule is applied,
one and only one of its entities sees its level chang-
ing to its potential next level. This means that the
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
98

level of an entity has to change in order to consider
that the rule was applied. Importantly, this also means
that a product cannot be updated simultaneously with
a reactant, and conversely. Similar ideas have been
firstly developed for discrete gene models by Thomas
and Snoussi (Snoussi, 1989; Thomas, 1991). This be-
haviour reflects the possibility for an entity to cross
a threshold without all the other entities levels doing
likewise.
In brief, starting from a given state, it is possible
to determine which rules of the system are applicable
at that state. The application of one of these rules then
changes the level of one entity, modifying the system
state. However, it is possible to stay indefinitely at a
same system state thanks to the rule Id (whose appli-
cation does not change the levels of the system entities
and that is always applicable).
It is then possible to establish a transition graph,
mapping all the possible transitions between the states
of a system. An infinite succession of transitions such
that the output state of a transition is the input state
of the next one is here called a path of the transition
graph.
Definition 8. [Transition graph] The transition
graph of an E -action network R is the labelled graph
whose set of vertices is the set of states ζ and the set
of edges T is the set of transitions of the form η
r
−→ η
0
such that one of the following condition is satisfied:
• r = Id and η
0
= η
• r ∈ R and there exists an entity e ∈ E such that
η
r
(e) is defined and:
– η
0
(e) = η
r
(e)
– ∀ e
0
∈ E r {e}, η
0
(e
0
) = η(e
0
).
Remark: The transition graph of an E-action net-
work R canonically defines a labelled Kripke struc-
ture L = (L, Σ, T ) as follows:
• L(η) = {α ∈ A | η α}.
• Σ = R ∪ {Id}.
• T can obviously be seen as the set of triplets
(η, r, η
0
) such that (η
r
−→ η
0
) is a transition of T .
A path (π ≡ η
0
r
0
−→ η
1
r
1
−→ . . .
r
i−1
−−→ η
i
r
i
−→ . . . ) is then
an infinite sequence of labelled transitions such that
the input state of r
i
is equal to the output state of r
i−1
for all i > 0. The set of paths is called Π
R
.
5 INTEGRATING
TOXICOLOGICAL
KNOWLEDGE
As the transition graph of a biological system includes
many biologically improbable paths, it is necessary
to filter out the irrelevant ones and only characterise
the interesting paths for toxicologists. Temporal logic
and model checking tools have been successfully ap-
plied to biological systems, either using Linear Tem-
poral Logic (Ito et al., 2014) or Computation Tree
Logic (Bernot et al., 2004). Here, since we seek to
filter paths, we need a logic able to express both state
and transition properties. We thus use the state/event
linear temporal logic (SE-LTL) developed by Chaki
(Chaki et al., 2004).
Since a path can be seen as an infinite alternance
between states and transitions, atomic temporal for-
mulas concern either a state or a transition. For states,
atomic temporal formulas are similar to atomic for-
mulas exposed in Definition 3. For transitions, atomic
temporal formulas only involve a rule identifier or the
identity operator.
Definition 9. [Temporal formula] Given an E-
action network R, the set T
R
of temporal formulas on
R is inductively defined by:
• (A ∪ R ∪ {Id}) ⊂ T
R
• if ϕ and ψ are formulas of T
R
, then ¬ϕ, ϕ ∧ ψ,
ϕ ∨ ψ, ϕ ⇒ ψ, Xϕ, Fϕ, Gϕ, ϕUψ are formulas of
T
R
.
Definition 10. [Temporal formula satisfaction]
Given an E -action network R and a path (π ≡ η
0
r
0
−→
η
1
r
1
−→ . . . ) ∈ Π
R
, the satisfaction relation ⊂ Π
R
×T
R
is inductively defined on the temporal formulas of T
R
by :
• π α where α ∈ A if and only if η
0
α,
• π r where r ∈ R ∪ {Id} if and only if r = r
0
,
• π ϕ ∧ ψ where (ϕ, ψ) ∈ T
2
R
if and only if π ϕ
and π ψ, other propositional logic connectives
are treated similarly,
• π Xϕ where ϕ ∈ T
R
if and only if (η
1
r
1
−→ η
2
r
2
−→
. . . ) ϕ,
• π Gϕ where ϕ ∈ T
R
if and only if for all i ∈ N,
(η
i
r
i
−→ η
i+1
r
i+1
−−→ . . . ) ϕ,
• π Fϕ where ϕ ∈ T
R
if and only if there exists
i ∈ N, (η
i
r
i
−→ η
i+1
r
i+1
−−→ . . . ) ϕ,
• π ϕ U ψ where (ϕ, ψ) ∈ T
2
R
if and only if there
exists j ∈ N, (η
j
r
j
−→ . . . ) ψ and for all 0 6 i < j,
(η
i
r
i
−→ . . . ) ϕ.
A Qualitative Framework Dedicated to Toxicology
99

Furthermore, for all r ∈ R of the form r : A
1
+···+
A
m
⇒ A
m+1
+ ··· + A
n
boost(ϕ) block(ψ), we note
app(r) the temporal formula (
V
m
i=1
A
i
> ε) ∧ ¬ψ stat-
ing that r is applicable at the current state (see Defini-
tion 6).
In addition, for all e ∈ E, we note ↓ e the temporal
formula stating that the level of the entity e decreases
in the next state:
W
l ∈ E (e)r{ε}
e = l ∧ X
e = decr
e
(l)
.
We proceed similarly for ↑ e.
For instance in our running example, the property
χ characterising paths where an excess of Pyr leads
to a future excess of oxaloacetate can be written as:
G(Pyr > ∆ ⇒ F(OAA > ∆)) and the formula ξ stating
that the rule r
A
is the first applied when Cit is absent
from the system can be written as: G(Cit = ε ⇒ r
A
).
In this situation, the path beginning with (η
0
r
B
−→ η
1
),
where η
0
= (∆, θ, ι, ∆, ε) and η
1
= (∆, θ, ι, θ, ε) satis-
fies χ but not ξ.
Finally, the association of the transition graph of a
system with a set of properties representing the rele-
vant biological pathways is called a constrained net-
work. This constrained network is actually a subset of
paths from the transition graph, with each path in this
subset satisfying all the expressed biological proper-
ties.
Definition 11. [Constrained network] An E-
constrained network is a couple N = (R, Ax) where
R is an E -action network and Ax is a set of temporal
formulas.
Definition 12. [Dynamics of a constrained net-
work] Given an E -constrained network N = (R, Ax),
the dynamics of N is the subset Π
N
of Π
R
such that
π ∈ Π
R
belongs to Π
N
if and only if π Ax.
Since properties filter out irrelevant paths from the
transition graph, it is thus possible to use them in con-
junction to formal methods to insure that the final con-
strained network respects basic biological and toxico-
logical properties as well as specific properties related
to the studied issue.
6 APPLICATION TO THE
ENERGY METABOLISM
To illustrate the formalism previously described, we
continue with the simplified energy metabolism de-
veloped in Section 3. The set of elements introduced
in Figure 1 is thus completed with the cell reducing
potential (pRed) and ethanol (Eth). The resulting sys-
tem signature is E
0
= {Gluc, Lac, Pyr, ACoA, OAA,
FA, pRed, Eth}.
In order to maximise the amount of possible paths,
let us consider that the set of admissible levels of the
endogenous entities (namely all entities except Eth)
is {ε, ι, ∆, θ}. In parallel, we will consider the ethanol
either absent, present moderately or present in excess
in the cell. Its set of admissible levels is thus {ε, ∆, θ}.
The role of each of these entities was developed
in Section 3. The reducing potential is here desig-
nated as pRed, but as previously described, it is an
abstraction of the balance between oxidised and re-
duced coenzymes. The rule ⇒ pRed thus abstracts
the rule OxidisedCoenzymes ⇒ ReducedCoenzymes.
As the amounts of oxidised and reduced coen-
zymes are interdependent, an increase in pRed means
both an increase in reduced coenzymes and a decrease
in oxidised coenzymes. This means that the rule
⇒ pRed cannot apply when there is an important
lack of oxidised coenzymes, namely when the reduc-
ing potential is in excess. This explains the presence
of block modulations linked to an excess of pRed in
some of the rules presented hereunder (GlyL, PyDC,
LDH, CC, βOx and EthOx).
The following rules summarise the interactions
described in Section 3 and constitute the E
0
-action
network R
0
:
GlyL : Gluc ⇒ Pyr + pRed block(pRed = θ)
PyDC : Pyr ⇒ ACoA+pRed block(pRed = θ∨ACoA > ∆)
PyC : Pyr ⇒ OAA
PyRed : Pyr + pRed ⇒ Lac
LDH : Lac ⇒ Pyr + pRed block(pRed = θ)
CC : ACoA ⇒ pRed block(pRed = θ ∨ OAA = ε)
MRC : pRed ⇒
GNG : OAA + pRed ⇒ Gluc
LipoG : ACoA + pRed ⇒ FA
βOx : FA ⇒ ACoA + OxPw block(pRed = θ)
EthOx : Eth ⇒ pRed block(pRed = θ) boost(Eth > ε)
Rule GlyL abstracts the whole glycolysis with its
transformation of Gluc into Pyr concomitant to the
production of pRed. The different futures of Pyr are
summarised in rules PyDC, PyC and PyRed. It should
be noted that the block modulation of PyDC repre-
sents possible conditions in which Pyr decarboxy-
lation is stopped while LDH is the reverse of rule
PyRed.
The rule CC represents the citrate cycle and
its production of pRed through the consumption of
ACoA. As OAA is an integral part of the cycle, this
rule is stopped by a lack of OAA. The rule MRC rep-
resent the ability for mitochondria to regenerate the
oxidised coenzyme (and therefore to decrease pRed).
Rules GNG and LipoG represent both carbohy-
drate (gluconeogenesis) and lipid (lipogenesis) an-
abolic pathways and their use of pRed. Conversely,
rule βOx abstracts β-oxidation and its production of
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
100
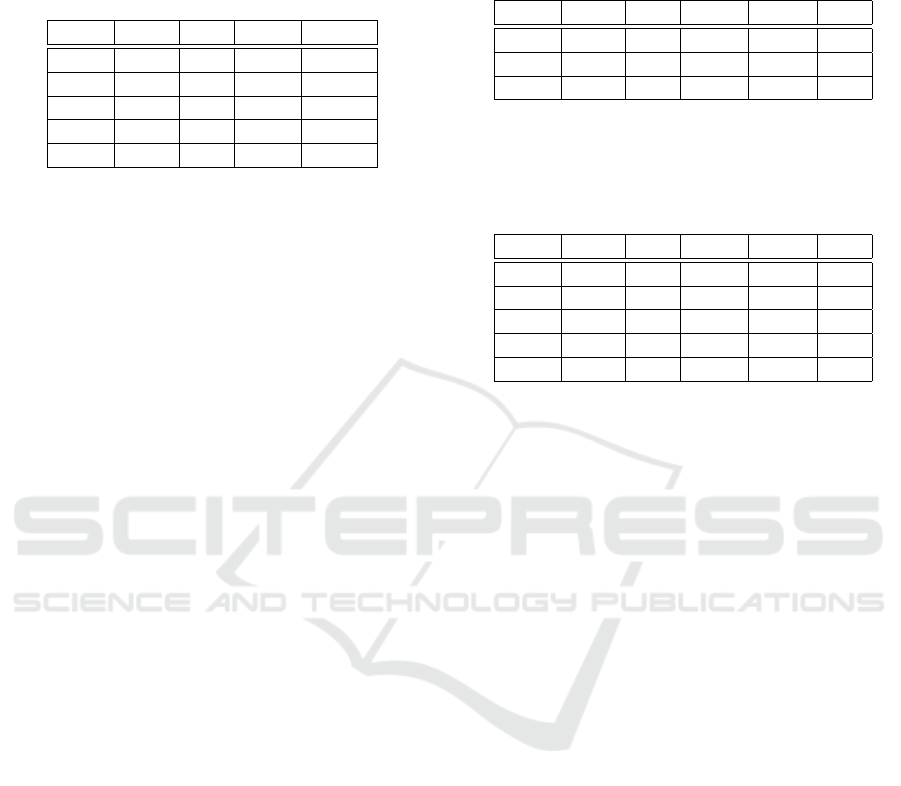
Table 1: The set of states present in path π
0
with rules ap-
plied at each step of the path. In all these states, FA and
pRed levels are normal (∆) while there is a lack of Lac (ι)
and a complete absence of Eth (ε). The level updated be-
tween each state is shown in bold.
State Gluc Pyr OAA ACoA
η
0
∆ ι ι ι
η
1
∆ ∆ ι ι
η
2
ι ∆ ι ι
η
3
ι ∆ ∆ ι
η
4
∆ ∆ ∆ ι
pRed.
Finally, the detoxification of Eth by the cell is de-
picted in EthOx, with the boost modulation represent-
ing the important amount of pRed possibly produced
by the detoxification. It should be noted that this
detoxification is known to be performed with high pri-
ority by the hepatic cell, but this kind of information
cannot appear in the rule.
Instead, it will be integrated in the model thanks to
temporal formulas as seen in Section 5. For instance,
it is known that Pyr can either be carboxylated (PyC)
or decarboxylated (PyDC), depending on the amount
of available ACoA. This can be summarized in the
temporal formula ϕ
0
:
G((ACoA < ∆ ∧ app(PyC) ∧ app(PyDC)) ⇒ ¬PyC)
This property states that when PyC and PyDC are
applicable, and that there is a lack of ACoA, the rule
PyC is not applied. Note that the G operator surround-
ing the formula indicates that the property remains
true at every step of the path. PyC thus never applies
when the previous conditions are satisfied.
It is also known that both decarboxylation and car-
boxylation of pyruvate prevail over pyruvate reduc-
tion (PyRed), as written in ϕ
1
:
G((app(PyRed) ∧ (app(PyC) ∨ app(PyDC))) ⇒ ¬PyRed)
This property is similar to the previous one and
litterally means that when PyRed and either PyC or
PyDC are applicable, PyRed never applies.
As previously said, these properties can be used to
characterise interesting paths allowed by R
0
. Let then
N
0
be the constrained network associating R
0
with the
two previous properties (ϕ
0
and ϕ
1
), and let us con-
sider the path π
0
beginning with the following prefix
(see Table 1):
η
0
GlyL
−−−→ η
1
GlyL
−−−→ η
2
PyC
−−→ η
3
GNG
−−−→ η
4
PyC
−−→ . . .
This prefix starts in state η
0
. This state can be as-
similated to a fasting state, where there is a normal
amount of energy supplies (Gluc, FA) and a lack of
every other metabolite. The first applied rule, GlyL
is the glycolysis, restablishing the normal level of Pyr
(η
1
). The rule is then triggered again, leading to a de-
Table 2: The set of states present in path π
1
with rules ap-
plied at each step of the path. In all these states, Lac, ACoA
and FA levels are normal (∆). The level updated between
each state is shown in bold.
State Gluc Pyr OAA pRed Eth
η
10
ι ∆ ι ∆ ε
η
11
ι ∆ ∆ ∆ ε
η
12
∆ ∆ ∆ ∆ ε
Table 3: The set of states present in path π
3
with rules ap-
plied at each step of the path. In all these states, Lac, ACoA
and FA levels are normal (∆). The level updated between
each state is shown in bold.
State Gluc Pyr OAA pRed Eth
η
20
ι ∆ ι ∆ θ
η
21
ι ∆ ι ∆ ∆
η
22
ι ∆ ι θ ∆
η
23
ι ι ι θ ∆
η
24
ι ι ι ∆ ∆
crease in Gluc level. The rule PyDC is then applied,
leading to an increase in OAA (η
3
). Finally, OAA fu-
els the gluconeogenesis (GNG), restablishing normal
glucose levels (η
4
).
Although the prefix of π
0
is allowed by the E
0
-
action network, it does not satisfy ϕ
0
. Indeed, the
priming of Pyr carboxylation (PyC) on Pyr decar-
boxylation (PyDC), namely the change from η
2
to
η
3
, in a fasting situation is very unlikely. As the pre-
fix does not satisfy one of the properties of the con-
strained network, the entire path is filtered out.
Properties are also important to express toxicolog-
ical knowledge. For instance, the priming of ethanol
detoxification (EthOx) over all the other rules can be
expressed by ψ
0
:
G(app(EthOx) ⇒ EthOx)
Furthermore, the detoxification reaction produce
a lot of reducing potential. This means that whenever
an excess of ethanol is detoxified through EthOx, the
reducing potential of the cell is impacted, hence the
formula ψ
1
:
G((Eth = θ ∧ EthOx) ⇒ (↓ Eth ∧ X(EthOx∧ ↑ pRed)))
Finally, if the cell struggles to decrease the pRed
level in presence of Eth, this means that the mitochon-
drial respiratory chain is saturated. In such cases, the
reduction of Pyr (PyRed) is triggered, hence the for-
mula ψ
2
:
G((Eth > ε ∧ ¬app(EthOx) ∧ app(PyRed)) ⇒ PyRed)
To illustrate the consequences of such constraints,
let us consider N
1
, the constrained network associat-
ing R
0
with the set of properties {ψ
0
, ψ
1
, ψ
2
}. Let us
also consider the path π
1
beginning with the follow-
A Qualitative Framework Dedicated to Toxicology
101

ing prefix (see Table 2):
η
10
PyC
−−→ η
11
GNG
−−−→ η
12
PyDC
−−−→ . . .
This path starts in the state η
10
, where there is no
ethanol, a lack of Gluc and OAA and a normal amount
of every other metabolite. This state corresponds to
an hypoglycemic state where the cell has the ability
to quickly recover its normal glucose level through
its supplies in FA and Pyr. Indeed, π
1
illustrates one
of the possible paths leading to the regeneration of
glucose, first with the application of PyC to restore
OAA levels, then with the application of GNG.
Let us now consider π
2
, a path similar to π
1
except
on the beginning state (see Tables 2 and 3):
η
20
PyC
−−→ η
11
GNG
−−−→ η
12
PyDC
−−−→ . . .
This path starts from η
20
, which is identical to η
10
except for the excessive presence of ethanol (see Ta-
ble 3). This path does not satisfy ψ
0
since ethanol
detoxification is not performed as soon as possible.
π
2
is thus not retained in N
1
.
Finally, let us take a look at path π
3
(see Table 3):
η
20
EthOx
−−−−→ η
21
EthOx
−−−−→ η
22
PyRed
−−−−→ η
23
PyRed
−−−−→ η
24
EthOx
−−−−→ . . .
Also starting from η
20
, π
3
then leads to the detox-
ification of a part of Eth through EthOx (η
21
), satis-
fying ψ
0
. The rule EthOx is then applied again, lead-
ing to the increase in pRed and satisfying ψ
1
. Rule
PyRed is then applied, decreasing the level of Pyr to
ι. As pRed is still in excess, PyRed is applied a second
time, leading to the recovery of normal levels of pRed
and satisfying ψ
2
. As π
3
satisfies ψ
0
, ψ
1
and ψ
2
, it
is retained in N
1
, illustrating the impaired ability for
a cell to regenerate normal glucose levels through the
gluconeogenesis in presence of ethanol.
7 CONCLUSION
In this article is presented a new formal framework
able to handle several specificities of the toxicology
domain not taken into account so far. This rule-based
modelling framework relies on the direct description
of equilibrium changes happening in a biological sys-
tem. This description does not model the difference
of reaction speed between the model rules, which can
affect the system equilibrium. It is however possi-
ble to integrate biological and toxicological knowl-
edge about rule kinetics through formulas expressed
in temporal logic.
As demonstrated on a simple model of the en-
ergy metabolism, its expressive power allows us to
describe both the equilibrium changes in the biologi-
cal system and knowledge about the prioritisation of
reactions. This knowledge is then used to filter out
irrelevant paths from the resulting model.
In the future, our formalism will be coupled with
formal methods with the purpose of generating the
comprehensive list of pathways of toxicity present in
a model. Indeed, through the use of biological prop-
erties, it is possible to define pathological states and
list all the paths leading to these states. The result-
ing paths shall finally be sorted thanks to additional
toxicological knowledge. Furthermore, filtering the
resulting paths could also highlight gaps in the cur-
rent toxicological knowledge and help toxicologists
in their design of new experiments.
Finally, this formalism will serve as a basis to
develop a software platform dedicated to toxicology.
This platform is currently under development and it is
already possible to run simulations on biological ac-
tion networks. In the future, the platform will also be
able to integrate the temporal formulas and to filter
out paths from the biological action networks that do
not satisfy the formulas. This will be achieved gen-
erating all the paths allowed by a system biological
action network and directly checking these paths for
their biological relevance thanks to expressed biolog-
ical properties. Finally, by defining states regarded as
pathologic, the platform will then be able to compute
all the paths leading to pathologic states and thus pro-
pose putative pathways of toxicity to toxicologists.
REFERENCES
Backhaus, T., Blanck, H., and Faust, M. (2010). Hazard and
risk assessment of chemical mixtures under REACH:
state of the art, gaps and options for improvement.
Swedish chemicals agency.
Bernot, G., Comet, J.-P., Richard, A., and Guespin, J.
(2004). Application of formal methods to biolog-
ical regulatory networks: extending thomas’ asyn-
chronous logical approach with temporal logic. Jour-
nal of theoretical biology, 229(3):339–347.
Brooks, G. A. (1998). Mammalian fuel utilization during
sustained exercise. Comparative Biochemistry and
Physiology Part B: Biochemistry and Molecular Bi-
ology, 120(1):89–107.
Calzone, L., Fages, F., and Soliman, S. (2006). Biocham:
an environment for modeling biological systems and
formalizing experimental knowledge. Bioinformatics,
22(14):1805–1807.
Chaki, S., Clarke, E. M., Ouaknine, J., Sharygina, N., and
Sinha, N. (2004). State/event-based software model
checking. In International Conference on Integrated
Formal Methods, pages 128–147. Springer.
Chance, B. and Williams, G. (1956). The respiratory chain
and oxidative phosphorylation. Adv Enzymol Relat Ar-
eas Mol Biol, 17:65–134.
Ciocchetta, F. and Hillston, J. (2009). Bio-pepa: A frame-
work for the modelling and analysis of biological sys-
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
102

tems. Theoretical Computer Science, 410(33):3065–
3084.
Colborn, T., Dumanoski, D., and Myers, J. P. (1996). Our
stolen future: are we threatening our fertility, intelli-
gence, and survival?: a scientific detective story. Dut-
ton.
Field, J. B., Williams, H. E., and Mortimore, G. E.
(1963). Studies on the mechanism of ethanol-induced
hypoglycemia. Journal of Clinical Investigation,
42(4):497.
Hellerstein, M., Christiansen, M., Kaempfer, S., Kletke, C.,
Wu, K., Reid, J., Mulligan, K., Hellerstein, N., and
Shackleton, C. (1991). Measurement of de novo hep-
atic lipogenesis in humans using stable isotopes. Jour-
nal of Clinical Investigation, 87(5):1841.
Ito, S., Hagihara, S., and Yonezaki, N. (2014). A qualita-
tive framework for analysing homeostasis in gene net-
works. In proceedings of 5th International Conference
on Bioinformatics Models, Methods and Algorithms
(BIOINFORMATICS2014), pages 5–16.
Kepner, J. (2004). Synergy: The big unknowns of pesticide
exposure. Pesticides and You, 23(4):17–20.
Owen, O. E., Kalhan, S. C., and Hanson, R. W. (2002).
The key role of anaplerosis and cataplerosis for citric
acid cycle function. Journal of Biological Chemistry,
277(34):30409–30412.
Pettit, F. H., Pelley, J. W., and Reed, L. J. (1975). Reg-
ulation of pyruvate dehydrogenase kinase and phos-
phatase by acetyl-coa/coa and nadh/nad ratios. Bio-
chemical and biophysical research communications,
65(2):575–582.
Pilkis, S. J. and Granner, D. (1992). Molecular physiology
of the regulation of hepatic gluconeogenesis and gly-
colysis. Annual review of physiology, 54(1):885–909.
Schulz, H. (1991). Beta oxidation of fatty acids. Biochim-
ica et Biophysica Acta (BBA)-Lipids and Lipid
Metabolism, 1081(2):109–120.
Snoussi, E. H. (1989). Qualitative dynamics of piecewise-
linear differential equations: a discrete mapping ap-
proach. Dynamics and stability of Systems, 4(3-
4):565–583.
Talcott, C. (2008). Pathway logic. In International
School on Formal Methods for the Design of Com-
puter, Communication and Software Systems, pages
21–53. Springer.
Thomas, R. (1991). Regulatory networks seen as asyn-
chronous automata: a logical description. Journal of
theoretical biology, 153(1):1–23.
A Qualitative Framework Dedicated to Toxicology
103
