
Automatic Algorithm for Extracting an Ontology for a Specific
Domain Name
Saeed Sarencheh and Andrea Schiffauerova
Concordia Institute for Information Systems Engineering (CIISE), Concordia University, Montreal, QC H3G 1M8, Canada
Keywords: Ontology, Web Mining, Data Mining, Crawling, Machine Learning, TF-IDF, NLP, Concepts, Taxonomy,
Non-taxonomy.
Abstract: Scientists use knowledge representation techniques to transfer knowledge from humans to machines.
Ontology is the well-known representation technique of transferring knowledge to machines. Creating a new
knowledge ontology is a complex task, and most proposed algorithms for creating an ontology from
documents have problems in detecting complex concepts and their non-taxonomic relationships. Moreover,
previous algorithms are not able to analyze multidimensional context, where each concept might have
different meanings. This study proposes a framework that separates the process of finding important concepts
from linguistic analysis to extract more taxonomic and non-taxonomic relationships. In this framework, we
use a modified version of Term Frequency – Inverse Document Frequency (TF-IDF) weight to extract
important concepts from an online encyclopedia. Data mining algorithms like labeling semantic classes are
used to connect concepts, categorize attributes, and label them and an online encyclopedia is used to create a
structure for the knowledge of the given domain. Part Of Speech tagging (POS) and dependency tree of
sentences are used to extract concepts and their relationships (i.e. taxonomic and non-taxonomic). We then
evaluate this framework by comparing the results of our framework with an existing ontology in the area of
“biochemy”. The results show that the proposed method can detect more detailed information and has better
performance.
1 INTRODUCTION
Knowledge-based systems use representation
techniques to process and analyze new knowledge or
update an existing ontology. Ontology is the well-
known knowledge representation technique used to
maintain, manage, and infer knowledge. Various
domain knowledge is being updated at a faster rate
than ever before and as a result, the current ontology
maintenance process and even the creation of new
emerging ontologies is being done automatically
rather than manually.
For this reason, techniques such as Text-To-Onto
(Maedche and Volz, 2001), PARNT (Serra et al.,
2013), and LASER (Li et al., 2012) have been
developed to create ontologies. Text-To-Onto
(Maedche and Volz, 2001) is a semi-automatic
algorithm that uses hierarchy clustering to extract
concepts and their taxonomic relationships from plain
text. Some scholars (Li et al., 2012; Fader et al., 2011)
use machine learning algorithms to extract an
ontology from plain texts. In the proposed algorithms,
frequent item sets and term frequency are used to
extract concepts and taxonomic relationships. These
studies use a technique known as supervised
algorithm which requires an ontology expert to label
a part of the data as a training dataset. Meanwhile, the
term frequency technique returns single word nouns
as concepts.
Zavitsanos et al., (2010) and Villaverde et al.,
(2009) use regular expression to extract ontology
elements (i.e. concept and taxonomy). In these
algorithms, a list of predefined patterns is used to
identify nouns as well as relationships between
concepts in sentences and label nouns as concepts.
These pattern-based algorithms neglect relationships
between words in terms of semantics because they
focus on the noun phrases of sentences only.
To overcome these problems of current
approaches, this study proposes a new framework that
considers a separate procedures for extracting
important concepts and identifying relationships
between concepts. This study uses a modified version
of the term frequency technique to extract complex
concepts from an online encyclopedia. Next, the POS
Sarencheh S. and Schiffauerova A.
Automatic Algorithm for Extracting an Ontology for a Specific Domain Name.
DOI: 10.5220/0006500400490056
In Proceedings of the 9th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (KEOD 2017), pages 49-56
ISBN: 978-989-758-272-1
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

tagging technique and dependency tree of sentences
are used to analyze the dependency relationships
between sentence components to identify the
taxonomic and non-taxonomic relationships between
concepts. Finally, the measured TF-IDF weight of
concepts and status of concept in dependency tree are
then used to create the ontology structure.
This paper is organized as follows: in Section 2,
we conduct a literature review of previous studies and
asses the gaps in research. In Section 3, we illustrate
our framework and explain the algorithms in detail.
We describe our experiment and implementations and
evaluate our method in Section 4 and present the
results of the experiment in Section 5 to compare it
with previous research. Finally, in Section 6, we
conclude by revisiting our research goals and discuss
the results of the experiment.
2 LITERATURE REVIEW
Many studies have looked at extracting ontology from
plain texts. SnowBall (Agichtein and Gravano, 2000),
Textrunner (Aroyo et al., 2002), OntoGen (Fortuna et
al., 2006), OntoLearn (Navigli and Velardi, 2004),
OntoLT (Buitelaar et al., 2004), and Mo’k (Bisson et
al., 2000), for example, all attempted to generate
domain ontology from plain texts, with some using
machine learning to identify concepts (i.e. OntoGen,
SnowBall, and OntoLearn). However, none of these
studies have focused on extracting the non-taxonomic
relationships of concepts.
Some studies have used the frequent-based
technique to extract concepts from plain texts.
Maedche et al., (2001) introduced a new framework
– “Text-To-Onto” – a semi-automatic algorithm, to
extract ontology from plain texts. In Text-To-Onto,
concepts are extracted using the term frequency
algorithm. In this framework, hierarchy clustering is
used to link related concepts and a modified version
of association rules algorithm is used to extract the
non-taxonomic relationships between concepts. In
their study, the TF-IDF algorithm was used to identify
concepts, but TF-IDF detects a single noun as concept
only. In a similar work, Anantharangachar et al.,
(2013) proposed a new approach for extracting an
ontology from unstructured texts. In their study,
Anantharangachar et al., (2013) use a Natural
Language Processing (NLP) technique to extract
concepts, the taxonomic, and non-taxonomic
relationships from documents. In NLP, the document
theme is extracted applying the equation below:
_∩
∩
This algorithm is not be able to detect the correct
theme for descriptive documents because most
writers explain the main topics in the first paragraph
and describe sub-topics in other paragraphs.
Moreover, in their study, Anantharangachar et al.,
(2013) also consider the noun as concept, which
decreases algorithm performance. Some nouns
phrases do address a concept but the proposed
algorithm extracts various concepts from all noun
phrases.
Zavitsanos et al., (2010) introduced a new
framework for extracting an ontology from plain text.
In this framework, stopwords are removed from
documents and feature vectors are created for the
remaining words. Afterwards, the Latent Dirichlet
Allocation (LDA) algorithm is applied to extract
latent topics from documents, and mutual information
rate is used to create a hierarchy structure in iterative
processing. This framework is not properly efficient
since in this case, document and paragraph length is
shorts.
Drymonas et al., (2010) proposed a new multi-
layer framework to extract an ontology from
unstructured text. In this framework, noun phrases are
extracted in the first layer. Then, association rule and
probabilistic techniques are applied to extract the
taxonomic and non-taxonomic relationships. The
technique proposed in this study has an ability to
extract more complex phrases.
Serra et al., (2013) developed an algorithm to
extract non-taxonomic relationships. They categorize
information into three different groups: the sentence
rule (SR), the sentence rule with verb phrase (SR),
and the apostrophe rule (AR). An intelligent
algorithm is used to detect noun or verb phrases
around concepts and refine extracted phrases and the
algorithm is used to specify the regular expression in
each step in order to extract non-taxonomic
relationships between concepts. An ontology
specialist has to evaluate the non-taxonomic
relationships, but it should be noted that this
algorithm cannot be used to create an ontology based
on the huge amount of documents and relationships
within the document. However, here, the non-
taxonomic relationship is extracted independent from
the verb, illustrating the type of relationship. As
Villaverde et al., (2009) have illustrated, two phrases
which do not have any similar words might be related
by one verb. Thus, the verb is an important factor in
identifying a non-taxonomic relationship when
creating an ontology that uses as an inferring
algorithm.
Villaverde et al., (2009) proposed a solution to
this problem. They extracted concepts from plain

texts using the NLP algorithm. They assign a triple
vector <
,,
> for each two consecutive concepts
using a regular expression method, where is a verb
between two concepts <
,
> in the same sentence.
Villaverde et al., (2009) extract the most powerful
non-taxonomic relationships by measuring the co-
occurrence of these triples in whole documents.
Meanwhile, Sanchez and Moreno (2008) use a
similar algorithm, creating triple vectors for noun
phrases and verb phrases. A statistic technique is used
for refining the vectors based on degree of
relatedness. Fader et al., (2011) also created similar
triple vectors for concepts of each phrase, but they use
a logistic regression classifier to select the most
important vectors. This approach has limitations in
that a specific number of co-occurrence has to be
detected in order to identify words as concepts.
Therefore, this algorithm depends on the quality of
contextual information in the documents. Moreover,
removing stopwords may influence the main
semantic of documents.
Li et al., (2012) proposed a new method for
extracting an ontology from domain specific websites
or texts. A text classifier method is used to extract
important words and cluster words in different groups
based on predefined patterns. To detect more
instances based on core seed patterns, they developed
an iterative pattern-based algorithm called LASER to
generalize patterns. LASER can detect more complex
noun phrases than previous algorithms since it
extracts noun phrases from text segments that either
surround the connectors, the modifiers, or so on.
LASER retrieves the relationships based on noun
syntax, but nouns can also have a semantic
relationships. LASER only extracts the taxonomic
relationships; however, non-taxonomic relationships
are also an important factor in building an ontology.
Generally, algorithms which use term frequency
methods (e.g. frequent item set) and that remove
stopwords to extract concepts from texts suffer from
neglecting relationships between words. For
example, take extracting an ontology of “car” from
texts related to cars. One document describes engine
characteristics, which consists of physical and
functional attribute definitions but another document
explains the car’s electric system. Here, we can see
how term frequency is not able to retrieve the deep
relationship between the car’s engine and electric
system.
In this study, we separate the process of extracting
complex concepts and identifying direct and indirect
relationships between concepts to increase algorithm
performance. In the following sections, our proposed
framework for analyzing plain text in order to create
an ontology is described.
3 CONCEPTUAL FRAMEWORK
Thus study proposes a new framework for extracting
an ontology from plain text or even a specific domain
name by combining text mining and web mining
techniques to generate a more comprehensive
ontology. This algorithm is unsupervised and has the
ability to analyze multidisciplinary text.
3.1 Solution Overview
As described earlier, ontology is a technique to
represent and transfer knowledge from humans to
machines. To date, various algorithms have been
developed to create an ontology from plain texts or
even domain specific texts. As mentioned in Section
2, most developed algorithms need expert human
interactions for evaluation. Also, they usually extract
a single word as concept and use a fixed number of
patterns to extract non-taxonomic relationships
between concepts.
We use an advanced machine learning algorithm
to detect concepts (complex concepts) and to link
concepts based on their status in sentences and
documents in this framework. A big picture of our
framework is shown in Figure 1.
Figure 1: Framework structure.
This framework has four components. In the web
mining component, the main webpage related to a
domain name is retrieved from Wikipedia and all
pages which connect to the main page are extracted.
In the machine learning component, all important
words or phrases are extracted using a modified
version of the TF-IDF algorithm. We then combine
the N-gram and TF-IDF algorithms to extract and
rank noun phrases and phrases from contexts. In the
next step, we analyze sentence structure and
relationships between words in the NLP component.
In this component, the dependency tree of each
sentence is created. In this tree, words connect to each
other based on their relationship (i.e. taxonomic or

non-taxonomic). Finally, these small dependency
trees are then connected to each other based on their
TF-IDF weights to create a comprehensive tree for
specific domains in the ontology extractor
component. Each component is described in
following sections.
3.2 Algorithm Description
In this framework, we use Wikipedia to create the
structure of knowledge in the given domain.
Afterwards, nouns phrases, taxonomic, and non-
taxonomic relationships are extracted by applying a
modified version of TF-IDF and POS tagging
analysis. Finally, TF-IDF weight is used to connect
concepts to create a knowledge schema.
3.2.1 Web Mining Algorithm
A general knowledge schema is created from an
online encyclopedia – Wikipedia. Wikipedia is a
well-known encyclopedia which has received more
than 22.2 million requests on 28th September, 2014
1
alone, and as of August 2015
2
contained more than
35.9 million articles. Wikipedia is a reliable source
for finding whole structures of concepts in specific
determined domains. In the proposed model,
Wikipedia pages categorized in a given domain name
are extracted. A graph –
,
is then created
based on the Wikipedia pages’ link structure, where
represents a set of nodes representing the web pages
and is a set of edges which connect the two nodes
if one web page contains a hyperlink to another page.
In addition, the degree of distance
for each page
is measured. As shown in formula below, is the main
page of the given domain in Wikipedia and is a
Wikipedia page which has a direct or indirect
connection to the main page
min
∈
,
∈
1,
1,
0,
Degree of distance (
) is used to give priorities to
concepts more related to domain topic. Therefore,
concepts inside documents that have small
are
categorized as the main subtopics in the knowledge
schema.
In the next step, we crawl and grab content of all
webpages retrieved from Wikipedia.
1
http://stats.wikimedia.org/EN/TablesPageViewsMonthlyCombined.htm
2
http://stats.wikimedia.org/EN/TablesArticlesTotal.htm
3.2.2 Machine Learning Algorithm
Wikipedia API, which was developed in Python, is
used to gain the main information related to the
webpages. After downloading Wikipedia’s
webpages, important concepts are extracted from the
downloaded documents. Scholars have proposed
various statistical methods for extracting main
keywords from documents. TF-IDF is a well-known
algorithm in this area. TF-IDF reflects how important
a word is to a document in a collection of documents.
Thus, a high TF-IDF weight means the word has high
term frequency in a document since it has a low
document frequency in a collection of documents.
TF-IDF has two main problems. First, TF-IDF weight
is measured by a specific word in a specific
document. This means that if a word occurs in two
different documents, two different TF-IDF weights
will be calculated for the same word. TF-IDF was
developed to measure the weight of a single word;
however, in our case, we need to extract key phrases,
which can consist of a simple word or be multi-word.
To overcome this problem, we use a modified version
of the TF-IDF algorithm. In this technique, a
modified R-precision algorithm is used to evaluate
key phrases and TF-IDF is applied on all extracted
phrases. We also calculate the average of all
calculated TF-IDF for each word (as shown in
following equation) and assign it as TF-IDF weight
of word.
∑
,∈
We apply the mentioned technique to extract a list of
concepts and measure their TF-IDF weights. This
step requires that main concepts be filtered from
others from which we can then compute a TF-IDF
weight threshold. All phrases that have a higher TF-
IDF weight in comparison with the threshold are the
main phrases of this domain. For this purpose, we
defined a new measurement –
– to evaluate the
position of a word in a document. This measure has
two parameters – , – where shows the position of
the sentence that contains the word and the
shows the position of the word in the sentence. For
calculating, the total number of sentences before
is calculated from the first line of the document and
is calculated by counting speech element parts until
, except for prepositions, conjunctions, and
interjections. For instance, the
""
6,5
means the word “hotel” appears in the 6th
sentence as the 5th word in that sentence.
To compute the threshold of the TF-IDF weight,
we use K-means clustering. All extracted phrases are
clustered based on TF-IDF weight, degree of distance
(
) of document that contain the word , and
the position of the words
in the document .
We assume K as the maximum distance between
extracted document and the main page of online
encyclopedia as shown below:
max
,∈
After clustering the words based on the mentioned
features, the TF-IDF threshold is calculated through
function as described in the
following equation:
,
,
,
∑
∈
|
|
Where as
is the cluster and j is member of
cluster
.
∑
Whereas R is the total number of levels of documents
in terms of distance weight.
Cluster
, one of the K-means clusters which
has the highest rate of diversity in terms of document
level (
) and the highest average of TF-IDF weights
(
) in comparison with the other clusters,
is considered the threshold.
Finally, the
is measured for each word.
Important concepts are extracted based on the
. In the next step, we find the
taxonomic and non-taxonomic relationships between
concepts.
3.2.3 Natural Language Processing
Algorithm
We use the NLP algorithm to detect concepts and
relationships from contexts. In this algorithm, nouns are
extracted as concepts. Therefore, all types of nouns are
extracted from sentences. We use the NLP to assign a POS
tag to each word and filter it based on below list:
[NN]: Noun, singular or mass
[NNS]: Noun, plural
[NNP]: Proper noun, singular
[NNPS]: Proper noun, plural
Only the main noun is captured as concept. For
example, “nanoprobe sequencing” is a combination
of two nouns. “nanoprobe” is tagged as [JJ] which
means adjective. In this study, a new structure has
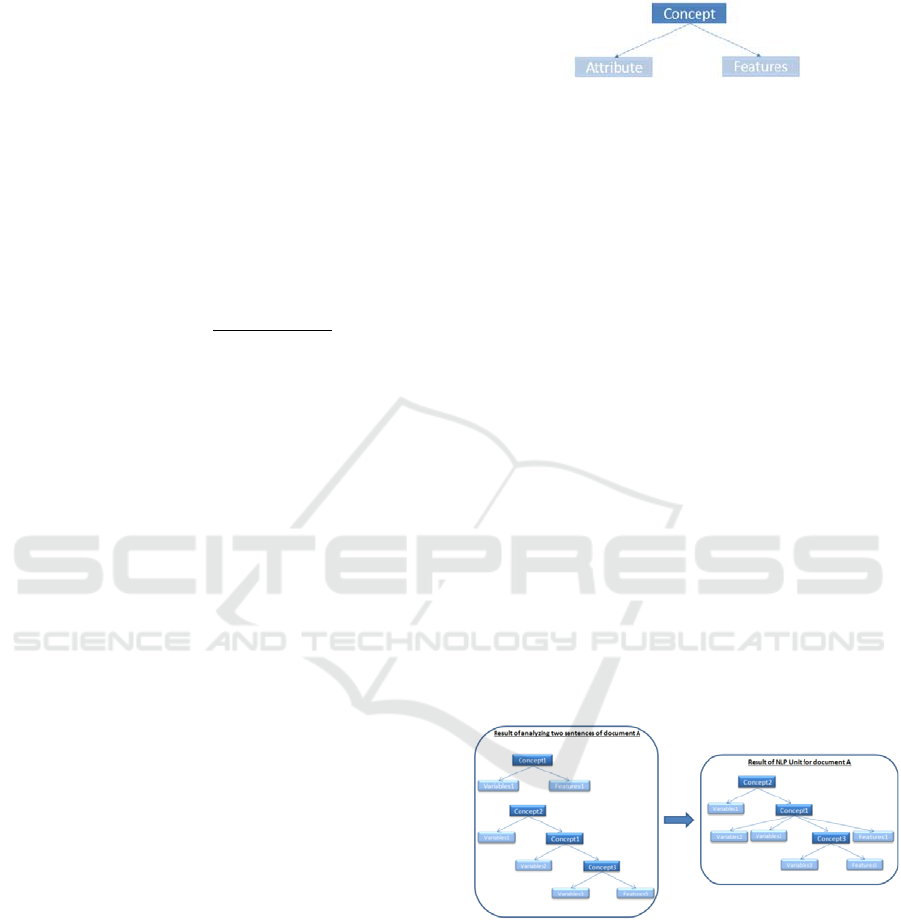
been proposed for each concept as shown in Figur.
Each concept has two parts: attribute and feature, as
shown in the Figure 2.
Figure 2: The structure of concept.
Attribute contains all the words which have a
direct impact on the concept such as adjectives and
complementary nouns. Accordingly, each attribute
explains a specific characteristic of a concept. For
instance, “red flower” has two parts “red [JJ]” and
“flower [NN]”. In this case, “flower” is labeled as a
concept that has a specific attribute, which is “red”.
This structure helps detect all the characteristics of a
concept. Feature are words that have non-taxonomic
relationships with the concept such as the object of
the sentence, nouns, adverbs, or even numbers.
In the next step, a concept is analyzed if it has a
higher
than the threshold. In the case that it
does not, despite it not having the proper
, it
will be processed if it has a non-taxonomic
relationships with another concept that has a higher
TF-IDF.
3.2.4 Ontology Extractor Algorithm
A tree for each sentence is created based on concept
dependencies subtrees. Afterwards, subtrees are
joined to each other in terms of taxonomic and non-
taxonomic relationships in each document (as shown
on Figure 3).
Figure 3: Building ontology structure.
In the next step, all document trees are combined
based on their distance and weight. Documents with
a low weight are processed earlier than others.
We use a labeling semantic class algorithm to
retrieve the name for every subclass of attributes. In
this algorithm, attributes are separated based on their
type and content (text, number, date and etc.).
Regular expression is used to analyze number, date,
and predefined texts. In addition, to find the name of

sub-classes, words are analyzed based on information
from WordNet. A WordNet graph structure is used to
measure the distance between each word.
4 EXPERIMENT DESIGN
The algorithm was evaluated by comparing the output
with an existing ontology. An ontology for
“biochemistry” was extracted and the results
compared with the provided ontology by Dumontier
Lab (Stanford University). As Figure 1 shows, all the
webpages which are related to biochemistry were
extracted using a web mining component. All
keywords were extracted from the downloaded
documents using a modified version of the TF-IDF
technique in the machine learning component.
Dependency subtrees were created for sentences, and
by joining subtrees, an ontology structure for
pharmacogenomics was created for evaluation by the
provided ontology by Dumontier Lab.
In first step, all pages related to “biochemistry”
were downloaded from Wikipedia. A crawler was
used to retrieve the hyperlink structure of this
Wikipedia page –
https://en.wikipedia.org/wiki/Biochemistry. Graph
,
was created where each node in the graph
represents the name of a category and each edge
illustrates that there is a hyperlink between these two
nodes in Wikipedia. We crawled Wikipedia
webpages until the shortest path between the
biochemistry webpage and other retrieved web pages
was less than four.
The number of pages which were retrieved in this
step is shown in Table 1.
Table 1: extracted dataset.
Number of pages: 9662
Level of tree: 3
Domain: Biochemistry
Source: www.wikipedia.org
In the machine learning component, stopwords
were removed while others were stemmed and
lemmatized. We measured the TF-IDF word weight
and used the K-means clustering method to determine
the threshold to filter the main keywords and then
others. The TF-IDF word weights for documents with
a distance weight of one were shown in Figure 4.
As shown in Figure 4, the minimum TF-IDF
weight value was for the word “protein” at 0.02165,
and the maximum value was for “pharmacogenom”
at 0.2032. In total, we extracted 550000 words from
9662 documents.
Figure 4: TF-IDF weight of words of documents with
distance weight of one.
We clustered words based on their TF-IDF
weight, distance of the weight of document, and the
location of words in the document (nth word in mth
sentence). In this case, the cluster considered best is
the one which has the highest
average and
contains words from most of the documents in all
levels (level is distance of document from main
webpage in Wikipedia). The threshold 0.016 is based
on the output of K-means, as shown in Figure 5.
Figure 5: K-means clustring.
We used the Stanford NLP engine to identify the
words’ POS tags in order to create the dependency
trees of sentences. We extracted 249 concepts from
the documents and created a dependency tree for each
concept and linked the concepts’ tree based on their
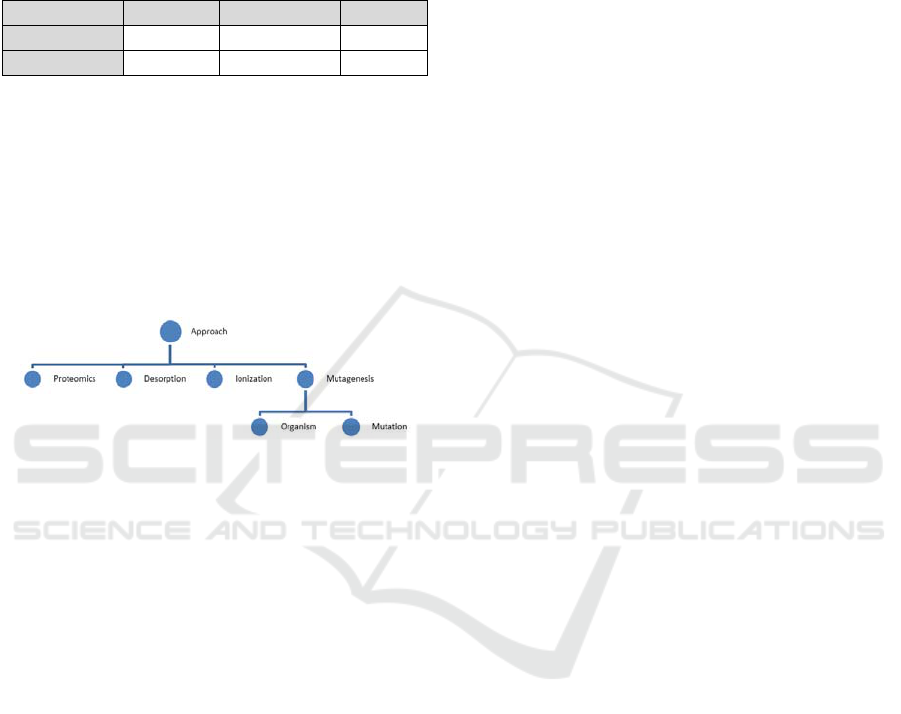
non-taxonomic relationships and TF-IDF weight. For
instance, concept “approach” has a dependency tree
as seen below.
We compared our framework output with the
ontology provided by the Dumontier Lab (Stanford
University) for the domain “pharmacogenomics”.
Pharmacogenomics is categorized as subclass of
biochemistry. The proposed ontology by Dumontier
Lab consists of 20 concepts. The ontology also
describes 37 taxonomy relationships between
concepts. However, the ontology created by our
framework illustrates nearly 242 concepts with 470
non-taxonomic and 240 taxonomic relationships as
shown in Table 2.
Table 2: Ontologies' structure.
Ontology #Taxonomy #Non-taxonomy # Concepts
Dumontier Lab 37 0 20
Our algorithm 470 240 242
The proposed ontology by Dumontier Lab is more
focused on technical and professional keywords and
relationships. For instance, their ontology does not
include “approach” as a concept. Our ontology
includes “approach” as a concept and clarifies which
type of “approach” is used in this field by adding
various attributes to the concept such as
“proteomics“, “desorption/ ionization”, and
“leaching”, as shown in Figure 6.
Figure 6: Concept structure of "Approach".
5 CONCLUSIONS
As discussed in above sections, various methods have
been developed to extract a specific domain
knowledge structure from unstructured text.
However, most of these use techniques that extract
single words as concept and moreover, extract only
the taxonomic relationships between concepts. In
addition, the knowledge structure of
multidimensional knowledge such as nanotechnology
is more complex because each concept might be
defined differently in various fields. Given these, we
developed a framework to create an ontology from
plain text documents that could include complex
concepts and non-taxonomic relationships. To
develop the framework, we used the online
encyclopedia Wikipedia and a lexical database. The
knowledge structure was built based on the the
information mentioned on Wikipedia webpages
related to the domain. We used a modified version of
the TF-IDF technique to extract complex concepts
from these documents. Meanwhile, an NLP technique
was used to extract POS noun tags and dependency
tree of sentences. In this study, we proposed a
structure for each concept. Each concept is explained
by two elements: feature and attribute. Concept
subtrees are connected to each other in terms of
and non-taxonomic relationships.
For validation purposes, we built an ontology for
the “pharmacogenomics” domain and compared it
with the proposed ontology by Stanford University
Dumontier Lab. The results show that our ontology
contains more detailed information such as higher
number of concepts, non-taxonomic relationships,
and taxonomic relationships. More detailed
information increases an ontology’s ability to
represents a multidiscipline domain more precisely.
Future studies should improve the proposed
framework to generate an ontology from a created
knowledge schema. In this study, the proposed
framework creates a knowledge schema for a given
domain in an online encyclopedia. To generate an
ontology, extracted concepts should be analyzed to
classify synonyms and taxonomic relationships
between words from WorldNet.
REFERENCES
Agichtein, E. and Gravano, L., 2000. Snowball. In
Proceedings of the fifth ACM conference on Digital
libraries - DL ’00. New York, New York, USA: ACM
Press, pp. 85–94. Available at: http://portal.
acm.org/citation.cfm?doid=336597.336644 [Accessed
August 24, 2017].
Anantharangachar, R., Ramani, S. and Rajagopalan, S.,
2013. Ontology Guided Information Extraction from
Unstructured Text. International Journal of Web and
Semantic Technology (IJWesT), 4(1), pp.19–36.
Aroyo, L. et al., 2002. A Layered Approach towards
Domain Authoring Support. ICAI 2002 (LAS VEGAS,
US) CSREA. Available at: http://citeseerx.ist.psu.edu/
viewdoc/summary?doi=10.1.1.18.8301 [Accessed
August 24, 2017].
Bisson, G. et al., 2000. Designing clustering methods for
ontology building: The Mo’K Workbench. IN
PROCEEDINGS OF THE ECAI ONTOLOGY
LEARNING WORKSHOP, pp.13--19. Available at:
http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.
1.1.35.6302 [Accessed August 24, 2017].
Buitelaar, P., Olejnik, D. and Sintek, M., 2004. A Protégé
Plug-In for Ontology Extraction from Text Based on
Linguistic Analysis. In Springer, Berlin, Heidelberg,
pp. 31–44. Available at: http://link.springer.com/
10.1007/978-3-540-25956-5_3 [Accessed August 24,
2017].
Drymonas, E., Zervanou, K. and Petrakis, E.G.M., 2010.
Unsupervised ontology acquisition from plain texts:
The OntoGain system. Lecture Notes in Computer
Science (including subseries Lecture Notes in Artificial
Intelligence and Lecture Notes in Bioinformatics), 6177

LNCS, pp.277–287.
Fader, A., Soderland, S. and Etzioni, O., 2011. Identifying
relations for open information extraction. Proceedings
of the Conference on Empirical Methods in Natural
Language Processing, pp.1535–1545. Available at:
http://dl.acm.org/citation.cfm?id=2145596 [Accessed
May 17, 2017].
Fortuna, B., Grobelnik, M. and Mladenič, D., 2006.
Background knowledge for ontology construction. In
Proceedings of the 15th international conference on
World Wide Web - WWW ’06. New York, New York,
USA: ACM Press, p. 949. Available at:
http://portal.acm.org/citation.cfm?doid=1135777.1135
959 [Accessed August 24, 2017].
Li, T. et al., 2012. Efficient Extraction of Ontologies from
Domain Specific Text Corpora. Proceedings of the 21st
ACM International Conference on Information and
Knowledge Management, (December), pp.1537–1541.
Available at: http://doi.acm.org/10.1145/2396761.
2398468.
Maedche, A. and Volz, R., 2001. The ontology extraction
and maintenance framework Text-To-Onto. Proc.
Workshop on Integrating Data …, pp.1–12. Available
at: http://users.csc.calpoly.edu/~fkurfess/Events/DM-
KM-01/Volz.pdf [Accessed May 17, 2017].
Navigli, R. and Velardi, P., 2004. Learning Domain
Ontologies from Document Warehouses and Dedicated
Web Sites. Computational Linguistics, 30(2), pp.151–
179. Available at: http://www.mitpressjournals.org/doi/
10.1162/089120104323093276 [Accessed August 24,
2017].
Sánchez, D. and Moreno, A., 2008. Learning non-
taxonomic relationships from web documents for
domain ontology construction. Data and Knowledge
Engineering, 64(3), pp.600–623. Available at:
http://www.sciencedirect.com/science/article/pii/S016
9023X07001838 [Accessed May 17, 2017].
Serra, I., Girardi, R. and Novais, P., 2013. PARNT: A
statistic based approach to extract non-taxonomic
relationships of ontologies from text. Proceedings of
the 2013 10th International Conference on Information
Technology: New Generations, ITNG 2013, pp.561–
566.
Villaverde, J. et al., 2009. Supporting the discovery and
labeling of non-taxonomic relationships in ontology
learning. Expert Systems with Applications, 36(7),
pp.10288–10294. Available at: http://www.
sciencedirect.com/science/article/pii/S0957417409000
943 [Accessed May 17, 2017].
Zavitsanos, E. et al., 2010. Learning subsumption
hierarchies of ontology concepts from texts. Web
Intelligence and Agent Systems, 8(1), pp.37–51.
