
Retrieving Similar X-ray Images from Big Image Data
using Radon Barcodes with Single Projections
Morteza Babaie
1,2
, H. R. Tizhoosh
1
, Shujin Zhu
3
and M. E. Shiri
2
1
KIMIA Lab, University of Waterloo, ON, Waterloo, Canada
2
SINA Lab, Mathematics & Computer Science Department, Amirkabir University of Technology, Tehran, Iran
3
School of Electronic & Optical Eng., Nanjing Univ. of Sci. & Tech., Jiangsu, China
Keywords: Radon Transform, Content-based Image Retrieval, Binary Barcode, Radon Barcodes, Big Data.
Abstract: The idea of Radon barcodes (RBC) has been introduced recently. In this paper, we propose a content-based
image retrieval approach for big datasets based on Radon barcodes. Our method (Single Projection Radon
Barcode, or SP-RBC) uses only a few Radon single projections for each image as global features that can
serve as a basis for weak learners. This is our most important contribution in this work, which improves the
results of the RBC considerably. As a matter of fact, only one projection of an image, as short as a single
SURF feature vector, can already achieve acceptable results. Nevertheless, using multiple projections in a
long vector will not deliver anticipated improvements. To exploit the information inherent in each projection,
our method uses the outcome of each projection separately and then applies more precise local search on the
small subset of retrieved images. We have tested our method using IRMA 2009 dataset a with 14,400 x-ray
images as part of imageCLEF initiative. Our approach leads to a substantial decrease in the error rate in
comparison with other non-learning methods.
1 INTRODUCTION
Nowadays, the role of computers has significantly in-
creased in our daily lives. As a result, most of the
computerized activities are stored as some sort of data
such as text, photos, videos, audio files and more.
Hence, it is not surprising that searching for data and
finding specific data in all these massive datasets is
not only a challenging task in many fields but also
quite often a necessary one (Rodríguez et al. 2015).
One of these challenges stems from the Content-
Based Image Retrieval (CBIR) which is considered as
an important task in “biomedicine, military, com-
merce, education, and Web image classification and
searching. In the biomedical domain, CBIR can be
used in patient digital libraries, clinical diagnosis,
searching of 2-D electrophoresis gels, and pathology
slides” (Wang 2001). CBIR is primarily concerned
with searching for and delivering similar images pro-
vided a query (input) image is given by a user.
One of the practical aspects of CBIR in medical
imaging is to assist clinicians for diagnostic purposes
by enabling them to compare the case they are
examining with previous (known) cases. It is
established pratcice that most hospitals do store their
patient data for a long periode of time; generally
images are stored in PACS (picture archiving and
communication system) and related documents such
as biopsy and treatment reports are stored in RIS
(radiology information system). Let us assume that a
diagnostic case is being inspected, by using a reliable
CIBR system clinicians can benefit from analogous
cases buried among millions of images, and hence
achieve higher diagnostic accuracy based on
comparative discrimination with previous (known)
cases (Kumar et al. 2013). A large number of CBIR
methods exist in the literature. Feature extraction,
learning, dictionary approaches, and binary
descriptions are among most commonly used
techniques to search for similar images.
In this paper, we propose compact features to fa-
cilitate fast image retrieval. Our method (Single Pro-
jection Radon Barcode, SP-RBC) benefit from the in-
formation inherent in single Radon projections, based
on the recently introduced Radon Barcodes (RBC)
that capture image information in short binary vec-
tors, or barcodes. We will report both cases where we
use the actual values of Radon projections as well as
their binary encodings known as Radon barcodes
(RBC).
The rest of this paper is organized as follows: Sec-
tion 2 provides a brief review of related works and
Babaie, M., Tizhoosh, H., Zhu, S. and Shiri, M.
Retrieving Similar X-ray Images from Big Image Data using Radon Barcodes with Single Projections.
DOI: 10.5220/0006202105570566
In Proceedings of the 6th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2017), pages 557-566
ISBN: 978-989-758-222-6
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
557

Radon transform. Section 3 will describe our pro-
posed method. The results of our experiments and
comparisons are reported in Section 4. The last sec-
tion provides some conclusions and suggestions for
the feature works.
2 RELATED WORK
Content-based image retrieval (CBIR) techniques au-
tomatically search for similar images in a database by
using visual features extracted to represent its
content, and not by a text description. In many cases,
textual descriptions may not be available, and in
many other cases, it is extremely difficult, if not im-
possible, to describe the image content (e.g., the
shape of an irregular tumor in a breast ultrasound
scan) adequately in all necessary details.
The major challenge for CBIR is to extract the rel-
evant image features based on relevant feature simi-
larity criteria and to organize the extracted features
into some sort of compact embeddings or representa-
tions for fast retrieval from big databases (Kumar et
al. 2013). The features or descriptors that represent
the properties/content of the images are often used in
CBIR systems. The choice of features or descriptors
should minimize the “semantic gap” between the ex-
tracted image features on one side and the human’s
interpretation of the image content on the other side.
Early CBIR systems often used the image fea-
tures/descriptor, such as histogram, shape and texture
descriptors (Gevers & Stokman 2004; Lee et al. 2003;
Saha et al. 2004). Gevers and Stokman proposed an
object recognition method based on the histograms
derived from photometric color invariants, which out-
performed the traditional color histogram scheme but
was very sensible to the noise (Gevers & Stokman,
2004). The edge histogram which contains the gen-
eral shape information and the moment that describes
the image pixel intensities were also used in early
CBIR systems (Shim et al. 2002; Zhu & Schaefer
2004).
The advanced features such as Scale Invariant
Feature Transform (SIFT) (Lowe 2004) and Speeded
Up Robust Features (SURF) (Bay et al. 2008) are
employed in CBIR systems to retrieve similar images
from different point of views and transformations (Do
et al. 2010; Velmurugan & Baboo 2011). As many of
these smart features are invariant to scale and rota-
tion, they are more robust than typical image trans-
forms. However, the features are typically large and
inefficient to conduct matching in big image data.
Ledwich et al. used the structure of typical indoor en-
vironments to reduce the need for rotational invari-
ance of the features, which has a minimal effect on
retrieval rate and significant improvement in effi-
ciency (Ledwich & Williams 2004). Velmurugan and
Babbo used the KD-tree with the Best Bin First (BBF)
(Beis & Lowe 1997) indexing method to accelerate
the similarity match of the SURF and color moments
combined features (Velmurugan & Baboo 2011).
With dramatic growth of image data in recent
years, one of the current trends in CBIR is to use
binary features such as Local Binary Patterns (LBP)
(Ojala et al. 2002), Binary Robust Invariant Scalable
Keypoints (BRISK) (Krig 2012), Binary Robust
Independent Elementary Features (BRIEF) (Calonder
et al. 2010), and Radon Barcodes (Tizhoosh 2015).
Comparing to non-binary features, the distance
computation between binary strings is much faster
for retrieving tasks. Bankar et al. proposed a CBIR
system based on LBP variance which characterized
the local contrast information into the one-dimen-
sional LBP histogram (Bankar et al. 2014). Subrah-
manyam et al. extended the LBP, which took ad-
vantage of the magnitude of the local difference be-
tween the center pixel and its neighbors which were
able to extract the edge information in the image, and
proposed the local maximum edge binary patterns
(LMEBP) descriptor (Subrahmanyam et al. 2012).
Other methods such as Deep Neural Networks
(DNNs), Convolutional Neural Networks (CNNs),
and Bag of Words (BoW) have been recently
developed for CBIR tasks. Learning from massive
annotated data in the deep learning networks, CNN
features would carry high-level and riche semantic in-
formation (Yan et al. 2016), which had been proved
to be successful in achieving state-of-the-art perfor-
mance (Babenko et al. 2014; Wan et al. 2014). Avni
et al. made use of SIFT descriptors to build the feature
dictionary with the bag of visual words approach, per-
forming outstandingly for the x-ray image retrieval
task in IRMA dataset (Avni et al. 2011). The best re-
sults so far have been reported by combining LBP and
saliency detection (Camlica et al. 2015).
However, most of CBIR methods face high com-
putational expenses and require considerable re-
sources during the learning phase. Besides, their im-
plementation requires sophisticated and complicated
design and data structures (Krig 2012).
In the medical field, CBIR systems can assist cli-
nicians to make more reliable clinical decisions by re-
trieving similar (proven) cases from the past stored in
their archives. But for the most of the afore-men-
tioned methods, they usually need to undergo param-
eter tuning to be applicable to medical image pro-
cessing (Huang et al. 2010). Not only because most
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
558
of the medical images have a known direction or
known scale (which means the global search would
not face related challenges), but also because at least
for the case of local descriptors in medical images,
experiments show they are generally not able to
achieve good results (Avni et al. 2011). It would be
acceptable if we use very simple content-based re-
trieval methods, rather than learning-based compli-
cated methods in order to save time for learning and
parameter tuning, even if we cannot reach top accura-
cies, but can provide comparable results.
As Radon transform can convert global detection
problem in the image domain into local peak detec-
tion problem in the parameter domain (Aundal &
Aasted 1996), it is widely applied in the medical field,
especially in Computed Tomography. Radon bar-
codes, RBC, are binary codes generated by Radon
Transform with projection binarization, proposed for
medical image retrieval system. RBCs have achieved
comparable results with many other methods from lit-
erature (Tizhoosh 2015) and are easy to implement
and efficient in matching and retrieving (via Ham-
ming distance) with low requirements for memory
and storage.
In this paper, a content-based image retrieval ap-
proach for big datasets is proposed. The multiple Ra-
don projections with selected angles are first ex-
tracted (considered as global features) for each image.
Each projection is used separately to search for simi-
lar images from the big dataset. The more precise lo-
cal features such as LBP and shifted Radon projec-
tions are then employed to refine the results. We use
the IRMA dataset of 14,400 x-ray images to validate
the approach.
2.1 Radon Transform
Radon introduced an integral transformation, which
calculates the sum of the values of an image along
parallel lines for various angels (Radon 1917). One
key factor of Radon transform is its ability to recon-
struct the main image from its transform. The Radon
transform has been applied in medical imaging, e.g.,
in computer tomography, for image reconstruction
(Gu & Sacchi 2009). The Radon transform is
generally given as for each given as follows:
R( , )= ( , )
f
xy xcos ysin dxdy
(1)
In this equation, , refers to grey-level inten-
sities of image f at position
,
, and . is a Dirac
delta operator.
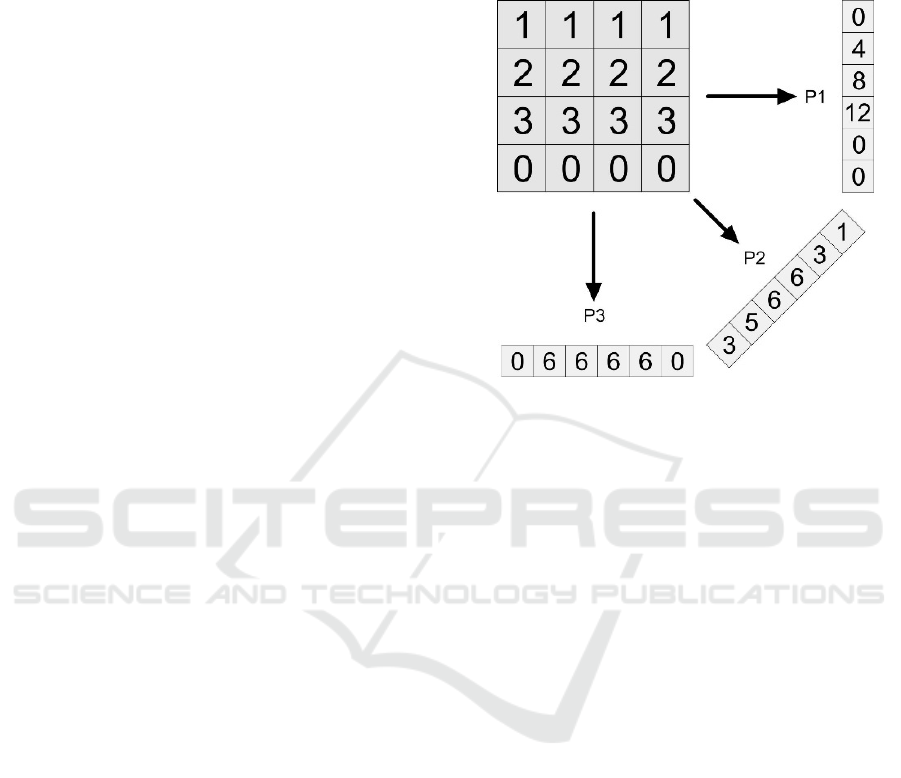
Figure 1 depicts an example for a small
matrix and three sample Radon projections which can
be used as image features and have been applied in
many fields of computer vision (Aundal & Aasted
1996). It also has been applied in form of Radon Bar-
codes for medical CBIR systems in large datasets
(Tizhoosh 2015).
Figure 1: Three Radon projections in 0, 45 and 90-degree
directions. Zero padding is applied to create same-length
vectors.
3 PROPOSED METHOD
In this section, we introduce the idea to use single Ra-
don projections for CBIR in its both forms, namely
real-valued (single projection Radon, SP-R) and bi-
nary (single projection Radon barcode, SP-RBC) im-
plementations. First, we describe the automated
preprocessing steps. In the next step, we explain how
we use Radon projections to reach top similarity for
each projection separately. We then introduce the ex-
ploitation method, which is used to find the most sim-
ilar images in the pre-selected set (Selection Pool).
Finally, we apply a binarization method to create SP-
RBC. For all our experiments, we use the IRMA da-
taset (Tommasi et al. 2009).
3.1 Pre-Processing Images
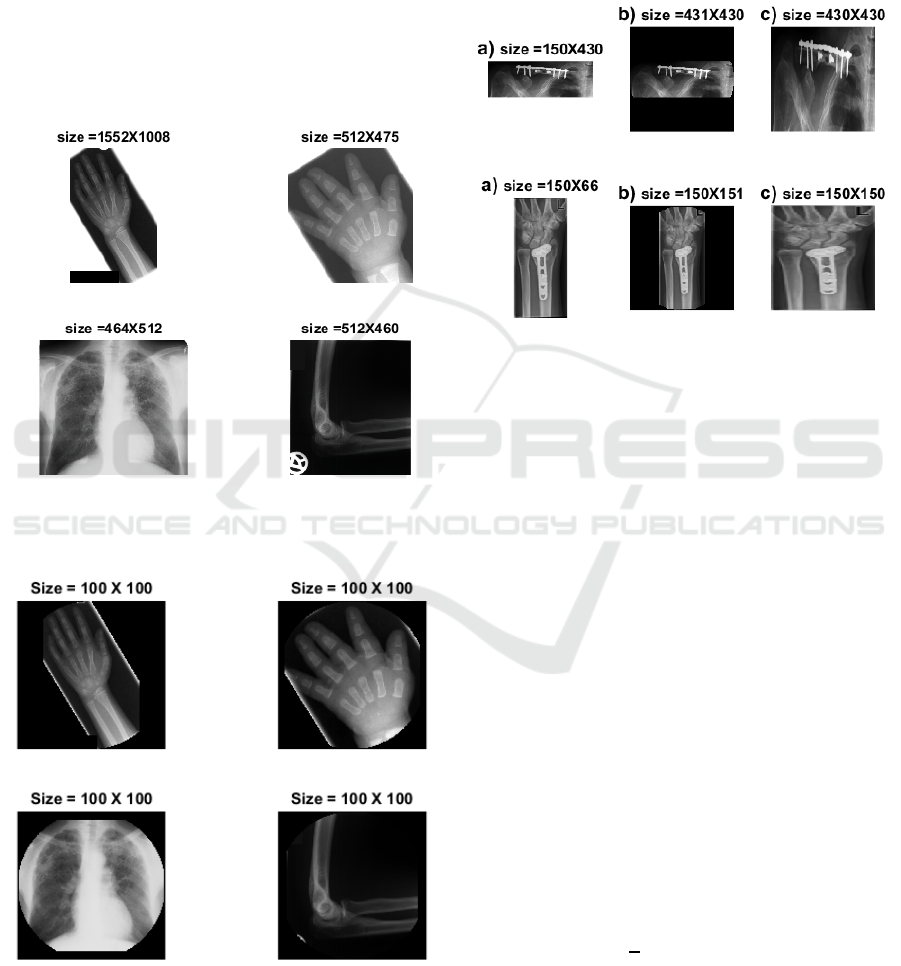
Data test images we use is quite challenging. For in-
stance, the imbalance in IRMA image dataset has
been noted as one of the most challenging aspects of
this dataset (Avni et al. 2011). There is major varia-
bility in IRMA images, not only in the term of a sam-
ple density in each category but also with respect to
image size, brightness, scale of body objects as well
as unrelated burned-in landmarks (Figure 2). To re-
duce the effects of these problems, we proposed a pre-
processing chain composed of three stages:
Retrieving Similar X-ray Images from Big Image Data using Radon Barcodes with Single Projections
559
1) resizing images to zero-padded squared im-
ages to avoid distortion due to necessary under-sam-
pling (most CBIR methods do require under-sam-
pling of images to reduce computational burden),
2) removing non-related parts such as burned-in
landmarks (e.g., letters) due to the digitization of x-
ray films, and
3) circular image margin suppression (super-im-
posing a circle on the image to eliminate the image
margin from the processing).
Figure 3 shows the output of our pre-processing
steps when applied on images from Figure 2.
Figure 2: Four samples from IRMA training set to illustrate
the variability of X-ray image data.
Figure 3: The pre-processing effect for same samples from
Figure 2 to create “normalized” inputs.
Figure 4 shows the difference between our
squaring method and simple image resize operations.
This operation preserves the original scale of image
whiteout distortion due to the resize operation which
shows significant improvements in the results (see
Table 4 in the result section) in comparison with pre-
vious works.
Figure 4: a) rectangular images, b) resized by our method
(squared with zero padding), and c) resized conventionally.
For the second pre-processing stage, we consider
the nature of summation operator in Radon vectors
which is clearly sensitive to bright parts of image. As
shown in Figure 2, most of the unrelated shapes
(letters, signs, landmarks etc.) in IRMA dataset are
depicted in (near-)white. To decrease their effect on
Radon projections, we remove these white parts by a
simple binary operation. Although this may
sometimes lead to eliminating some relevant
characteristics of some images. In general, however,
our results have shown that this stage of pre-
processing helps to deliver better results (see Table 4
in result section). In this stage, all images are
binarize by a threshold at 98% of maximum image in-
tensity to locate (near-)white image regions (Figure
5). Subsequently, these parts are replaced (filled) with
the median intensity value of their neighbors (we ac-
cess the neighbors by using some morphological op-
erators). Also, since artificial marks/signs appear
mostly close to the image border, replacing (near-
)white pixels is strictly restricted to the image mar-
gins.
Radon projections change their length based on
projection angel. All Radon operators consider the
maximum length
√
2
(where N is the largest of im-
age width/height) and use zero-padding for this pur-
pose. Based on (Tsai & Chiang 2010), we discard all
pixels out of circumference area from the image cen-
ter within a diameter of N/2.
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
560
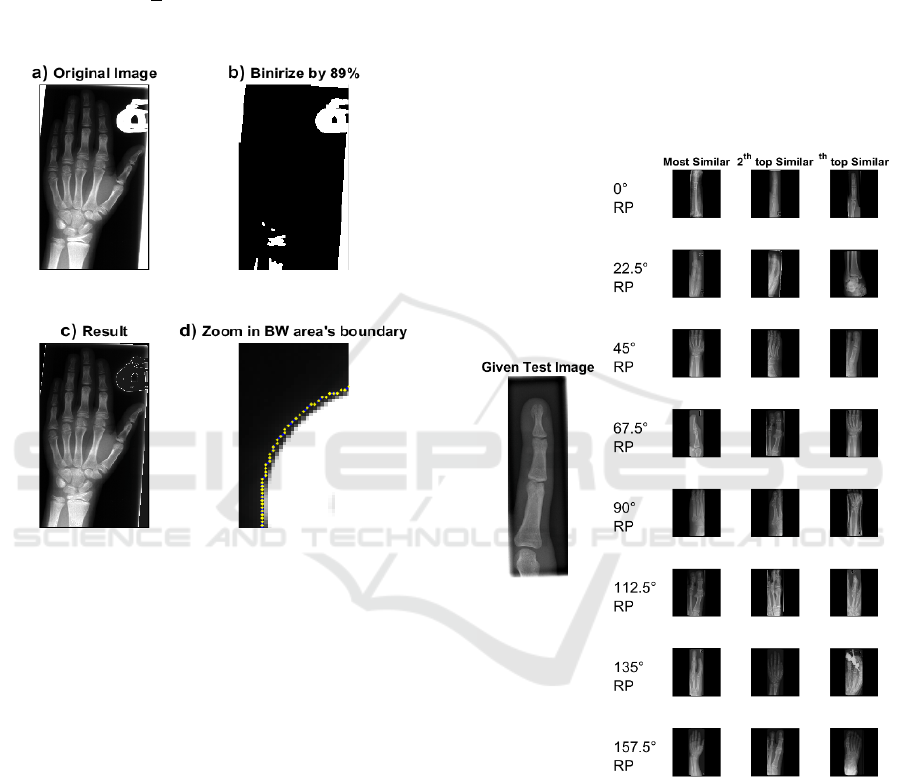
Figure 3 shows sample images with superimposed
circles. This process helps to achieve shorter vectors;
the assumption, of course, is that image margins do
not offer much diagnostic information, hence we can
eliminate them. Our experiments confirm the
assumption for x-ray images in IRMA dataset. We
also can perform this step by selecting just N center
elements out of
√
2
elements in all Radon projec-
tions.
Figure 5: Removing irrelevant (near-)white landmarks: a)
original image, b) binary image with 98% thresholding, c)
the result after removing the irrelevant pixels by replacing
them with the median value of their neighborhood, d) mag-
nification of upper region of the image to show the bound-
aries which are used for median calculation.
3.2 Radon Projections
Using several Radon projections simultaneously has
been used successfully in many CBIRs woks (Zhu &
Tizhoosh 2016; Tizhoosh 2015; Liu et al. 2016). It
may be seen as obvious that using more projections
in all possible directions is associated with better re-
sults and naturally a more time-consuming search. In
our experiments, we examine a new approach by us-
ing only one projection for each retrieval attempt.
Obviously, we did not anticipate very promising
retrieval results for just one projection. However,
there was an interesting point in the results. If we, for
the sake of analysis, select the minimum IRMA error
for each projection, then the total IRMA error de-
creases dramatically (much better than concatenating
them into one vector). It means that although individ-
ual projection results are slightly better than of ran-
dom results (the error of each projection results is var-
ying between 570 to 640, and random search error is
around 900, see Table 3). This observation confirms
that separated projection results may have little over-
lap with each other. As a result, while one projection
(for example the projection at zero degree) fails to
find the most similar image, other projection (say the
45-degree direction) might be able to find it. By
choosing a certain number of “best matches” (say the
top three matches) for each Radon projection, we can
create a “Selection Pool” (see Figure 6).
Figure 6: A sample of Selection Pool for a test image and
top three images for each Radon projection. The final re-
sult(s) can be extracted from the Selection Pool after more
computationally demanding search methods are applied.
The surprising point is that the minimum error
rate for each test image in the Selection Pool consid-
erably decreases (e.g., error=196 for 8 projections).
Figure 7 motivates our idea of using single projec-
tions separately, where the error of the first hit (image
with the highest similarity to the query image) for 8
projections are depicted. The right side plot in
Retrieving Similar X-ray Images from Big Image Data using Radon Barcodes with Single Projections
561
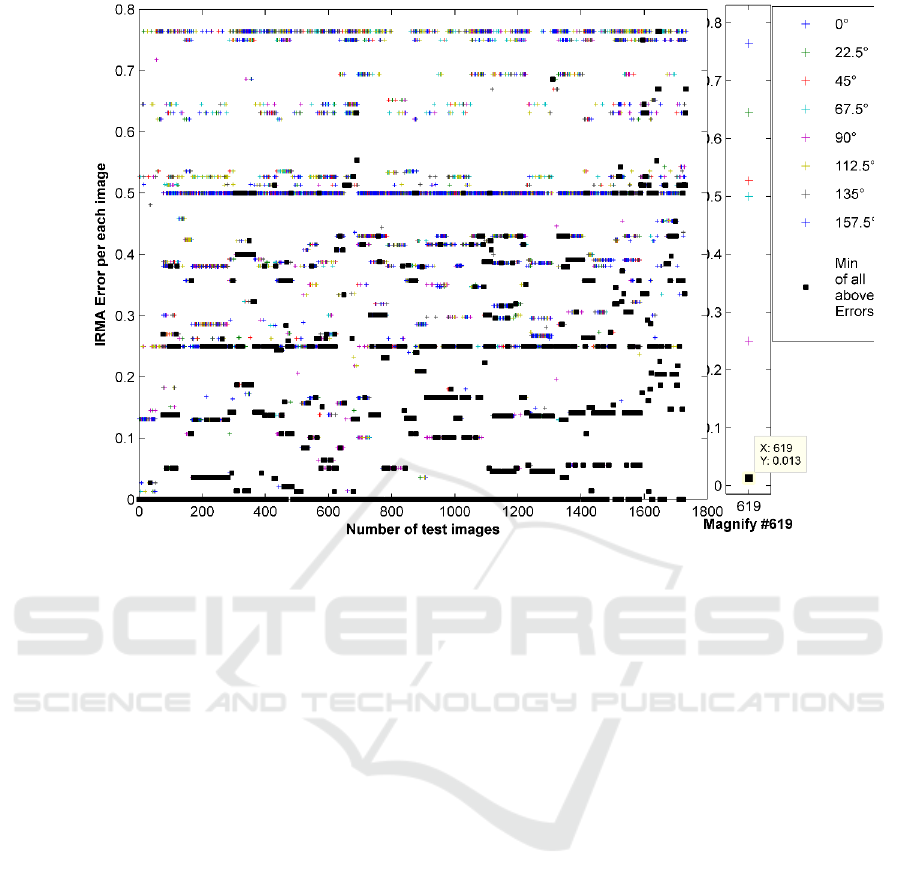
Figure 7: The IRMA error of 1733 images for 8 single equidistant Radon projections. Black squares are the minimum error
for each test image. The sum of black squares amounts to 196. The right section magnifies an arbitrary test image results
(#619) for better visualization.
Figure 7 is the magnification of the errors of one spe-
cific test image (the image #619 in IRMA dataset). As
shown, each projection can be used to retrieve an im-
age as the first hit with different IRMA error. If we
record the lowest error for each projection, we can
reach a total error of 196 which, looking at the re-
ported numbers in literature, is quite low. For 10 top
images for each projection, the error even decreases
to 65, an error level not yet reached by any method in
literature. It means if we had an algorithm to identify
the best Radon projection for each image, we could
achieve the outstanding results. However, reliable
learning methods to select the best projection is ap-
parently quite challenging and subject to future
works. Hence, we attempt to exploit the discrimina-
tion power of single projections in a Selection Pool
via an exploitative approach.
3.3 Exploitation Search
After we have a small group of candidate images (Se-
lection Pool), retrieved from thousands of images in
training set, we can now apply a refined and more ex-
ploitative search to choose the best image from within
the Selection Pool. As mentioned before, if we man-
ually pick the best image from the top ones, we can
achieve the best scores. In this section, two methods
are combined to search the Selection Pool; Shifted
Radon and LBP.
Shifted Radon is proposed to eliminate the effect
of translation in images or image regions. Using eight
equi-distanced projections and shifting each Radon
projection in the test image to align with its counter-
part projection in the Selection Pool makes the algo-
rithm robust against translation (Figure 8 shows this
process for two sample images). We use the smallest
distance between the two projections using cross-
correlation (shifting by ± 10% the length of the
projection). By looking at cross-correlation between
each pair of Radon projections of two images, all
eight minimum distances (we use eight projections)
are summed up. After calculating the distance for all
images in the Selection Pool, we normalize this error
in [0,1].
LBP (local binary patterns) have been used in
many CBIRs based on thier power and speed (Avni et
al. 2011)(Nanni et al. 2010). In our method, LBP and
Shifted Radon are used together to improve the re-
sults. The LBP error rate is calculated by the
normalized sum of their absolute values. Final deci-
sion making is done by using the smallest value of the
sum of these two error vectors. Since both vectors
(Shifted Radon and LBP) are normalized between
zero and one, they are comparable to each other.
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
562
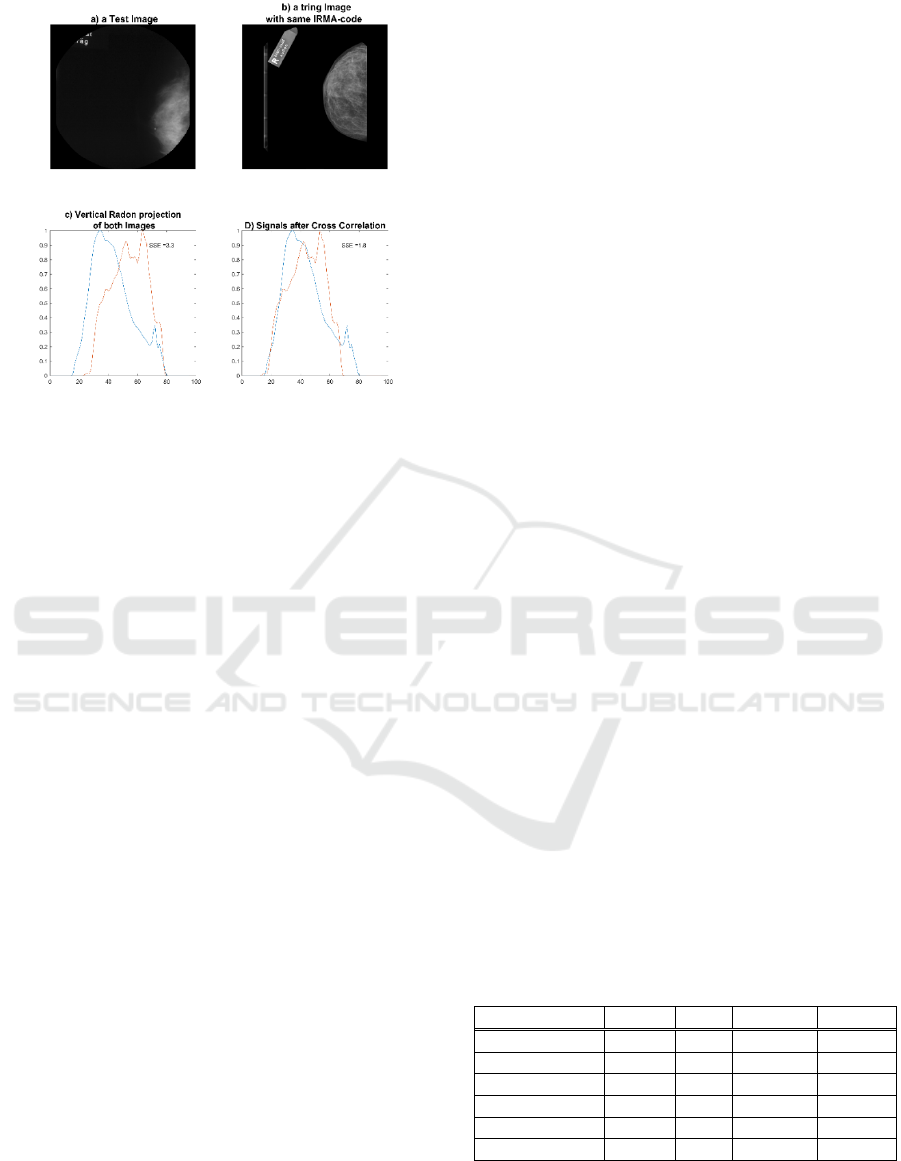
Figure 8: Two sample images with vertical subject shift and
their zero-degree Radon projections before and after cross-
correlation-based shifting.
3.4 Binarization of Projections
Computational complexity is one of the critical
features of any retrieval method. Our approach is very
light in comparison with most reported descriptors.
We just search the dataset using eight vectors as short
as the length of the resized image (under-sampled to
64x64 pixels in our experiments). Additionally, to
further shorten the search time, we have created the
binary version of projections to test SP-RBC which
employs two methods to create binary codes from Ra-
don projections. The MinMax and Median methods
have already been applied to threshold Radon projec-
tions (Tizhoosh 2015, Tizhoosh et al. 2016).
The Median method uses the median value of
each projection as a threshold, all elements below the
threshold are set to zero (Tizhoosh 2015). On the
other hand, the MinMax method sets zeros and ones
depending on the locations of minimums and maxi-
mums of each projection (Tizhoosh et al. 2016).
4 EXPERIMENTS AND RESULTS
This section introduces the data set used in our
research. We then detail our results and compare our
method with other methods.
4.1 Image Dataset: IRMA
The Image Retrieval in Medical Application, short
IRMA, is a challenging dataset, which has composed
of 12,677 images for training and 1,733 images for
evaluating any proposed retrieval method. Every
IRMA image is associated with a 13-digit code, and
each code is divided into four parts:
IRMA Code: TTTT-DDD-AAA-BBB
The first four digits describe the imaging modal-
ity, the next three represent the body orientation in the
image, the next four describe the body region and fi-
nally, the last four indicate the biological system ex-
amined. IRMA creators have also introduced a sys-
tem for measuring the error between two IRMA codes
and return an error number between zero and one
(Tommasi et al. 2009). So, retrieval algorithms
should return the most similar image for all 1733 test
images using the training dataset which supposed to
contain similar cases for every query image although
due to imbalance the easiness of finding similar cases
strongly varies.
4.2 Results
In this section, we first compare our best-achieved re-
sult with the results of other methods. After that, we
discuss the impact of each part of implementation on
achieved results. Finally, we show the results for each
Radon projection separately.
For comparing with other methods, we consider
approaches in two different groups, non-learning
methods and learning-based methods. The results for
RBC
4
, RBC
8
, and LBP are used from literature
(Tizhoosh 2015). However, since there is a different
way for error calculation in IRMA database than in
some papers, we recalculate the error for all men-
tioned methods based on (Tommasi et al. 2009) to
have consistent and fair comparisons.
In non-learning comparison, we consider L as the
length of descriptors, T as the type of searching (B for
Binary search and F for Floating point search), E
Total
as total IRMA error, and N
0
indicates the percentage
of zero-error cases (in case consider the classification
problem).
Table 1: IRMA error for non-learning methods (L=length,
T=type [B=binary; F=float], E=error, N
0
=percentage of re-
trieved cases with zero error).
Method L T E
Total
N
0
SP-R 8×64 F 311.80 45.76%
SP-RBC
Min-Max
8×64 B 356.57 42.30%
SP-RBC
Median
8×64 B 419.86 34.16%
LBP 1×135 F 365.23 38.26%
RBC
8
8×64 B 580.68 25.39%
RBC
16
16×64 B 564.54 23.54%
Table 1 shows that the proposed method SP-RBC
does loose some information in exchange for some
increase in speed compared to SP-R (single projection
Retrieving Similar X-ray Images from Big Image Data using Radon Barcodes with Single Projections
563

Radon) which simply uses the floating-point projec-
tions values without thresholding. But it can be
observed that the MinMax method is significantly
better than Median thresholding.
We also compared our results with the most suc-
cessful methods, which are applied on IRMA dataset.
Because all of them use some notion of learning, they
may only use labeled IRMA test images (94 images
in IRMA 2009 are not labelled; some works just ig-
nore them). Hence, their error might increase around
5-6%.
In the next part of this section, we have analyzed
the parameter tuning and the details of our observa-
tions. Firstly, we discuss the size of the Selection
Pool, which can affect the error rate significantly.
Figure 9 reflects the relationship between the error
rate and the number of top images per projection. As
it can be seen, the error rate has dropped substantially
between the first hit and the top five, while the de-
creasing rate tends to remain constant after number of
top choices reaches 10 for SP-R. The error rate seems
to continue to improve for SP-RBC beyond consider-
ing more than 10 top choices. The best answer is
reached by looking at top 14 images per projection. It
means we search among 112 images in the Selection
Pool (we use 8 separate projections).
Figure 9: Total IRMA error based on number of top similar
images for each projection.
Table 2: Best learning-based methods. All result marked
with * are reported in (Tommasi et al. 2009).
Learning method E
Total
(Camlica et al. 2015)
146.55
TAUbiomed
*
169.50
diap
*
178.93
VPA
*
242.46
FEITIJS
*
261.16
SP-R
311.8
MedGIFT
*
317.53
VPA
*
320.61
SP-RBC
356.57
IRMA
*
359.29
MedGIFT
*
420.91
Table 2
shows the results compared to learning-
based methods. The best results reported by Camlica
et al. is based on extensive saliency detection. The
second-best solution uses a dictionary approach ac-
companied by PCA application.
The results for each separate Radon projection, as
well as their binary version are provided in Table 3.
Each projection has a relatively high error.
Table 3: Results for SP-R and SP-RBC.
Learner SP-R
E
Total
SP-RBC
E
Total
0-degree Radon
567.67 644.52
22.5degree Radon
598.7 687.71
45-degree Radon
613.46 700.15
67.5-degree Radon
642.25 719.29
90-degree Radon
561.38 649.21
112.5-degree Radon
629.77 710.43
135-degree Radon
618.41 721.03
157.5-degree Radon
575.43 676.69
In Table 4, we share the results of concatenated Ra-
don projections as one vector. We also provide infor-
mation about the impact of preprocessing steps.
Normalization in general improves the results about
5%. Table 4 shows that there is some improvement
(approx. 10%) in white part removal and zero-padded
square resizing.
Table 4: Eight concatenated Radon projections.
Type of preprocessing E
Total
Radon whiteout preprocessing 439.93
Normal Radon 420.82
Normal Radon + resize method 389.10
Normal Radon + resize method +criclize 384.68
Normal Radon + removing white spots 385.75
All preprocessing steps 383.41
5 SUMMARY
In this paper, we proposed the idea of using single
Radon projections for medical image retrieval in large
archives. This can be considered an improvement of
previous works (Tizhoosh 2015) which introduced
the idea of Radon Barcodes by binarization of a se-
lected number of Radon projections.
In our method, a Selection Pool can be assembled
when multiple single Radon projections are applied
separately to retrieve many images from the database.
The single projection Radon Barcodes (SP-RBCs)
may lose some information due to the thresholding
process but they are compact and fast for retrieval in
big image data. Subsequently, more time consuming
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
564

local search can be performed on the Selection Pool
to retrieve the most similar cases. In this paper, we
employed LBP and Shifted Radon but many other al-
ternatives could be investigated.
In our future work, we will focus on improving
the exploitative search in the Selection Pool and
learning method to find the best projection to search
data set by just one projection.
REFERENCES
Aundal, P. & Aasted, J., 1996. The Radon Transform
Theory and Implementation Peter Toft Department of
Mathematical Modelling Section for Digital Signal
Processing Technical University of Denmark.
Avni, U. et al., 2011. X-ray categorization and retrieval on
the organ and pathology level, using patch-based visual
words. IEEE Transactions on Medical Imaging, 30(3),
pp.733–746.
Babenko, A. et al., 2014. Neural Codes for Image Retrieval.
CoRR, abs/1404.1. Available at: http://arxiv.org/
abs/1404.1777.
Bankar, R.T. et al., 2014. CBIR Representation In Terms of
Rotation Invariant Texture using LBP Variance.
International Journal of Emerging Science and
Engineering (IJESE), (5), pp.75–77.
Bay, H. et al., 2008. Speeded-Up Robust Features (SURF).
Computer Vision and Image Understanding, 110(3),
pp.346–359.
Beis, J.S. & Lowe, D.G., 1997. Shape indexing using
approximate nearest-neighbour search in high-
dimensional spaces. In Proceedings of IEEE Computer
Society Conference on Computer Vision and Pattern
Recognition. pp. 1000–1006.
Calonder, M. et al., 2010. BRIEF: Binary robust
independent elementary features. In Lecture Notes in
Computer Science (including subseries Lecture Notes
in Artificial Intelligence and Lecture Notes in
Bioinformatics). pp. 778–792.
Camlica, Z., Tizhoosh, H.R. & Khalvati, F., 2015. Medical
image classification via SVM using LBP features from
saliency-based folded data. Proceedings - 2015 IEEE
14th International Conference on Machine Learning
and Applications, ICMLA 2015, pp.128–132.
Do, T. et al., 2010. Deluding Image Recognition in SIFT-
based CBIR Systems. Analysis, (1), pp.7–12.
Gevers, T. & Stokman, H., 2004. Robust Histogram
Construction from Color Invariants for Object
Recognition. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 26(1), pp.113–118.
Gu, Y.J. & Sacchi, M., 2009. Radon transform methods and
their applications in mapping mantle reflectivity
structure. Surveys in Geophysics, 30(4–5), pp.327–354.
Huang, W., Gao, Y. & Chan, K.L., 2010. A review of
region-based image retrieval. Journal of Signal
Processing Systems, 59(2), pp.143–161.
Krig, S., 2012. Computer vision metrics,
Kumar, A. et al., 2013. Content-based medical image
retrieval: a survey of applications to multidimensional
and multimodality data. Journal of digital imaging,
26(6), pp.1025–1039.
Ledwich, L. & Williams, S., 2004. Reduced SIFT features
for image retrieval and indoor localisation. Australian
conference on robotics and automation.
Lee, D.-J., Antani, S. & Long, L.R., 2003. Similarity
measurement using polygon curve representation and
fourier descriptors for shape-based vertebral image
retrieval. In Medical Imaging 2003. pp. 1283–1291.
Liu, X., Tizhoosh, H.R. & Kofman, J., 2016. Generating
Binary Tags for Fast Medical Image Retrieval Based on
Convolutional Nets and Radon Transform.
International Joint Conference on Neural Networks,
(Ijcnn). Available at: http://arxiv.org/abs/1604.04676.
Lowe, D.G., 2004. Distinctive image features from scale-
invariant keypoints. International Journal of Computer
Vision, 60(2), pp.91–110.
Nanni, L., Lumini, A. & Brahnam, S., 2010. Local binary
patterns variants as texture descriptors for medical
image analysis. Artificial Intelligence in Medicine,
49(2), pp.117–125.
Ojala, T., Pietikäinen, M. & Mäenpää, T., 2002.
Multiresolution gray-scale and rotation invariant
texture classification with local binary patterns. IEEE
Transactions on Pattern Analysis and Machine
Intelligence, 24(7), pp.971–987.
Radon, J., 1917. On determination of functions by their
integral values along certain multiplicities. Ber. der
Sachische Akademie der Wissenschaften
Leipzig,(Germany), 69, pp.262–277.
Rodríguez, F., Lecumberry, F. & Fernández, A., 2015.
Pattern Recognition Applications and Methods. Lecture
Notes in Computer Science (including subseries
Lecture Notes in Artificial Intelligence and Lecture
Notes in Bioinformatics), 9443, pp.196–205.
Saha, S.K., Das, A.K. & Chanda, B., 2004. CBIR using
perception based texture and colour measures. In
Proceedings of the 17Th International Conference on
Pattern Recognition, Vol 2. pp. 985–988.
Shim, S., Choi, T. & Member, S., 2002. Edge color
histogram for image retrieval. Science, pp.957–960.
Subrahmanyam, M., Maheshwari, R.P. &
Balasubramanian, R., 2012. Local maximum edge
binary patterns: A new descriptor for image retrieval
and object tracking. Signal Processing, 92(6), pp.1467–
1479. Available at: http://dx.doi.org/10.1016/
j.sigpro.2011.12.005.
Tizhoosh, H.R., 2015. Barcode annotations for medical
image retrieval: A preliminary investigation. In
Proceedings - International Conference on Image
Processing, ICIP. pp. 818–822.
Tizhoosh, H.R. et al., 2016. MinMax Radon Barcodes for
Medical Image Retrieval. In To appear in proceedings
of the 12th International Symposium on Visual
Computing. Available at: http://arxiv.org/abs/
1610.00318.
Retrieving Similar X-ray Images from Big Image Data using Radon Barcodes with Single Projections
565

Tommasi, T. et al., 2009. Overview of the CLEF 2009
medical image annotation track. In CEUR Workshop
Proceedings.
Tsai, H.-H. & Chiang, J.K., 2010. A Rotation Invariant
Image Descriptor based on Radon Transform Yudong.
In International Conference on New Trends in
Information Science and Service Science (NISS). pp.
671–676.
Velmurugan, K. & Baboo, S.S., 2011. Content-Based
Image Retrieval using SURF and Colour Moments.
Global Journal of Computer Science and Technology,
11(10), pp.1–4.
Wan, J. et al., 2014. Deep Learning for Content-Based
Image Retrieval. Proceedings of the ACM International
Conference on Multimedia - MM ’14, pp.157–166.
Wang, J.Z., 2001. Integrated Region-Based Image
Retrieval, Norwell, MA, USA: Kluwer Academic
Publishers.
Yan, K. et al., 2016. CNN vs. SIFT for Image Retrieval:
Alternative or Complementary? In Proceedings of the
2016 ACM on Multimedia Conference. MM ’16. New
York, NY, USA: ACM, pp. 407–411. Available at:
http://doi.acm.org/10.1145/2964284.2967252.
Zhu, S. & Tizhoosh, H.R., 2016. Radon Features and
Barcodes for Medical Image Retrieval via SVM.
Available at: http://arxiv.org/abs/1604.04675.
Zhu, S.Y. & Schaefer, G., 2004. Thermal Medical Image
Retrieval by Moment Invariants. In J. M. Barreiro et al.,
eds. Biological and Medical Data Analysis: 5th
International Symposium, ISBMDA 2004, Barcelona,
Spain, November 18-19, 2004. Proceedings. Berlin,
Heidelberg: Springer Berlin Heidelberg, pp. 182–187.
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
566
