
GenesLove.Me: A Model-based Web-application for Direct-to-consumer
Genetic Tests
Jos
´
e Fabi
´
an Reyes Rom
´
an
1,2
, Carlos I
˜
niguez-Jarr
´
ın
1,3
and
´
Oscar Pastor L
´
opez
1
1
Research Center on Software Production Methods (PROS), DSIC,
Universitat Polit
`
ecnica de Val
`
encia, Camino Vera s/n. 46022, Valencia, Spain
2
Department of Engineering Sciences, Universidad Central del Este (UCE),
Ave. Francisco Alberto Caama
˜
no De
˜
n
´
o, 21000, San Pedro de Macor
´
ıs, Dominican Republic
3
Departamento de Inform
´
atica y Ciencias de la Computaci
´
on,
Escuela Polit
´
ecnica Nacional, Ladr
´
on de Guevara E11-253, Quito, Ecuador
Keywords:
GLM, Direct-to-consumer Genetic Tests, Genetic Test, BPMN, Conceptual Models.
Abstract:
The objective of this work was to enhance personalized medicine through the development and implementation
of Genomic Information Systems (GeIS). For this, a web application called ”GenesLove.Me” (GLM) was
developed to provide direct-to-consumer genetic tests (DCGT). This paper focuses on the development of the
business processes (BPMN) and a conceptual model (CM) for GLM, designed to analyze and improve the
processes involved in this type of service and provide a model-based platform to manage genetic diagnoses
in a scalable, secure and reliable way. Software Engineering (SE) approaches applied to the genomic context
play a key role in the advancement of personalized and precision medicine.
1 INTRODUCTION
The current availability of direct-to-consumer genetic
tests (DCGT) has a great number of advantages for
the genomic domain, making it easier for end-users
to access early genetic-origin diseases diagnosis ser-
vices.
Romeo-Malanda (Romeo-Malanda, 2009) defines
“direct-to-consumer genetic analysis” as a term
which is used to describe analytic services offered to
detect polymorphism and health-related genetic vari-
ations. Although this type of analysis is available
through direct sales systems in pharmacies or other
health care bodies, the Internet has become the main
selling channel for direct-to-consumer genetic analy-
ses. The usual procedure is to take a biological sam-
ple at home and send it to an analysis laboratory. The
findings (results) of the analysis are communicated to
the client by telephone, mail or electronic mail, or
through secure access to an Internet portal (UNESCO,
2004).
The heterogeneity and dispersion of the data
sources represent a great challenge, known today as
”genomic chaos” (Le
´
on et al., 2016). As the man-
agement of genomic repositories offers many bene-
fits to the biomedical community, it is necessary to
carry out studies and analyses to support the imple-
mentation of mechanisms to improve aspects related
to data integrity, consistency and homogeneity (Reyes
Rom
´
an, 2014). In this way it will be possible to gen-
erate more reliable results for end-users by guarantee-
ing true Precision Medicine (PM).
The genomic data repositories (e.g., NCBI (NCBI
Resource Coordinators, 2013), OMIM (Hamosh
et al., 2005) and Ensembl (Cunningham et al., 2015)
contain a very extensive set of information capable of
being analyzed to extract concise data and generate
results with greater accuracy. The efficient use of ad-
vances in genomic research allows the patient to be
treated in a more direct way, which is reflected in re-
sults such as:”better health” and ”quality of life”.
In order to treat the data that will be used in
the proposed application, we implemented the SILE
(Search-Identification-Load-Exploitation) methodol-
ogy (Reyes Rom
´
an and Pastor L
´
opez, 2016) which
was conceived in the PROS’s Research Center in or-
der to improve selective loading processes of our Hu-
man Genome Database (HGDB).
Our aim in this work was twofold:
• To provide “GenesLove.Me” (GLM) as a web ap-
plication designed to generate direct-to-consumer
genetic tests (DCGT) supported by Business
Román, J., Iñiguez-Jarrín, C. and López, Ó.
GenesLove.Me: A Model-based Web-application for Direct-to-consumer Genetic Tests.
DOI: 10.5220/0006340201330143
In Proceedings of the 12th International Conference on Evaluation of Novel Approaches to Software Engineering (ENASE 2017), pages 133-143
ISBN: 978-989-758-250-9
Copyright © 2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
133

Process modeling (BPMN) (Object Management
Group, 1997) and conceptual modeling (MC)
(Oliv
´
e, 2007) techniques to study and analyze the
essential elements of the processes involved in the
genomic domain and improve the development of
Genomic Information Systems (GeIS).
• To apply Software Engineering (SE) knowledge
to improve Precision Medicine (PM). Ensuring
PM for end-users involves implementing tools for
studying, treating, exploring and exploding the
genomic data generated and stored for several
years to generate genetic diagnosis. The current
DCGT demand was the reason for studying and
analyzing the processes and stakeholders involved
in this domain. The genetic tests are the result of
the progress in the genomic environment with the
aim of exploiting PM.
The genomic domain requires methodologies and
modeling techniques capable of integrating innova-
tive ideas into data management, process improve-
ment and the inclusion of quality standards.
The paper is divided into the following sections:
Section 2 reviews the present state of the art. Section
3 describes BPMN applied to genetic testing. Section
4 contains the representation of the domain by a con-
ceptual model. Section 5 describes a case study with
the GLM application, and Section 6 summarizes the
lessons learned and outlines future work.
2 RELATED WORK
Bioinformatics now play an important role in con-
tributing advances to the medical and technological
sector. Genetic testing reveals existing knowledge
about ”genes” and ”variations” in the genomic do-
main, which is used to diagnose diseases of genetic
origin in order to prevent or treat them. This brings
precision medicine closer to end-users (i.e., clients /
patients).
The study of genomics (i.e., data repositories, ge-
netic variations, diseases, treatments, etc.) is con-
stantly growing and is increasingly seeking to ensure
the application of precision medicine. DNA sequenc-
ing began in 1977 and since then software tools have
been developed for its analysis. Thanks to NGS Tech-
nologies (Mardis, 2010), it is now possible to ma-
nipulate files (e.g., VCF) in order to generate genetic
diagnoses in a more agile and efficient way (Reyes
Rom
´
an, 2014).
PM is a way of treating patients that allows doc-
tors to identify an illness and select the treatment most
likely to help the patient according to a genetic con-
cept of the disease in question (this is why it has also
been called Personalized Medicine) (Aguilar Carta-
gena, 2015).
Figure 1: From Genomics to Precision Medicine.
As shown in Figure 1, this approach is based on
a detailed knowledge of the genomic domain and on
the information derived from the large amount of data
generated in recent years. This information iscon-
stantly growing as research provides more and more
findings every day. Fowler et. al. (Jim
´
enez, 2014)
describe the advances in genomics that will provide
practical information on diseases, those who are most
likely to suffer them, and how most successful treat-
ments can be applied, thus reducing the uncertainty
and stress of patients and their families (Jim
´
enez,
2014), (Instituto Nacional del C
´
ancer, 2015).
The advantages of genetic tests are innumerable
and allow us to identify mutations or alterations in
the genes and are of great use and interest in clinical /
personalized medicine and the early diagnosis of dis-
eases (Fisiotraining C
´
ordoba, 2016), (Grupo RETO
Hermosillo, 2016). Bioinformatics supports the tasks
of: management of biological databases; metabolic
processes and population genetics; artificial intelli-
gence and others. The exploitation of genomics is
of interest to various branches of research, such as:
a) sequence analysis, b) genome annotation, c) evo-
lutionary computational biology, d) gene expression
analysis, e) protein expression analysis, f) cancer mu-
tation analysis, g) comparative genomics, h) model-
ing of biological systems, i) protein-protein coupling,
etc. (Franco et al., 2008)
By 2008 there were around 1,200 genetic tests
available around the world (Fisiotraining C
´
ordoba,
2016), (Grupo RETO Hermosillo, 2016), but they had
some limitations (e.g., data management, genome se-
quencing, etc.) and their cost was quite high. For this
reason, companies were interested in reducing costs
and providing services to end-users in the comfort of
their own homes. Technological advances played a
fundamental role in the genomic environment, since
the introduction of the NGS for sequencing samples
made it possible to obtain sequences more quickly
and cheaply (Metzker, 2010), (Voelkerding et al.,
2009).
23andMe (23andMe, 2016a), in this sector. This
American company offers a wide range of services.
The type of information obtained from genetic sam-
ples is oriented to: genetic history (ancestors) and
ENASE 2017 - 12th International Conference on Evaluation of Novel Approaches to Software Engineering
134

personal health (risk of diseases), and is presented
mostly in probabilistic terms (23andMe, 2016b). In
the same way, in Spain companies of this type have
emerged (e.g., Genotest
1
or IMEGEN
2
), all with the
aim of providing genetic tests to end-users, simply
and in the form of providing a diagnosis that allows
end-users to take reactive or corrective actions (e.g.,
prevention and treatment) to improve their quality of
life.
Genetic tests contain a large amount of sensitive
information, so that before offering these services it is
necessary to evaluate all the elements involved, such
as: ethical, moral, legal, etc. It is important to be
aware that the studies carried out must always become
prevention instruments oriented to helping end-users.
Companies within this business context treat sensitive
information, so it is necessary to implement security
(high quality) and data protection mechanisms, in ad-
dition to including all relevant legal measures. Span-
ish legislation requires these services to be provided
under the following laws:
1. Organic Law 15/1999, of December 13, on the
Protection of Personal Data (Consolidated text
dated March 5, 2011) (BOE, 1999).
2. Periods of Conservation of Personal Data in
Biomedical Research (BOE, 2011).
3. General Health Law 86 (BOE, 2015).
For the implementation of DCGTs it is necessary
to consider the aforementioned elements. In future
years we will continue to improve our understanding
of the human genome.
3 BPMN:
DIRECT-TO-CONSUMER (DTC)
GENETIC TEST
The GemBiosoft company is a spin-off of the Univer-
sitat Polit
`
ecnica de Val
`
encia (UPV), founded in 2010.
The main objective of this company is to de-
fine the Conceptual Model of the Human Genome
(CMHG) (Reyes Rom
´
an et al., 2016), (Reyes Rom
´
an
et al., 2016) to obtain a precise schema to manage,
integrate and consolidate the large amount of ge-
nomic data in continuous growth within the genomic
domain. To achieve this objective, Gembiosoft has
extensive experience in Model Driven-development
(MDD) (Pastor et al., 2008) and an interdisciplinary
team of people -engineers and Ph.D.s- trained to im-
plement solutions aimed at i+D companies applying
1
http://www.trkgenetics.com/genotest
2
https://www.imegen.es/
information technologies in the bioinformatics and
health fields. GemBiosoft’s collaborators include:
PROS Research Center (UPV), PrincipiaTech and
IMEGEN (Instituo de Medicina Gen
´
omica).
3.1 Case Description
GemBiosoft has a web application called
”GenesLove.Me” which offers direct genetic
testing to the consumer. The information provided
by the genetic tests is accessible online to all users
without prior registration (anonymous users). Figure
2 depicts the general use case of the interaction
between actors and the web application, i.e. the ap-
plication’s functionality. For example, non-registered
users of the web application are able to consult all
information related to the diagnosis of rare diseases
of genetic origin, their characteristics, treatment,
tutorials and videos of the way in which the process
is performed.
Access security in GenesLove.Me is controlled by
profiles. Users can access GLM under 3 profiles: (1)
clients (patients), (2) provider and (3) administrator.
An authenticated user with a certain access profile is
authorized to carry out the operations corresponding
to the access profile.
(1) Clients (patients): Users with this profile are
able to contract the services offered by selecting the
services (direct-to-customer genetic test) they are in-
terested in and then paying the fee. The user is able to
monitor the notifications and messages related to the
diagnoses, besides consulting the histories of all the
studies and treatments carried out and updating the
information associated with his profile.
(2) Supplier: Users with this profile are able to
generate notifications about the change of status in
the treatment of samples. After receiving the genetic
sample, the user activates the sample by entering its
code number. They can then track the sample until
the sequence file is generated. They can also update
their profiles and consult all the activated samples (in
progress and finalized).
(3) Administrator: A user with administration
privileges performs administration and maintenance
tasks of the web application, such as: a) publishing
online results (the administrator uploads the result-
ing diagnoses from the analysis performed on sam-
ples sequenced by the VarSearch
3
tool. The applica-
tion automatically notifies the user when his/her re-
sults have been published); b) publishing advertise-
ments; c) publishing new diagnostic services to diag-
3
VarSearch is a web application that seeks variation (or
variant) files in a genomic database based on a Conceptual
Model of the Human Genome (CMHG).
GenesLove.Me: A Model-based Web-application for Direct-to-consumer Genetic Tests
135
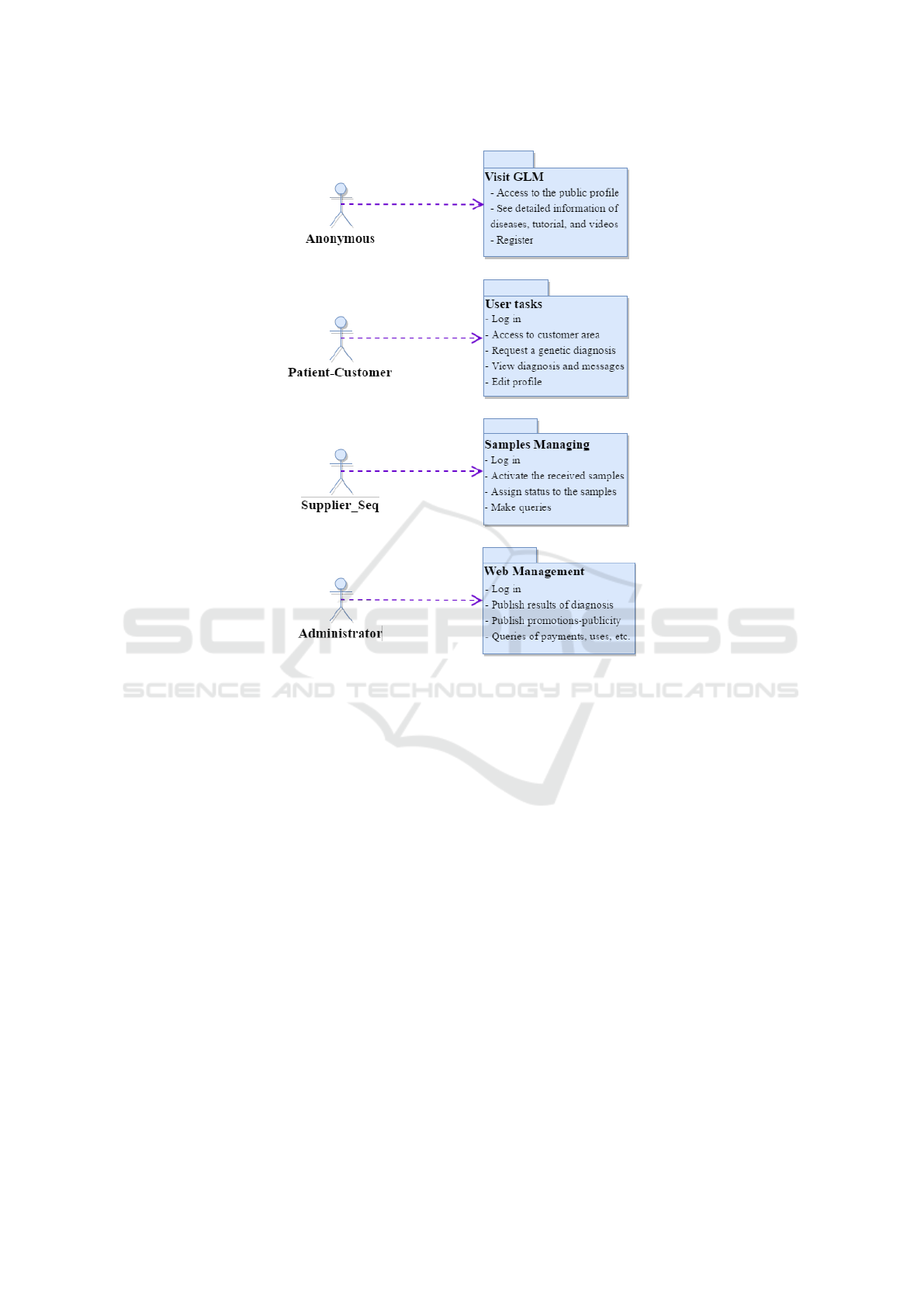
Figure 2: General view of the system.
nose new diseases; d) consulting payment reports and
the application usage report (custom time period).
3.2 Genomic Diagnosis Process
Genetic tests are currently offered with the aim of de-
tecting a person’s predisposition to contracting a dis-
ease of hereditary origin (U. S. National Library of
Medicine, 2017). The bioinformatics domain seeks
to provide the necessary mechanisms and means to
generate genetic diagnoses that allow the end-users
(patients) to obtain these results to facilitate a person-
alized prevention treatment.
Figure 3 shows a BPMN diagram describing the
genetic diagnosis process (from the end-user’s ser-
vice request until he/she receives his/her genetic test
report). In this process, the three actors/users spec-
ified in Section 3.1 are involved: 1) The client (pa-
tient) who requests the service to determine whether
or not he / she has a disease of genetic origin; 2) The
company, in this case GemBiosoft, which is in charge
of managing and performing the Genomic Diagnosis;
and 3) the Suppliers, who in this case prepare the file
containing the reference of the patient involved in the
genetic test.
The general process begins when the end-user (pa-
tient) enters the web application and requests the ge-
netic analysis (t1: task 1). The company (Gem-
Biosoft) processes this request and proceeds to send
the sample container to the client (t2). When the client
receives the container, he must activate it by regis-
tering its identifier in the web application (t3), then
place the sample in the container and send it back to
the company (t4). Upon receipt of the sample, the
company confirms that it meets the necessary require-
ments for the study and notifies the customer of its
receipt (t5). The next step is to determine the sup-
plier who will be responsible for sequencing the sam-
ples and send him the sample (t6). The selected sup-
plier receives the sample and notifies its reception to
the company (t7). Sequence preparation is initiated
through the sequencing technology used by the sup-
plier (t8). The supplier sends the resulting sequence
of the sample (file) to the company (t9). The company
confirms its reception to the supplier and proceeds to
analyze the sequenced sample as part of the genetic
diagnosis (t10). The definitive diagnosis report (t11)
is then generated. The company (in this case the ad-
ENASE 2017 - 12th International Conference on Evaluation of Novel Approaches to Software Engineering
136

Figure 3: Genetic diagnosis process.
ministrator/user) proceeds to publish the genetic diag-
nosis (result) in the web application and the end-users
are automatically notified of the results (t12). To end
the process, the end-user accesses the web application
to obtain the diagnosis and make any queries (results)
(t13).
The BPMN (Business Process Model and Nota-
tion) gives companies the ability to understand their
internal business procedures in graphical notation and
the ability to communicate these procedures in a stan-
dard way (Chinosi and Trombetta, 2012). Through
the model shown in Figure 3, it facilitates the un-
derstanding of commercial collaboration and transac-
tions between organizations. In this figure we can see
the interactions between end-user, company and sup-
pliers (Reyes Rom
´
an, 2014). The company is inter-
ested in providing a web application that allows end-
users to obtain a quality genetic test in a simple way
that aids the treatment and prevention of diseases of
genetic origin.
4 REPRESENTATION OF THE
DOMAIN: CONCEPTUAL
MODEL
It is widely accepted that applying conceptual mod-
els facilitates the understanding of complex domains
(like genetics) (Reyes R et al., 2016). In our case
we used this approach to define a model represent-
ing the characteristics and the processes of direct-to-
consumer genetic tests (DCGT).
One of the leading benefits of CM is that it accu-
rately represents the relevant concepts of the analyzed
domain (Reyes Rom
´
an et al., 2016). After perform-
ing an initial analysis of the problem domain, the next
step is to design a domain representation in the form
of a CM. Our CM (Figure 4) evolved with the new dis-
coveries made in the field of genomics (Reyes Rom
´
an
et al., 2016) in order to improve data processing to en-
sure effective precision medicine (PM). We can thus
see how CM gives positive support to the knowledge
in which precision medicine plays a key role (Reyes
Rom
´
an, 2014). It is important to highlight that the ad-
vantage of CM for representing this domain is that it
eases the integration of new knowledge into the model
(Reyes Rom
´
an et al., 2016).
After an analysis of the requirements required for
this work, important decisions were taken to arrive at
an adequate representation of the basic and essential
concepts in the understanding of the domain under
study. Figure 4 presents the conceptual model pro-
posed, which can be classified into three main parts:
a) Stakeholders, b) Genetic diagnostics and c) Sales
management.
The first part of our conceptual model represents
all the participants involved in the web application.
To represent the ”Users” class of the inherit type we
have the ”Administrator”, ”Supplier Seq”, ”Patient
Client” and ”Anonymous” classes respectively
The DCGTs are initialized when the end-user
(”patient customer”) accesses the application. If the
interaction does not have user credentials, it remains
an ”anonymous” user, otherwise, it becomes a client.
The ”client customer” contracts one or more services,
which cover one or more diseases, represented in the
model by the ”Services” and ”Diseases” classes, re-
spectively.
Diseases are studied using the SILE methodology
(Reyes Rom
´
an and Pastor L
´
opez, 2016), and when
complete, the study is offered as a service. The end-
user adds the services of interest to his shopping cart
(represented by the ”Shopping Cart” class) and then
GenesLove.Me: A Model-based Web-application for Direct-to-consumer Genetic Tests
137
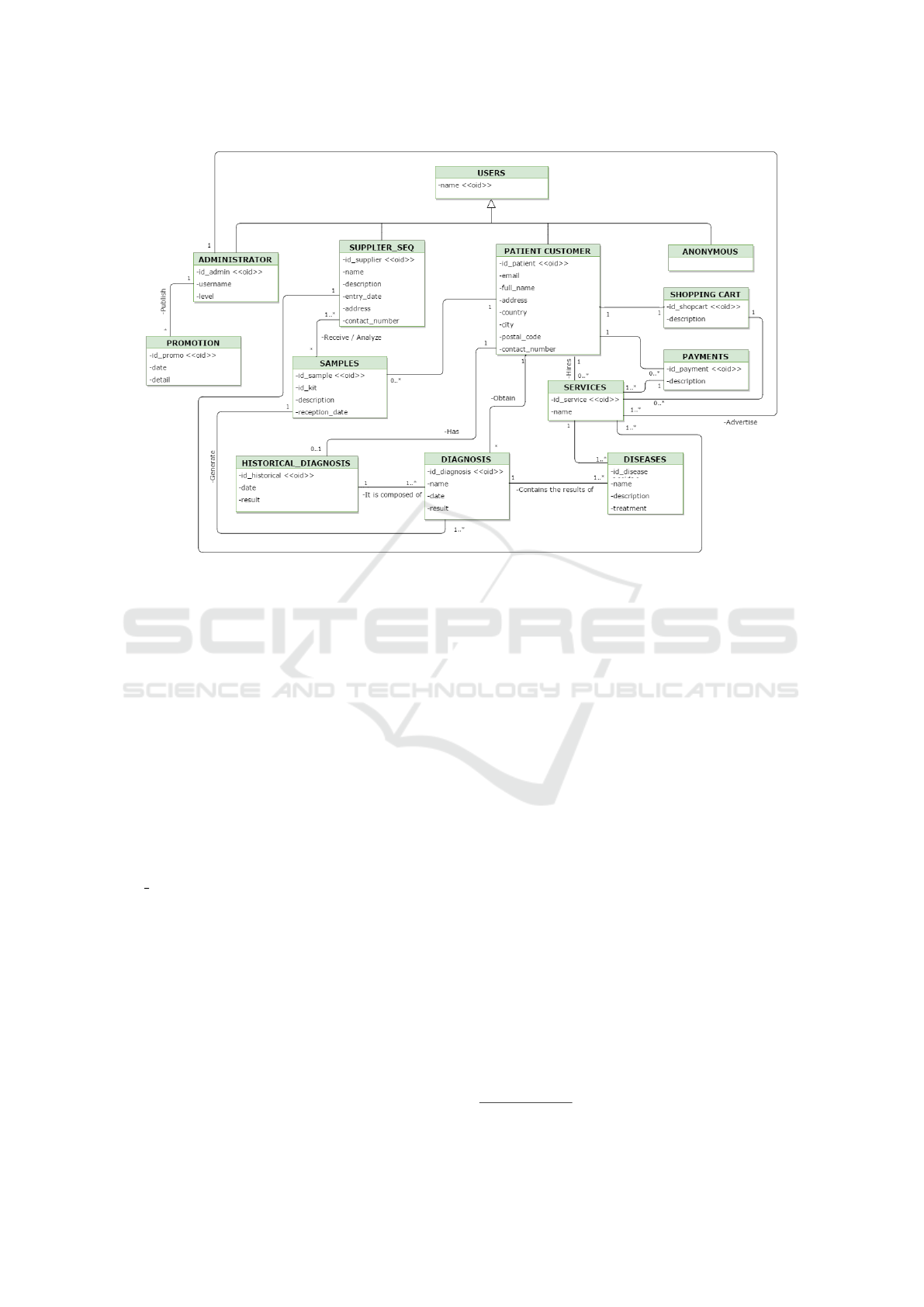
Figure 4: GLM conceptual model.
performs the payment process. For this, all the billing
information, shipping address and acceptance of the
purchase contract (which explains the rights and du-
ties of the end-user and the company) is entered in the
CM through the ”Payments” class.
The management of genetic diagnoses is repre-
sented in the conceptual model through the ”Diagnos-
tic” and ”Historical diagnosis” classes. The ”Diag-
nosis” class shows the results obtained after the anal-
ysis of the sequence and a list of the diseases con-
tracted. The user has the ability to consult his history
associated with all the diagnoses requested by the ap-
plication.
The diagnosis of genetic data begins when the
company receives the file containing the sample
sequenced by the supplier (represented by ”Sup-
plier Seq” class). The application administrator is re-
sponsible for publishing the diagnosis results on the
web and automatically notifies the end-user of the
availability of his report. Other activities of the ad-
ministration consist of the publication of advertising
related to the implementation of new services and
control of sales of the application.
Through our CM we incorporate genetic data cur-
rently used in the PM, achieving a conceptual repre-
sentation that meets the needs of the bioinformatics
domain. As we mentioned above, this model aims to
improve the conceptual definition of the treatment re-
lated to genomic diagnosis, and thus leave a concep-
tual framework for further improvements.
5 CASE STUDY: GenesLove.Me
SERVICE FOR END-USERS
In this section, we describe the design and implemen-
tation of GenesLove.Me
4
(GLM), an online web ap-
plication based on the business process described in
Section 3 of this work. The application becomes the
point of interaction between three actors: the clients
of genetic tests, the company that performs the genetic
diagnosis and the suppliers who sequence the genetic
samples. The main objective is to operate the DCGT
service through an accessible online and easy-to-use
platform for the actors involved in the process.
The platform offers a wide range of genetic tests
on its main page (Figure 5), from which users are
able to contract one or several genetic testing services
(e.g., lactose intolerance, androgenetic alopecia, al-
cohol sensitivity, etc.) according to their needs. The
company in charge of genetic analysis and diagno-
sis manages all the service orders received and mon-
itors the change of order status throughout the ser-
vice delivery process. For instance, the status of the
order changes to ”KIT SENT TO SUPPLIER” when
the company (in charge of the analysis and diagno-
4
http://www.geneslove.me
ENASE 2017 - 12th International Conference on Evaluation of Novel Approaches to Software Engineering
138
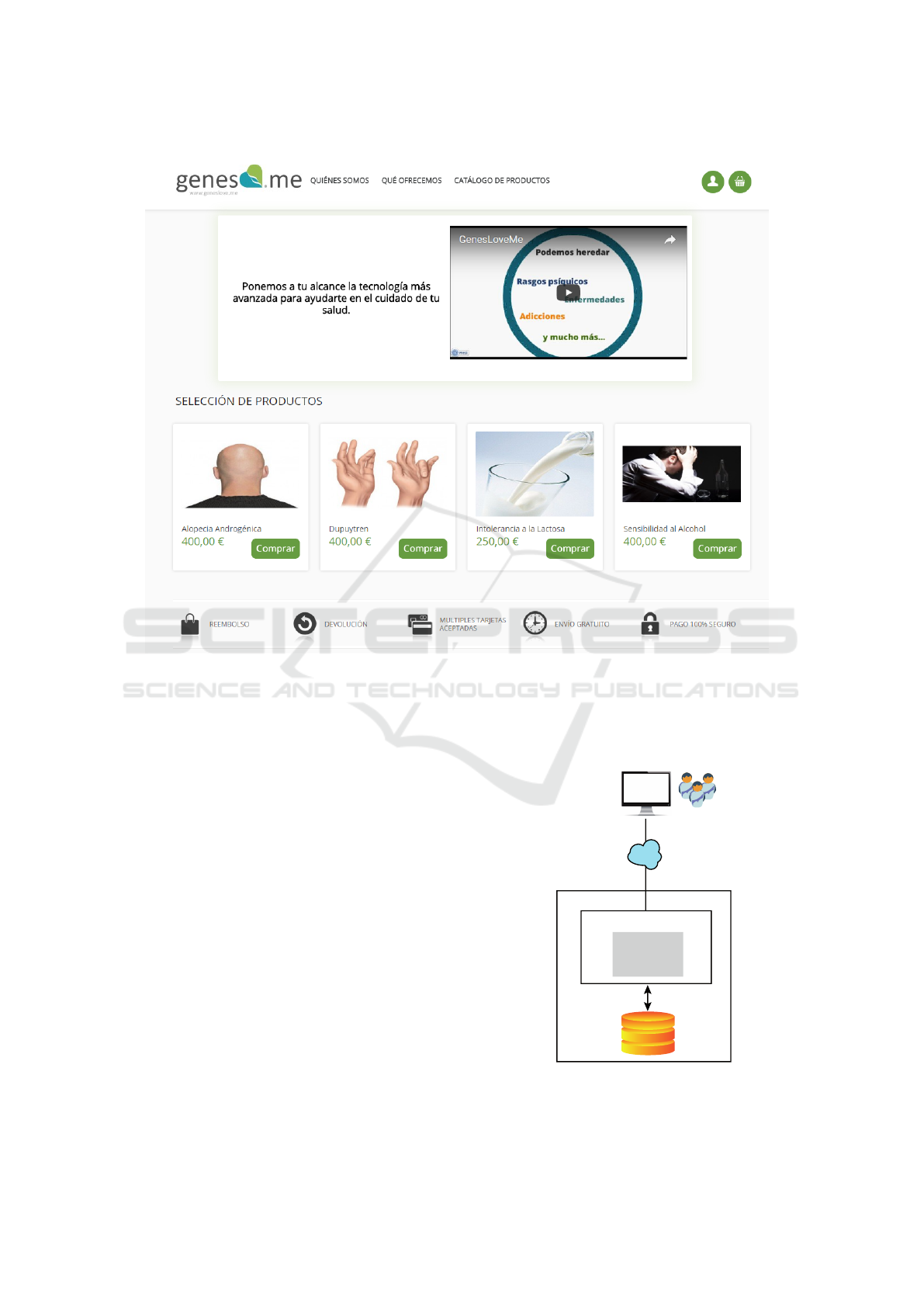
Figure 5: GenesLove.Me’s home web page showing the available direct-to-customer genetic tests.
sis) sends the genetic sample to the suppliers respon-
sible for sequencing the genetic samples and once the
genetic samples have been sequenced and received
from the supplier, the order is assigned ”ANALYSING
SAMPLE” status. Each status change is notified to
the end-user via email and can be further visualized
on the web user interface.
GLM is a web application implemented under a
client-server architecture as is shown in Figure 6. The
client side, the user’s browser (e.g., Firefox (Mozilla,
2017), Chrome (Google, 2017), Internet Explorer
(Microsoft, 2017), etc.) serves as the interaction point
between the user and application. The end-user in-
teracts with an easy-to-use graphical web interface to
request genetic testing services available in the ap-
plication. The server side is hosted on the Internet
and contains: a) the Apache 2.2 web server (The
Apache Software Foundation, 2017) with the applica-
tion logic implemented with PHP (W3schools, 2017)
programming language and b) the data stored in the
MySQL5.5 (Oracle Corporation, 2017) database en-
gine.
The GLM design allows customers to access the
online product range (i.e., clinical tests) from any-
where and at any time. In the same way, the adminis-
trator users in charge of site management and internal
business tasks can access the GLM through an Inter-
net connection.
Database
(MySQL)
Web Server (Apache)
Server
http
http
internet
browser’s
user
Geneslove.me
Application
(PHP)
Figure 6: GenesLove.Me architecture.
GenesLove.Me: A Model-based Web-application for Direct-to-consumer Genetic Tests
139
GLM is implemented under Prestashop
5
, an open
source eCommerce CMS platform, which facilitates
the implementation of customized solutions oriented
to the marketing of products framed in a simple
purchase-sale process. The platform incorporates
both modules and website templates in order to pro-
vide, respectively, specific functionality and graphic
style customized according to the business’s needs.
There is a great variety of free and commercial mod-
ules
6
and website templates created by web develop-
ers available on the Internet to be down- loaded and
used.
The default Prestashop download package avail-
able on the official site includes modules of basic
functionality (e.g., customers, products, orders, etc.)
which are sufficient to create and manage a basic
e-commerce platform. However, Prestashop allows
complex functionality modules to be incorporated to
tailor sites to particular needs. For instance, it is pos-
sible to install a module that deals with the complexity
associated with the credit card payment process.
GLM takes full advantage of the range of func-
tionalities offered by a CMS e-commerce platform
such as Prestashop. Indeed, GLM offers a variety
of on- line genetic tests, keeps the customer database
and orders the genetic test sales process thanks to the
available payment methods, such as bank transfer,
credit card payments through PayPal (Jackson, 2012)
or payments by electronic check.
The GLM client module represents the database of
registered clients interested in the genetic tests. The
ordering module lists the requests made by the cus-
tomers, which consists of a reference code useful for
order tracking, the name of the customer, the total
value, the payment method and the current status of
the order within the business process. The applica-
tion’s administrator manages the orders and updates
the state of the orders. Updating the order state means
a status change that automatically notifies the end user
via e-mail. GLM facilitates the management of orders
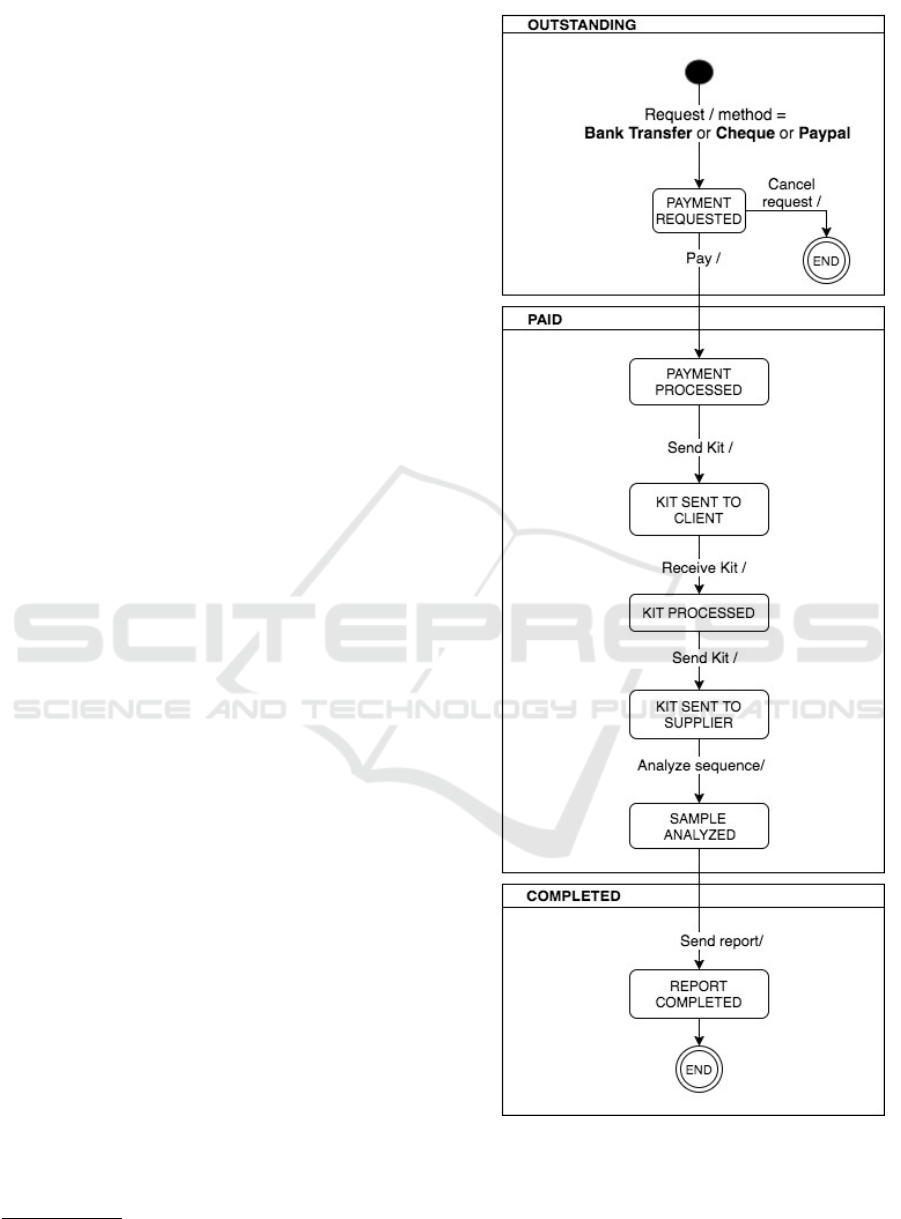
through the sequence of 13 states shown in Figure 7.
Every state order change generates an email notifica-
tion to keep customers informed of the processing of
their orders. It is important to note that GLM includes
interaction with VarSearch (see task 10 and 11 of Fig-
ure 3), an application developed by PROS Research
Center to automatically identify the relevant informa-
tion contained in the genomic databases and directly
related to the genetic variations of the sequenced sam-
ple. VarSearch relies heavily on a Conceptual Model
of the Human Genome (CMHG), which makes inte-
gration of external genomic databases feasible. How-
5
https://www.prestashop.com
6
(http://addons.prestashop.com/en/2modules)
Figure 7: GLM’s states sequence to manage orders.
ever, due to the large amount of information available,
the data loaded in VarSearch are the result of a selec-
tive loading process (Reyes Rom
´
an and Pastor L
´
opez,
2016) where the selected data correspond to the rele-
vant information on the disease to be analyzed.
ENASE 2017 - 12th International Conference on Evaluation of Novel Approaches to Software Engineering
140

The GLM user administrator has access to the ad-
ministration panel (back-office) to manage the entire
business process as well as the security and function-
ality of the site. The management screen has relevant
information and direct access to the configuration and
maintenance zones. For example, a strategically vis-
ible part of the main management screen displays in-
formation on the sales indicators and useful informa-
tion for the management of the business accounting.
By accessing the access security module, it is possi-
ble to configure profiles and access permissions for
all users. It is also possible to manage the modules
of users, products, orders, as well as the rules for dis-
counts on purchases and customer service (e.g., fo-
rums, e-mail notifications). Thanks to the web plat-
form on which the solution is implemented, site man-
agement can be carried out from anywhere and at any
time.
Each module installed adds a functionality that
supports to a greater or lesser degree the main rea-
son of the platform: buying and selling products.
The buying-and-selling process establishes the buyer-
seller relationship through a simple process consisting
of three states: Pending, Paid and Sent. ”Pending” in-
dicates that the user has requested a product, stored
it in the shopping basket but still has to generate the
payment. The ”Paid” status indicates that the user
has registered the payment for any of the options con-
figured and available (e.g., bank transfer) and ”Sent”
indicates that the seller has shipped the product and is
in the process of delivery to the customer.
5.1 Process Validation
In order to validate the process proposed in Section 3,
test cases were performed with the implemented solu-
tion. The validation scenario consisted of a group of
five (5) users, who made requests for genetic testing
for ”lactose intolerance”. To begin the process, each
user involved in the case study authorized the proce-
dure through an ”informed consent” (Reyes Rom
´
an,
2014), (de Galicia, 2001) which becomes a legal sup-
port that establishes the rights and obligations of the
service offered and its expected scope.
6 LESSONS LEARNED AND
FUTURE WORK
This paper describes a study and analysis of the
implementation of a web application to facilitate
DCGT, which informs end-users on their predispo-
sition to suffer certain genetically based illnesses.
Through the development of our web application
”GenesLove.Me” we seek to provide end-users with
a genomic diagnosis in a secure and reliable way.
The use of BPMN and Conceptual Modeling-
based approaches for this type of service aids the
understanding of the participants in the processes in
the genomic domain and improves the processes in-
volved. Through the proposed models we were able
to evaluate different points:
1. Process evaluation: the processes involved in
the direct-to-consumer genetic tests (DCGT) were
studied, and a business model was developed to
define the structures and relationships with the
stakeholders.
2. Improvement in data management: a conceptual
model (CM) was proposed for the definition of
the relevant concepts in the domain with the ob-
jective of guaranteeing reliable and personalized
medicine.
The validation of -GenesLove.Me- included the
study of four disorders of genetic origin: Alcohol
Sensitivity, Androgenic Alopecia, Lactose Intoler-
ance, and Dupuytren. The research process was car-
ried out using the SILE (Search-Identification-Load-
Exploitation) Methodology, and after obtaining the
variations/genes associated with the diagnoses, we
made the selective loading of the data to our HGDB
(Human Genome Database).
This web application was developed to provide
end-users with an early genetic diagnosis through an
attractive and easy-to-use distribution channel. Bioin-
formatics is a domain that is constantly evolving, and
with the application of conceptual models we can ex-
tend our genomic knowledge and conceptual repre-
sentation accurately and simply.
Future research work will focus on three main
goals:
• The study and treatment of new diseases of ge-
netic origin and continue expanding the list of ill-
nesses available in the web application.
• To extend our Conceptual Model of the Human
Genome (CMHG) by integrating new genomic in-
formation , to improve the generation of genetic
diagnoses.
• Implementation of data management mechanisms
to enhance the quality of personalized medicine.
ACKNOWLEDGEMENTS
The author thanks to the members of the PROS Cen-
ter’s Genome group for fruitful discussions. This
work was supported by the Ministry of Higher
GenesLove.Me: A Model-based Web-application for Direct-to-consumer Genetic Tests
141

Education, Science and Technology (MESCyT) of
Santo Domingo, Dominican Republic and the Sec-
retar
´
ıa Nacional de Educaci
´
on, Ciencia y Tecnolog
´
ıa
(SENESCYT) and the Escuela Polit
´
ecnica Nacional
de Ecuador. The project also had the support of
the Generalitat Valenciana through Project IDEO
(PROM- ETEOII/2014/039) and the Spanish Ministry
of Science and Innovation through Project DataME
(ref: TIN2016-80811-P).
The author thanks Alberto Garc
´
ıa S., Mercedes
R. Fern
´
andez Alcal
´
a, Vicente Mart
´
ınez Perell
´
o and
Ver
´
onica Burriel Coll for their collaboration with this
this project.
REFERENCES
23andMe (2016a). 23andMe. https://www.23andme.com/.
[Online; accessed 19-December-2016].
23andMe (2016b). How it works.
https://www.23andme.com/howitworks/. [Online;
accessed 19-December-2016].
Aguilar Cartagena, A. (2015). MEDICINA PERSON-
ALIZADA, MEDICINA DE PRECISI
´
ON, ¿CU
´
AN
LEJOS ESTAMOS DE LA PERFECCI
´
ON? Carci-
nos, 5:1–2.
BOE, A. e. B. O. d. E. (1999). Disposiciones Generales.
BOE, A. e. B. O. d. E. (2011). Ley 14/2007 de Investigaci
´
on
Biom
´
edica.
BOE, A. e. B. O. d. E. (2015). Ley 14/1986 General de
Sanidad.
Chinosi, M. and Trombetta, A. (2012). BPMN: An intro-
duction to the standard. Computer Standards and In-
terfaces, 34(1):124–134.
Cunningham, F., Amode, M. R., Barrell, D., Beal, K., Billis,
K., Brent, S., Carvalho-Silva, D., Clapham, P., Coates,
G., Fitzgerald, S., Gil, L., Gir
´
on, C. G., Gordon, L.,
Hourlier, T., Hunt, S. E., Janacek, S. H., Johnson,
N., Juettemann, T., K
¨
ah
¨
ari, A. K., Keenan, S., Mar-
tin, F. J., Maurel, T., McLaren, W., Murphy, D. N.,
Nag, R., Overduin, B., Parker, A., Patricio, M., Perry,
E., Pignatelli, M., Riat, H. S., Sheppard, D., Taylor,
K., Thormann, A., Vullo, A., Wilder, S. P., Zadissa,
A., Aken, B. L., Birney, E., Harrow, J., Kinsella, R.,
Muffato, M., Ruffier, M., Searle, S. M. J., Spudich, G.,
Trevanion, S. J., Yates, A., Zerbino, D. R., and Flicek,
P. (2015). Ensembl 2015. Nucleic Acids Research,
43(D1):D662–D669.
de Galicia, C. A. (2001). Ley 3/2001, reguladora del con-
sentimiento informado y de la historia cl
´
ınica de los
pacientes.
Fisiotraining C
´
ordoba (2016). Test Gen
´
eticos.
http://www.fisiotraining.com/cordoba/
analisisgeneticos
quees.htm. [Online; accessed
19-December-2016].
Franco, M. L., Cediel, J. F., and Pay
´
an, C. (2008). Breve
historia de la bioinform
´
atica. Colombia M
´
edica,
39:117–120.
Google (2017). Personalizar Chrome.
Grupo RETO Hermosillo, A. (2016). El c
´
ancer de mama.
http://gruporetohermosilloac.com/index.php. [Online;
accessed 19-December-2016].
Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A.,
and McKusick, V. A. (2005). Online Mendelian Inher-
itance in Man (OMIM), a knowledgebase of human
genes and genetic disorders. Nucleic Acids Research,
33(DATABASE ISS.):D514–D517.
Instituto Nacional del C
´
ancer (2015). Qu
´
e es la
medicina de precisi
´
on en el tratamiento del c
´
ancer.
https://www.cancer.gov/espanol/cancer/tratamiento/
tipos/medicina-de-precision. [Online; accessed 19-
December-2016].
Jackson, E. M. (2012). The PayPal Wars: Battles with eBay,
the Media, the Mafia, and the Rest of Planet Earth.
WND Books, 2nd edition.
Jim
´
enez, N. (2014). Una medicina nueva , m
´
as inteligente
y menos invasiva. http://www.lifescienceslab.com.
Le
´
on, A., Reyes, J., Burriel, V., and Valverde, F. (2016).
Data quality problems when integrating genomic in-
formation. In International Conference on Conceptual
Modeling, pages 173–182. Springer.
Mardis, E. R. (2010). The $1,000 genome, the $100,000
analysis? Genome Medicine, 2(11):84.
Metzker, M. L. (2010). Sequencing technologies - the next
generation. Nature reviews. Genetics, 11(1):31–46.
Microsoft (2017). Internet Explorer - Microsoft Download
Center.
Mozilla (2017). Firefox.
NCBI Resource Coordinators (2013). Database resources
of the National Center for Biotechnology Information.
Nucleic Acids Research, 41(D1):D8–D20.
Object Management Group (1997). Business Process
Model and Notation.
Oliv
´
e, A. (2007). Conceptual Modeling of Information Sys-
tems. Springer Berlin Heidelberg, Berlin, Heidelberg,
1 edition.
Oracle Corporation (2017). MySQL :: MySQL 5.7 Refer-
ence Manual.
Pastor, O., Espa
˜
na, S., Panach, J. I., and Aquino, N.
(2008). Model-driven development. Informatik-
Spektrum, 31(5):394–407.
Reyes R, J. F., Pastor, O., Valverde, F., and Rold
´
an, D.
(2016). Including haplotypes treatment in a Ge-
nomic Information Systems Management. In Pro-
ceedings of the XIX Ibero-American Conference on
Software Engineering (CIbSE 2016), pages 27–29,
Quito, Ecuador.
Reyes Rom
´
an, J. F. (2014). Integraci
´
on de haplotipos al
modelo conceptual del genoma humano utilizando la
metodolog
´
ıa sile. http://hdl.handle.net/10251/43776.
Reyes Rom
´
an, J. F., Pastor,
´
O., Casamayor, J. C., and
Valverde, F. (2016). Applying conceptual modeling
to better understand the human genome. In Comyn-
Wattiau, I., Tanaka, K., Song, I.-Y., Yamamoto, S.,
and Saeki, M., editors, Conceptual Modeling: 35th
International Conference, ER 2016, Gifu, Japan,
November 14-17, 2016, Proceedings, pages 404–412.
Springer International Publishing, Cham.
ENASE 2017 - 12th International Conference on Evaluation of Novel Approaches to Software Engineering
142

Reyes Rom
´
an, J. F., Pastor Lopez, O., Valverde, F., and
Rold
´
an, D. (2016). How to deal with Haplotype data :
An Extension to the Conceptual Schema of the Human
Genome. CLEI ELECTRONIC JOURNAL, 19(2).
Reyes Rom
´
an, J. F. and Pastor L
´
opez,
´
O. P. (2016). Use
of GeIS for Early Diagnosis of Alcohol Sensitivity.
In Proceedings of the 9th International Joint Confer-
ence on Biomedical Engineering Systems and Tech-
nologies, pages 284–289.
Romeo-Malanda, S. (2009). An
´
alisis gen
´
eticos direc-
tos al consumidor: cuestiones
´
eticas y jur
´
ıdicas.
http://www.institutoroche.es/legalactualidad/85/
analisis. [Online; accessed 19-December-2016].
The Apache Software Foundation (2017). Welcome! - The
Apache HTTP Server Project.
U. S. National Library of Medicine (2017). What is genetic
testing? - Genetics Home Reference.
UNESCO (2004). Declaraci
´
on Internacional
sobre los Datos Gen
´
eticos Humanos.
http://unesdoc.unesco.org/images/0013/001361/
136112so.pdf. [Online; accessed 19-December-
2016].
Voelkerding, K. V., Dames, S. A., and Durtschi, J. D.
(2009). Next-generation sequencing:from basic re-
search to diagnostics. Clinical Chemistry, 55(4):641–
658.
W3schools (2017). PHP 5 Tutorial.
GenesLove.Me: A Model-based Web-application for Direct-to-consumer Genetic Tests
143
