
Mapping Food Composition Data from Various Data Sources to a
Domain-Specific Ontology
Gordana Ispirova
1,2
, Tome Eftimov
1,2
, Barbara Koroušić Seljak
1
and Peter Korošec
1,3
1
Computer Systems Department, Jožef Stefan Institute, Jamova cesta 39, 1000 Ljubljana, Slovenia
2
Jožef Stefan International Postgraduate School, Jamova cesta 39, 1000 Ljubljana, Slovenia
3
Faculty of Mathematics, Natural Science and Information Technologies, Glagoljaška ulica 8, 6000 Koper, Slovenia
Keywords: Semantic Web, Food Domain Ontology, Food Composition Data, Text Similarity, Text Normalization.
Abstract: Food composition data are detailed sets of information on food components, providing values for energy and
nutrients, food classifiers and descriptors. The data of this kind is presented in food composition databases,
which are a powerful source of knowledge. Food composition databases may differ in their structure between
countries, which makes it difficult to connect them and preferably compare them in order to borrow missing
values. In this paper, we present a method for mapping food composition data from various sources to a
terminological resource-a food domain ontology. An existing ontology used for the mapping was extended
and modelled to cover a larger portion of the food domain. The method was evaluated on two food
composition databases: EuroFIR and USDA.
1 INTRODUCTION
Food composition data (FCD) are detailed sets of
information on the nutritional components of foods,
providing values for energy and nutrients, food
classifiers and descriptors. This type of data is
presented in Food Composition Databases (FCDBs)
(Greenfield, Southgate, 2003). Nowadays, FCDBs
tend to be compiled using a variety of methods,
including: chemical analysis of food samples carried
out in analytical laboratories, imputing and
calculating values from data already within the
database and estimating values from other sources,
including manufacturers food labels, scientific
literature and FCDBs from other countries.
The three main limitations of FCDBs are:
variability in the composition of foods between
countries, age of data (limited resources mean that,
inevitably, some values are not current) and
incomplete coverage of foods or nutrients leading to
missing values. Foods, being biological materials,
exhibit variations in composition. Therefore, a
database cannot accurately predict the composition of
any given single sample of a food. Further, FCDBs
cannot predict accurately the nutrient levels in any
food and the composition of a given food may change
with time. Predictive accuracy is also constrained by
the ways in which data are maintained in a database
(as averages, for example). FCDBs frequently cannot
be used as literature sources for comparison with
values obtained for the food elsewhere. Values from
one country should be compared with values obtained
in other countries by reference to the original
literature. Despite major efforts on harmonizing food
descriptions, nutrient terminology, analytical
methods, calculation and compilation methods,
values from existing food composition tables and
databases are not readily comparable across
countries. The description of food composition data-
nutrient terminology in different FCDBs can differ
(e.g. beta carotene, carotene-beta). To harmonize
them there is a need of text normalization methods.
Normalizing text means converting it to a more
convenient, standard form. Text normalization is the
process of transforming text into a single canonical
form. This process requires awareness of the type of
text being normalized and how it will be processed
afterwards, there is no all-purpose normalization
procedure. The main idea of normalizing a text is to
map a text description to an already existing
description contained in a domain specific
terminological resource, which can be a classical
dictionary, thesauri or a domain specific ontology.
A domain ontology represents concepts which
belong to a specific domain. Food domain ontologies
Ispirova G., Eftimov T., KorouÅ ˛aiÄ
˘
G Seljak B. and KoroÅ ˛aec P.
Mapping Food Composition Data from Various Data Sources to a Domain-Specific Ontology.
DOI: 10.5220/0006504302030210
In Proceedings of the 9th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (KEOD 2017), pages 203-210
ISBN: 978-989-758-272-1
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

model and represent the domain of food. Having
different food related datasets, it is possible to match
an entity mention from the datasets to a concept in the
terminological resource i.e. the domain specific
ontology. Being more specific, the data from a FCDB
can be matched to a food domain ontology, and to
each of the entities an ontology tag can be assigned,
thus linking the dataset to the ontology. This type of
data mapping is an ontology-based data integration
(Leida, Ceravolo, Damiani, Cui, Gusmini, 2010;
Kerzazi, Navas-Delgado, F.Aldana-Montes, 2009).
This linking opens up a whole window of new
opportunities. Major problem in FCDBs are missing
values of components. One of the solutions to this
problem is borrowing data from other FCDBs. This
can be accomplished from this type of linking. By
linking datasets to a domain specific ontology, the
linked datasets can borrow missing values
interchangeably.
In this paper we compare the results for text
normalization of short text segments, specifically
names or descriptions of nutrients, obtained using two
different approaches: standard text similarity
measures and a modified version of Part of Speech
(POS) tagging probability weighted method
(Eftimov, Koroušić-Seljak, 2017). Starting with an
overview of related work concerning text
normalization methods and food domain ontologies
in Section 2, we continue with explaining the two
datasets and the methods used in our experiments
(Section 3 and Section 4). In Section 5 we give the
results obtained from the experimental work, and a
comparison of the methods. The last section is an
overall discussion of the problem, the method used
and the obtained results as a conclusion to our work.
2 RELATED WORK
In this section an overview of related work is
presented. Starting from existing text normalization
methods and food domain ontologies. To the best of
our knowledge there is no text normalization method
specifically developed for the food domain.
2.1 Text Normalization
The aim of text normalization methods is mapping
same concepts coming from different sources,
described in different ways to a concept from
terminological resource, which will imply that the
information contained in these concepts is the same.
The majority of normalization methods are based on
matching entity mentions to concept synonyms listed
in a terminological resource (Aronson, 2001; Savova,
Masanz, Ogren, Zheng, Shon, Kipper-Schuler, et al.,
2010; Friedman, Shagina, Socratous, Zeng, 1996;
Friedman, 2000; Garla, Brandt, 2013). More
sophisticated methods combine or rank the results
obtained using a number of different terminological
resources (Collier, Oellrich, Groza, 2015; Fu, Batista-
Navarro, Rak, Ananiadou, 2014). Pattern-matching
or regular expressions approaches (Fan, Sood, Huang,
2013; Ramanan, Broido, Nathan, 2013; Wang,
Akella, 2013) can account for frequently occurring
variations not listed in the terminological resource.
Methods based on machine learning, or hybrid
methods combining rules and machine learning, have
also been proposed (Goudey, Stokes, Martinez, 2007;
Leaman, Doğan, Lu, 2013).
String similarity methods have been employed in
a number of normalization efforts (Doğan, Lu, 2012;
Kate, 2015). These methods assign a numerical score
representing the degree of similarity between an
entity mention and a concept synonym, which means
that, unlike the limited types of variations that can be
handled by rules or regular expressions, string
similarity methods can handle a virtually unlimited
range of variations.
Character-level methods consider the number of
edits (e.g., insertions, deletions or substitutions)
required to transform one phrase into another (Jaro,
1995), or look at the proportion and/or ordering of
characters that are shared between the phrases being
compared (Jaro, 1995; Winkler, 1999; Kondrak,
2005). This can help to account for the fact that
concepts may be mentioned in text using words that
have the same basic root but many different forms,
including different inflections (e.g., reduced vs.
reduce), alternative spellings (e.g. fiber vs. fibre) and
nominal vs. verbal forms (e.g., reduce vs. reduction).
Word-level similarity metrics (Jaccard, 1912) can
be more appropriate when the phrases to be compared
consist of multiple words. Such metrics make it
possible to ensure that a match is only considered if a
certain proportion of words is shared. Weights may
be applied to the individual words (as is the case for
TF-IDF (Term Frequency-Inverse Document
Frequency) (Moreau, Yvon, Cappe, 2008)), to ensure
that greater importance is placed on matching words
with high relevance to the domain, than function
words like: the, of, etc.
Hybrid methods (e.g. SoftTFIDF (Cohen,
Ravikumar, Fienberg, 2003)) also operate at word
level, but use a character based similarity method to
allow matches between words that closely resemble
each other, even if they do not match exactly. This
helps to account for the fact that concepts may be
mentioned in text using multi-word terms whose
exact forms may vary from synonyms listed in the
terminological resource. Such methods can also help
to address the problem of normalizing entity mentions
containing spelling errors. The accuracy of string
similarity methods could be improved by integrating
semantic-level information.
In the paper (Alnazzawi, Thompson, Ananiadou,
2016) the authors present a method, called
PhenoNorm. It was developed using the PhenoCHF
corpus, which is a collection of literature articles and
narratives in Electronic Health Records, annotated for
phenotypic information relating to congestive heart
failure (CHF). This method links CHF-related
phenotype mentions to appropriate concepts in the
UMLS Metathesaurus, using a version of PhenoCHF.
However, in the food domain, where we
concentrate our research, this type of work has not
been previously done.
2.2 Food Domain Ontologies
There are several food ontologies: FoodWiki (Celik,
2015), AGROVOC (Caracciolo et al., 2012), Open
Food Facts (Open food facts ontology, 2017), Food
Product Ontology (Kolchin, Zamula, 2013), FOODS
(Diabetics Edition) (Snae, Bruckner, 2008) and
FoodOn Ontology (FoodOn Ontology, 2017). In the
paper (Boulos, Yassine, Shirmohammadi, Namahoot,
Bruuckner, 2015), the authors provided a review of
the mentioned food ontologies.
Despite the attempts of building an ontology for
wider uses, thus the attempt of FoodOn for
generalized ontology for the food domain, all of the
mentioned ontologies are developed for very specific
uses. To overcome the limited scope of food
ontologies an ontology that covers wider domain is
needed.
Not scientifically validated data in the systems
providing data about food and nutrition is the main
cause of having invalid data in FCDBs. This problem
has been mostly solved with the project QuaLiFY
(Qualify, 2017). In this project a new food ontology
for harmonization of food-and nutrition-related data
and knowledge, called Quisper (Eftimov, Koroušić-
Seljak, 2015), has been developed. We have updated
and extended this ontology with additional concepts.
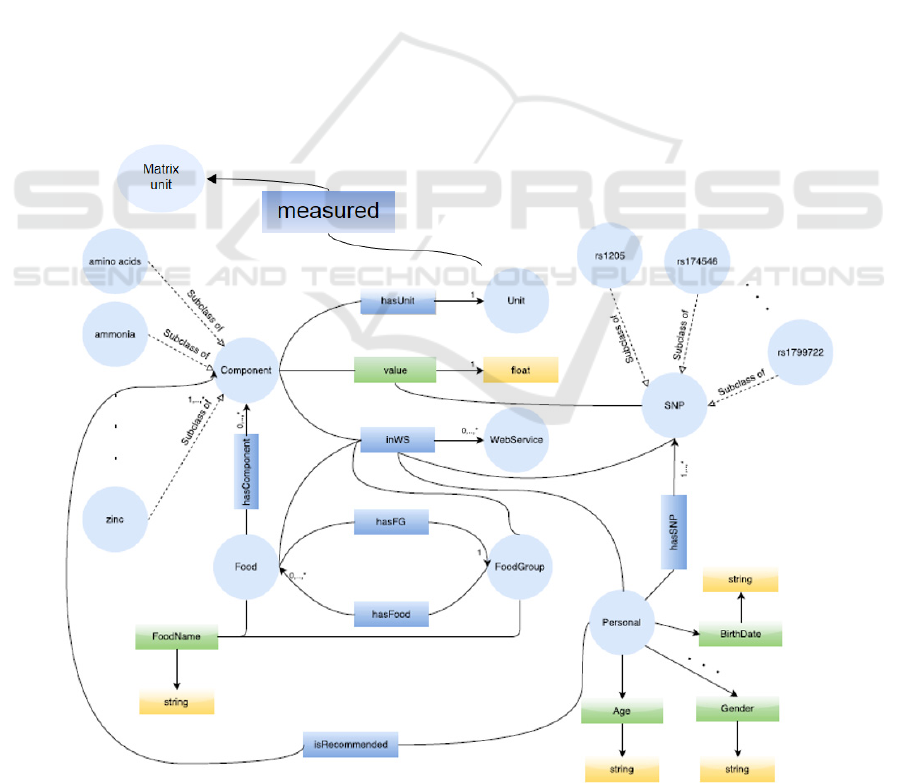
In Figure 1 the updated structure is shown. In the
Figure 1: Updated structure of Quisper ontology.

developing process of this ontology first, using the
POS tagging-probability weighted method, the
similar terms provided from the web services are
extracted, then an initial taxonomy with the similar
terms is created, to which terms that are typical only
for one web service are added. At the end, using the
extracted terms in the taxonomy and the relations
between the terms in the web services, an ontology
from scratch is created using the software Protégé
(Protégé, 2016), which is available to the human
experts. The proposed approach could be also used in
other domains simply by modifying the probability
model in order to fit the purposes of the domain of
interest.
3 DATA
In this section we describe the data used in our
experiments, which comes from two sources. The
purpose is to link the nutrient information from both
sources presented on a different way to a food domain
ontology. For example: “fatty acids 18:1-11 t (18:1t
n-7)” needs to be linked to “fatty acid 18:1 n-7 trans”;
“Tocotrienol, gamma” needs to be linked to “gamma-
tocotrienol”; etc.
3.1 EuroFIR Dataset
European Food Information Resource Network
(EuroFIR) AISBL is an international, non-profit
association under the Belgian law. As an organization
its purpose is developing, publishing and exploiting
food composition information and promoting
international cooperation and harmonization of
standards to improve data quality, storage and access
(European Food Information Resource Network,
2017). The EuroFIR data interchange uses files in
XML format, which follow a nested structure.
EuroFIR presents a data model for FCD management
and data interchange. The EuroFIR format for FCD
starts with “Foods” element which holds separate
“Food” elements that report the data for each
individual food item. Within each “Food” element,
together with the elements describing the food, are
nested collections of “Component” records, each with
its set of “Value” records. For the purposes of this
project, an XML file from EuroFIR Component
Thesaurus version 1.3 is extracted. There are 997
components in total and for each component a short
abbreviation, the full name/description of the
component, the date when it was added to the
database and the date when it was last updated are
listed.
3.2 USDA Dataset
The United States Department of Agriculture
(USDA), is the federal executive department of the
U.S., whose responsibility is developing and
executing federal laws related to farming, agriculture,
forestry, and food. Its aims are meeting the needs of
farmers and ranchers, promoting agricultural trade
and production, assuring food safety, protecting
natural resources, foster rural communities and
ending hunger in the United States and internationally
(USDA, Food Composition Database, 2017).
This department has produced the USDA
National Nutrient Database, which is a database that
provides the nutritional content of many generic and
proprietary-branded foods. New releases occur about
once per year. The database can be searched online,
queried through a REST API (NDB API, 2017), or
downloaded. For the needs of this project we accessed
the nutrients list from the USDA FCDB through the
REST API.
The obtained file is in XML format, following a
nested structure. There are 190 nutrients in the list,
and for each nutrient an identification number and the
nutrient’s name is listed.
4 METHOD
In this section, we describe the pre-processing of the
datasets and the two approaches of text normalization
used in our experiments.
4.1 Pre-processing
After obtaining the XML files from both datasets the
relevant information is extracted. From the EuroFIR
dataset for each component we extracted the short
abbreviation and the full name/description of the
component in a CSV file. The same is applied for the
USDA dataset, where the names of the nutrients
alongside with their identification numbers are
extracted in a CSV file. From the OWL file of the
extended Quisper ontology, all the sub-concepts of
the concept “Component” and their corresponding tag
from the ontology are extracted. The conversion from
XML to CSV is made using simple parsing in R
(Development Core Team, 2008).
4.2 Normalization of FCD
Having the CSV files from the three sources the next
step was to match the names of the nutrients from
both food composition databases to the names of the

nutrients from the ontology. The matching is made by
using two different methods: using text similarity
measures and using a modified version of POS
tagging combined with probability theory.
4.2.1 Normalization using Text Similarity
Measures
The first method of normalization is performed in
RStudio IDE (RStudio Team, 2015), using the
package ‘stringdist’ (Van der Loo, Van der Laan,
Logan, 2016). A total of eight text similarity
measures are applied:
1. Optimal string alignment (OSA), (restricted
Damerau-Levenshtein distance) - Levenshtein
distance is the number of deletions, insertions
and substitutions necessary to turn string into
string . OSA is like the Levenshtein distance but
also allows transposition of adjacent characters.
Here, each substring may be edited only once.
2. Full Damerau-Levenshtein distance is like the
OSA distance except that it allows multiple edits
on substrings.
3. Longest common substring distance is defined as
the longest string that can be obtained by pairing
characters from string and string while
keeping the order of characters intact. This
distance is defined as the number of unpaired
characters, and it is equivalent to the edit distance
allowing only deletions and insertions, each with
weight one.
4. -gram distance - A -gram is a subsequence of
consecutive characters of a string. If () is
the vector of counts of -gram occurrences in
string (), the -gram distance is given by the
sum over the absolute differences |
−
|.
The computation is aborted when is larger than
the length of any of the strings. In that case
is returned.
5. Cosine distance between -gram profiles is
computed as:
1−∙/(
|
|
|
|
) (1)
Where and were defined above.
6. Jaccard distance between -gram profiles - Let
be the set of unique -grams in and the set of
unique -grams in . The Jaccard distance is
given by:
1−
|
∩
|
/
|
∪
|
(2)
7. Jaro, or Jaro-Winker distance - The Jaro distance,
is a number between 0(exact match) and 1
(completely dissimilar) measuring dissimilarity
between strings. It is defined to be 0 when both
strings have length 0, and 1when there are no
character matches between and . Otherwise,
the Jaro distance is defined as:
1 −(1/3)(
/||+
/||+
( − )/) (3)
Here, || indicates the number of characters in ,
is the number of character matches and t the
number of transpositions of the matching
characters. The
are weights associated with
the characters in , characters in and with
transpositions. Two matching characters are
transposed when they are matched but they occur
in different order in string and . The Jaro-
Winkler distance adds a correction term to the
Jaro-distance. It is defined as:
−·· (4)
Where is the Jaro-distance. Here, is obtained
by counting, from the start of the input strings,
after how many characters the first character
mismatch between the two strings occurs, with a
maximum of four. The factor is a penalty
factor, which in the work of Winkler is often
chosen 0.1.
8. Distance based on soundex encoding - This text
similarity measure translates each string to a
soundex code. The distance between strings is 0
when they have the same soundex code,
otherwise 1.
All eight text similarity measures are applied two
times on the data, without any previous pre-
processing, and with an additional pre-processing
step. The pre-processing step is removing the
punctuation from the names of the nutrients from both
datasets and from the names of the nutrients from the
ontology.
4.2.2 Normalization using POS and
Probability Theory
The second method of normalization is also
performed in RStudio IDE and it includes using POS
tagging combined with probability theory. This
particular method has been previously used (Eftimov,
Koroušić-Seljak, Korošec, 2017; Eftimov, Korošec,
Koroušić-Seljak, 2017; Eftimov, Koroušić-Seljak,
2017) and for the requirements of this project it is
modified.
Because we are working with terms related to
chemical names, on each description of the nutrient,
using POS tagging, nouns, adjectives and numbers
are extracted. These three morphological tags or
categories are selected with previously examining the
datasets. The descriptions of the nutrients can be
consisted of:
• only nouns (Example: copper; gluten;
sodium; …)
• nouns and adjectives (Example: acetic acid;
amino acids, total aromatic; pentoses in
dietary fibre …)
• nouns and numbers (Menaquinone-10;
vitamin_B1; …)
• nouns, adjectives and numbers (Example:
10-formylfolic acid; fatty acid 10:0 (capric
acid; starch, resistant_RS4; …)
The nouns carry the most information about the
term’s description, the adjectives explain the terms in
most specific form and the numbers are in most cases
related to the chemical nomenclature.
If:
•
= {nouns extracted from the -th
dataset}
•
= {adjectives extracted from the -th
dataset}
•
= {numbers extracted from the -th
dataset}
Correspondingly is the similarity between the
nouns extracted in one of the nutrient datasets
(EuroFIR or USDA) and the nouns extracted from the
names of the nutrients from the ontology. Same
implies for and . Having that these events are
independent from each other:
(
)
=
(
)
×
(
)
×
(
)
(5)
The formula for calculating each of the
probabilities is:
() = (
∩
+1) (
∪
+2) (6)
Where ={,,}. The probability that
two strings match is obtained with replacing, equation
(6) for each of the probabilities in equation (5).
5 RESULTS
The results from both methods are exported in CSV
format files.
In order to compare the methods, we manually
label all the instances in the result files. The labels
assigned are the following:
• 0 -Either no match is found (which is a case
only with the second method) or the match
found is not the correct one.
• 1 - A match is found.
• 2- Multiple matches are found, one of them
being the correct (most suitable) one.
• 3 - Multiple matches are found with none of
them being suitable or correct.
After labelling the instances, simple statistics are
performed, counting the instances from each
category. Because the Quisper ontology is
constructed based on an ontology-learning method
where one of the initial sets is the EuroFIR dataset,
when matching the nutrient names from EuroFIR and
the nutrient names from the ontology we obtained
perfect matches and 100% accuracy, and with this the
goal of assigning an ontology label to each nutrient is
met. However, for the nutrient names from USDA
dataset we obtained different results.
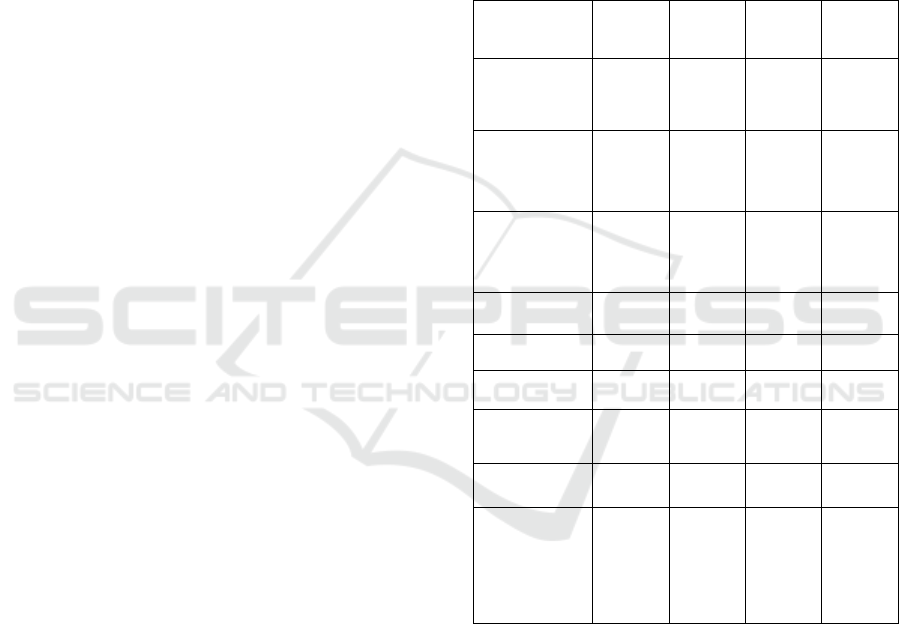
Table 1: Results from text normalization on USDA dataset.
Measure
Label
‘0’
Label
‘1’
Label
‘2’
Label
‘3’
Optimal
string
alignment
20 114 22 34
Full
Damerau -
Levenshtein
21 113 24 32
Longest
common
substring
33 127 13 17
- gram
36 112 24 18
Cosine
56 127 3 4
Jaccard
45 94 34 17
Jaro -
Winker
52 131 1 6
Soundex
2 48 110 30
POS
tagging
with
probability
theory
23 161 6 0
Judging from the results shown in Table 1, the
POS tagging method with probability theory gives the
best results. For a total of 167 instances it gives the
correct matches and only for 23 instances either it
cannot find a match or it returns the incorrect match,
which makes it have an accuracy of 87.9%. There are
no instances that belong to label′3′, and only 6
instances that belong to label ‘2’, which implies that
this method gives multiples choices only for a few
instances. By writing a simple code in R we
determined the threshold of the probability:
• if P < 0.067, then label ′0′ is assigned

The second best is Soundex text similarity
measure, with 158 instances with correct matches
and 32 instances with incorrect matches. It is clear
that this method works with giving a lot of options,
thus the number of instances with label′2′ is much
larger than the number of instances with label′1′, and
the number of instances with label ′3′ is also larger
than the number of instances with label′0′.
From further observation of the results we are
able to see that 19 out of those 23 instances for which
the best method is giving either no matches or the
incorrect matches the other eight measures also do not
give correct matches. From this we've come to the
conclusion that the other eight measures cannot be
applied to these instances in a cascade type of method
for improvement. For 4out of the 23 instances for
which this method doesn't give matches: “Ash”,
“Fiber, total dietary”, “Lutein + zeaxanthin” and
“Thiamin”, the other eight methods give correct
matches: “ash, total”, “fibre, total dietary”, “lutein
plus zeaxanthine” and “thiamine”. From looking into
this problem we have come to the conclusion that this
is because of the fact that the POS tagging method
does not recognize them as part of any morphological
class and cannot assign morphological tags to them.
In order to improve this results the second best
(Soundex) and third best measure (Longest common
substring) are applied separately on these instances
and the correct matches are obtained, with the
difference that the Soundex measure, again, for some
of the instances, gives more than one option, but the
Longest common substring measure gives only one
option for each, thus making it the better measure in
this case. After this step the total number of correctly
matched instances is 171 which is an
accuracyof90%.
6 CONCLUSIONS
This paper focuses on the problem of mapping food-
related information from different FCDBs to a
domain specific ontology that covers a large portion
of the food domain. Our work focuses on using text
normalization methods for linking short text
segments, in this case nutrient terminology, to a
concept from a domain specific terminological
resource, in this case a food domain ontology. The
implementation of this work allows the same nutrient
data represented on different ways in various data
sources to be linked to a concept from food domain
ontology, which makes sharing, combining and
reusing this kind of data easier. So far, we have linked
the largest two FCDBs to the Quisper ontology on
nutrient level. This concept can be modified and
further extended on food level.
With this work a certain level of harmonizing
FCD is achieved. If the principles of this work are
further followed by existing and newly constructed
FCDBs, the quality of the data and the database will
improve significantly.
ACKNOWLEDGMENTS
This work is supported by the project RICHFIELDS,
which received funding from the European Union’s
Horizon 2020 research and innovation programme
under grant number 654280.
REFERENCES
Greenfield, H., Southgate, D. A. T., 2003. Food
Composition Data: Production, Management, and Use,
Food & Agriculture Org, 2
nd
edition.
Celik, D., 2015. Foodwiki: Ontology-driven mobile safe
food consumption system, The Scientic World Journal.
Caracciolo, C., et al., 2012. Thesaurus maintenance,
alignment and publication as linked data: the agrovoc
use case. In International Journal of Metadata, Seman-
tics and Ontologies. Inderscience Enterprises Ltd.
Open food facts, accessed April 2017. http://world.open
foodfacts.org/who-we-are.
Kolchin, M., Zamula, D., 2013. Food product ontology:
Initial implementation of a vocabulary for describing
food products. In: Proceeding of the 14th Conference of
Open Innovations Association FRUCT.
Snae, C., Bruckner, M., 2008. Foods: a food-oriented
ontology-driven system. In Second IEEE International
Conference on Digital Ecosystems and Technologies.
FoodOn Ontology, accessed April 2017. http://food
ontology.github.io/foodon/.
Qualify, accessed April 2017. http://quisper.eu.
European Food Information Resource Network-EuroFIR,
accessed April 2017. http://www.eurofir.org/.
Eftimov, T., Koroušić-Seljak, B., 2015. QOL - Quisper
Ontology Learning using personalized dietary services.
In IJS delovno poročilo, 11985, confidential.
Boulos, M. N. K., Yassine, A., Shirmohammadi, S.,
Namahoot, C. S., Bruuckner, M., 2015. Towards an
internet of food: Food ontologies for the internet of
things. In Future Internet 7.
Protégé, accessed March 2017. http://protege.stanford.edu/.
Aronson, A., 2001. Effective Mapping of Biomedical Text
to the UMLS Metathesaurus: The MetaMap Program.
In Proceedings of the AMIA Annual Symposium.
Savova, G., Masanz, J., Ogren, P., Zheng, J., Shon, S.,
Kipper-Schuler, K., et al., 2010. Mayo clinical Text
Analysis and Knowledge Extraction System
(cTAKES): architecture, component evaluation and

applications. In Journal of the American Medical
Association.
Friedman, C., Shagina, L., Socratous, S. A., Zeng, X., 1996.
A WEB-based version of MedLEE: A medical
language extraction and encoding system. In:
Proceedings of the AMIA Annual Fall Symposium.
Friedman, C., 2000. A broad-coverage natural language
processing system. In Proceedings of the AMIA
Symposium. American Medical Informatics
Association.
Garla, V. N., Brandt, C., 2013. Knowledge-based
biomedical word sense disambiguation: an evaluation
and application to clinical document classification. In
Journal of the American Medical Informatics
Association.
Collier, N., Oellrich, A., Groza, T., 2015. Concept selection
for phenotypes and diseases using learn to rank. In
Journal of Biomedical Semantics.
Fu, X., Batista-Navarro, R, Rak, R, Ananiadou, S., 2014. A
strategy for annotating clinical records with phenotypic
information relating to the chronic obstructive
pulmonary disease. In Proceedings of Phenotype Day
at ISMB.
Fan, J., Sood, N., Huang, Y., 2013. Disorder concept
identification from clinical notes an experience with the
ShARe/CLEF 2013 challenge. In Proceedings of the
ShARe/CLEF Evaluation Lab.
Ramanan, S., Broido, S., Nathan, P. S., 2013. Performance
of a Multi-class Biomedical Tagger on Clinical
Records. In Proceedings of the ShARe/CLEF
Evaluation Lab.
Wang, C., Akella, R., 2013. UCSC's System for CLEF
eHealth. In: Proceedings of the ShARe/CLEF
Evaluation Lab.
Goudey, B., Stokes, N, Martinez, D., 2007. Exploring
Extensions to Machine Learning-based Gene
Normalisation. In Proceedings of the Australasian
Language Technology Workshop.
Leaman, R., Doğan, R. I., Lu, Z., 2013. DNorm: disease
name normalization with pairwise learning to rank. In
Bioinformatics.
Doğan, R. I., Lu, Z., 2012. An inference method for disease
name normalization. In Proceedings of the 2012 AAAI
Fall Symposium Series.
Kate, R. J., 2015. Normalizing clinical terms using learned
edit distance patterns. In Journal of the American
Medical Informatics Association.
Jaro, M. A., 1995. Probabilistic linkage of large public
health data files. In Statistics in medicine.
Winkler, W. E., 1999. The state of record linkage and
current research problems. In Statistical Research
Division, US Census Bureau.
Kondrak, G., 2005. N-gram similarity and distance. In
String Processing and Information Retrieval. Springer,
Berlin, Heidelberg.
Jaccard, P., 1912. The distribution of the flora in the alpine
zone. In New Phytologist.
Moreau, E., Yvon, F., Cappe, O., 2008. Robust similarity
measures for named entities matching. In Proceedings
of the 22nd International Conference on Computational
Linguistics.
Cohen, W., Ravikumar, P, Fienberg S., 2003. A comparison
of string metrics for matching names and records. In
Proceedings of the KDD workshop on data cleaning
and object consolidation.
Alnazzawi, N., Thompson, P., Ananiadou, S., 2016.
Mapping Phenotypic Information in Heterogeneous
Textual Sources to a Domain-Specific Terminological
Resource. In PLOS ONE 11.
USDA, Food Composition Database, accessed April 2017.
https://ndb.nal.usda.gov/ndb/.
NDB API, accessed April 2017. https://ndb.nal.usda.gov/
ndb/doc/index.
Development Core Team, 2008. R: A language and
environment for statistical computing. http://www.R-
project.org.
RStudio Team, 2015. RStudio: Integrated Development for
R. https://www.rstudio.com/. RStudio Inc., Boston. R
Foundation for Statistical Computing, Vienna.
Eftimov, T., Koroušić-Seljak, B., Korošec, P., 2017. A rule-
based Named-entity Recognition Method for
Knowledge Extraction of Evidence-based Dietary
Recommendations. In PLOS ONE.
Eftimov, T., Korošec, P., Koroušić-Seljak, B., 2017.
StandFood: Standardization of Foods Using a Semi-
Automatic System for Classifying and Describing
Foods According to FoodEx2. In Nutrients.
Eftimov, T., Koroušić-Seljak, B., 2017. POS Tagging-
probability Weighted Method for Matching the Internet
Recipe Ingredients with Food Composition Data. In
Proceedings of the 7th International Joint Conference
on Knowledge Discovery, Knowledge Engineering and
Knowledge Management (IC3K 2015).
Van der Loo, M., Van der Laan, J., Logan, N., 2016.
Approximate String Matching and String Distance
Functions.
Leida, M., Ceravolo, P., Damiani, E., Cui, Z., Gusmini, A.,
2010. Semantics-aware matching strategy (SAMS) for
the Ontology meDiated Data Integration (ODDI). In
Int. J. Knowledge Engineering and Soft Data
Paradigms, Vol. 2, No. 1.
Kerzazi, A., Navas-Delgado, I., F.Aldana-Montes, J., 2009.
Towards an Ontology-based Mediation Framework for
Integrating Biological Data. In SWAT4LS.
