
Automated Diagnosis of Breast Cancer and Pre-invasive Lesions on
Digital Whole Slide Images
Ezgi Mercan
1
, Sachin Mehta
2
, Jamen Bartlett
3
, Donald L. Weaver
3
, Joann G. Elmore
4
and Linda G. Shapiro
1
1
Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, WA, U.S.A.
2
Department of Electrical Engineering, University of Washington, Seattle, WA, U.S.A.
3
Department of Pathology, University of Vermont, Burlington, VT, U.S.A.
4
Deparment of Medicine, University of Washington, Seattle, WA, U.S.A.
Keywords:
Breast Pathology, Automated Diagnosis, Histopathological Image Analysis.
Abstract:
Digital whole slide imaging has the potential to change diagnostic pathology by enabling the use of computer-
aided diagnosis systems. To this end, we used a dataset of 240 digital slides that are interpreted and diagnosed
by an expert panel to develop and evaluate image features for diagnostic classification of breast biopsy whole
slides to four categories: benign, atypia, ductal carcinoma in-situ and invasive carcinoma. Starting with a
tissue labeling step, we developed features that describe the tissue composition of the image and the struc-
tural changes. In this paper, we first introduce two models for the semantic segmentation of the regions of
interest into tissue labels: an SVM-based model and a CNN-based model. Then, we define an image feature
that consists of superpixel tissue label frequency and co-occurrence histograms based on the tissue label seg-
mentations. Finally, we use our features in two diagnostic classification schemes: a four-class classification,
and an alternative classification that is one-diagnosis-at-a-time starting with invasive versus benign and ending
with atypia versus ductal carcinoma in-situ (DCIS). We show that our features achieve competitive results
compared to human performance on the same dataset. Especially at the critical atypia vs. DCIS threshold, our
system outperforms pathologists by achieving an 83% accuracy.
1 INTRODUCTION
The importance of early detection in breast can-
cer is well understood and has been emphasized for
decades. Today, regular screenings for certain pop-
ulations are recommended and conducted especially
in developed countries. However, there is a grow-
ing concern in the medical community that the fear
of under-diagnosing a patient leads to over-diagnosis
and contributes to the ever-increasing number of pre-
invasive and invasive cancer cases. Recent findings
indicate that DCIS cases might be over-treated with-
out significantly better outcomes, making diagnostic
errors even more critical for patient care (Park et al.,
2017). Diagnostic errors are alarmingly high, espe-
cially for pre-invasive lesions of the breast. A recent
study showed that the agreement between patholo-
gists and experts for the atypia cases is only 48% (El-
more et al., 2015).
Digital whole slide imaging provides researchers
with an opportunity to study the diagnostic errors and
(a) benign (b) atypia (c) DCIS (d) invasive
cancer
Figure 1: Example images from four diagnostic categories
of our dataset.
develop image features for accurate and efficient di-
agnosis. An automated diagnosis system can assist
pathologists by highlighting diagnostically relevant
regions and image features associated with the malig-
nancy, therefore providing unbiased and reproducible
feedback. The success of a diagnostic support sys-
tem depends on the descriptive power of the image
features considering the complexity that the full spec-
trum of diagnoses that the breast biopsies present.
The majority of breast cancers are ductal, i.e.
cancer of the breast ducts. The breast ducts are
60
Mercan, E., Mehta, S., Bartlett, J., Weaver, D., Elmore, J. and Shapiro, L.
Automated Diagnosis of Breast Cancer and Pre-invasive Lesions on Digital Whole Slide Images.
DOI: 10.5220/0006550600600068
In Proceedings of the 7th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2018), pages 60-68
ISBN: 978-989-758-276-9
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

pipe-like structures that are responsible for produc-
ing and delivering the milk. Breast biopsies are two-
dimensional cross-sections of the breast tissue, and a
healthy duct appears as a circular arrangement of two
layers of epithelial cells. These cells can proliferate
in different degrees and change the structure of the
ducts. The diagnosis of the biopsy depends on the
correct interpretation of these cellular and structural
changes. Figure 1 shows example breast biopsy im-
ages from our dataset with different diagnoses, that
range from benign tissue to invasive cases.
We propose an automated diagnosis system for
breast tissue using digital whole slides images (WSIs)
of breast biopsies. To this end, we used a dataset of
breast WSIs with a wide range of diagnoses from be-
nign to invasive cancer. The first step of our diagnosis
pipeline is the semantical segmentation of biopsy im-
ages into different tissues. We then extracted features
from the semantic masks that describe the distribution
and arrangement of the tissue in the image. Finally,
we trained classifiers with our tissue-label-based fea-
tures for automated diagnosis of the biopsy images.
Our experimental results suggest that the features ex-
tracted from the semantic masks have high descriptive
power and helps in differentiating a wide range of di-
agnoses, from benign to invasive.
Our work on automated diagnosis of breast biopsy
images considers the full spectrum of diagnoses en-
countered in clinical practice. We designed a novel
semantic segmentation scheme with a set of tissue
labels developed for invasive cancer as well as pre-
invasive lesions of the breast. The semantic seg-
mentation gives us a powerful abstraction for diag-
nostic classification, so that even simple features ex-
tracted from the segmentation masks can achieve re-
sults comparable to the diagnostic accuracy of the ac-
tual pathologists.
2 RELATED WORK
Automated Diagnosis. Automated malignancy de-
tection is a well-studied area in the histopathological
image analysis literature. Most of the related work
focuses on the detection of cancer in a binary classi-
fication setting with only malignant and benign cases
(Chekkoury et al., 2012; Doyle et al., 2012; Tabesh
et al., 2007). These methods do not take pre-invasive
lesions or other diagnostic categories into account,
which limits their use in real-world scenarios. There
is also limited research on analyzing images for sub-
type classification (Kothari et al., 2011) or stromal de-
velopment (Sertel et al., 2008) using only tumor im-
ages.
Recently, some researchers have begun to study
the pre-invasive lesions of the breast: (Dong et al.,
2014) reports promising results in discrimination of
benign proliferations of the breast from malignant
ones. They extract 392 features corresponding to
the mean and standard deviation in nuclear size and
shape, intensity and texture across 8 color channel,
and apply L1-regularized logistic regression to build
discriminative models. Their dataset contains only
usual ductal hyperplasia (UDH), which maps to be-
nign in our dataset, and ductal carcinoma in-situ
(DCIS) cases. To our best knowledge, there is no
study that considers the full spectrum of pre-invasive
lesions of the breast including atypia (atypical duc-
tal hyperplasi and atypical lobular hyperplasia). Our
study is the first of its kind to attempt a diagnostic
classification with categories from benign to invasive
cancer.
CNNs for Medical Image Analysis. Convolutional
neural networks (CNNs) have been successfully ap-
plied for medical image analysis. Most notable
among is: classifying WSI into tumor subtypes and
grades (Hou et al., 2016), segmenting EM images
(Ronneberger et al., 2015), segmenting gland im-
ages (Chen et al., 2017), and segmenting brian im-
ages (Fakhry et al., 2017). (Hou et al., 2016) apply
a sliding-window approach to reduce the WSI size
and combine predictions made on patches to classify
WSIs. Their work exploits WSI characteristics such
as the heterogeneity of tissue in terms of tumor grades
and subtypes. (Ronneberger et al., 2015), (Chen et al.,
2017), and (Fakhry et al., 2017) follow an encoder-
decoder network approach with skip-connections for
segmenting medical images. For semantic segmenta-
tion using CNN, we use the recently proposed resid-
ual encoder-decoder network by (Fakhry et al., 2017).
3 DATASET
3.1 Breast Biopsy Whole Slide Images
240 breast biopsies were selected from the Breast
Cancer Surveillance Consortium (http://www.bcsc-
research.org/) archives in New Hampshire and Ver-
mont for our studies. The final dataset spans a wide
spectrum of breast diagnoses that are mapped to four
categories: benign, atypia, ductal carcinoma in-situ
(DCIS) and invasive cancer.
The original H&E (heamatoxylin and eosin)
stained glass slides were scanned to produce whole
slide images using an iScan CoreoAu
R
in 40X mag-
nification. Quality control was conducted by a tech-
Automated Diagnosis of Breast Cancer and Pre-invasive Lesions on Digital Whole Slide Images
61

Table 1: Distribution of the diagnostic categories based on
expert consensus.
Diagnostic Category # of Cases # of ROIs
Benign 60 102
Atypia 80 128
DCIS 78 162
Invasive cancer 22 36
Total 240 428
nician and an experienced breast pathologist to obtain
the highest quality. The final average image size for
the 240 digital slides was 90,000 × 70,000 pixels.
3.2 Expert Consensus Diagnoses and
Regions of Interest
Each digital slide was first interpreted and diagnosed
by three expert pathologists individually. Each ex-
pert provided a diagnosis and a region of interest
(ROI) that supports the diagnosis for each digital
slide. Following the individual interpretations, sev-
eral in-person and webinar meetings were held to pro-
duce an expert-consensus diagnosis and one or more
expert consensus ROIs for each case. Since some
cases had more than one ROI per WSI, the final set
of expert consensus ROIs includes 102 benign, 128
atypia, 162 DCIS and 36 invasive samples. Table 1
summarizes the data. For a detailed explanation of
the development of the cases and the expert consen-
sus data, please see (Oster et al., 2013) and (Allison
et al., 2014).
3.3 Tissue Labels
To describe the structural changes that lead to can-
cer in the breast tissue, we produced a set of eight
tissue labels in collaboration with an expert patholo-
gist: background, benign epithelium, malignant ep-
ithelium, normal stroma, desmoplastic stroma, secre-
tion, blood, and necrosis.
The epithelial cells in the benign and atypia cat-
egories were labeled as benign epithelium, whereas
the epithelial cells from the DCIS and invasive can-
cer categories were labeled with the malignant ep-
ithelium label. Compared to benign cells, the cells in
the malignant epithelium are bigger and irregular in
shape. Stroma is a term used for the connective tissue
between the ductal structures in the breast. In some
cases, stromal cells proliferate in response to cancer.
We used desmoplastic stroma and normal stroma la-
bels for the stroma associated with the tumor and reg-
ular breast stroma, respectively. Since breast ducts
are glands responsible for producing the milk, they
are sometimes filled with molecules discharged from
the cells. The secretion label was used to mark this
benign substance filling the ducts. The label necrosis
was used to mark the dead cells at the center of the
ducts in the DCIS and invasive cases. The blood label
was used to mark the blood cells, which are rare but
have a very distinct appearance. Finally, the pixels
that do not contain any tissue, including the empty
areas inside the ducts, were labeled as background.
Figure 2 illustrates the eight tissue labels marked by
the pathologist.
Although some of the labels are not important in
diagnostic interpretation, our tissue labels were in-
tended to cover all the pixels in the images. Due to
the expertise needed for labeling and the large sizes of
the WSIs, we randomly selected a subset of 40 cases
(58 ROIs). These 58 ROIs are annotated by a pathol-
ogist into eight tissue labels. Figure 2. shows three
example images and the pixel labels provided by a
pathologist.
4 METHODOLOGY
4.1 Semantic Segmentation
Segmenting WSIs of breast biopsies into building
blocks is crucial to understand the structural changes
that lead to diagnosis. Semantic segmentation masks
can provide important information about the distribu-
tion and arrangement of different tissue types.
We used a supervised machine learning approach
to semantically segment H&E images into eight tis-
sue labels. We trained and evaluated two models:
(1) an SVM-based model that starts with a super-
pixel segmentation and assigns each superpixel a tis-
sue label based on color and texture features, and (2)
a CNN-based model with a sliding-window approach
that classifies each pixel in a sliding-window into a
background
benign epithelium
malignant epithelium
normal stroma
desmoplastic stroma
secretion
blood necrosis
Figure 2: The set of tissue labels used in semantic segmen-
tation: (top row) three example cases from the dataset and
(bottom row) the pixel labels provided by a pathologist.
ICPRAM 2018 - 7th International Conference on Pattern Recognition Applications and Methods
62

label class simultaneously.
4.1.1 Segmentation using SVM
We used the SLIC algorithm (Achanta et al., 2012) to
segment H&E images into superpixels of size 3000
pixel. From each superpixel, we extracted L*a*b*
color histograms and LBP texture histograms (He and
Wang, 1990). We calculated the texture histograms
from the H&E channels, which were obtained through
a color deconvolution algorithm (Ruifrok and John-
ston, 2001).
The size of the superpixels, 3,000 pixels, was se-
lected to have approximately one or two epithelial
cells in one superpixel so that the detailed structures
of the ducts could be captured. Our preliminary ex-
periments showed that some individual superpixels
were misclassified, while their neighbors were cor-
rectly classified. To improve the classification, we
included two circular neighborhoods around each su-
perpixel in feature extraction. The color and texture
histograms calculated from the superpixels and circu-
lar neigborhoods were concatenated to produce one
feature vector for each superpixel. Figure 3 illustrates
the two circular neighborhoods from which the same
features were extracted and appended to superpixel
feature vector.
(a) superpixel segmentation (b) neighborhoods
Figure 3: Initial superpixel segmentation and the circular
neighborhoods used to increase the superpixel classification
accuracy for supervised segmentation.
We used the concatenated color and texture his-
tograms to train an SVM model that classifies super-
pixels into eight tissue labels. To address the non-
uniform distribution of the tissue labels and ROI size
variation, we sampled 2000 superpixels for each of
the eight labels (if possible) from each ROI. We eval-
uated the performance of the SVM-based model in
10-fold cross-validation experiments on the subset of
ROIs labeled by the pathologist (N=58). For the di-
agnostic classification, we trained a final SVM model
with the samples from all folds and applied the model
to the full dataset (N=428 ROIs) to obtained tissue la-
bel segmentations.
4.1.2 Segmentation using CNN
The CNN experiments were an attempt to improve
the semantic segmentation performance achieved by
the feature-based SVM methodology. Rather than us-
ing features, the CNN learns to recognize the patterns
from the image itself. Following the work of (Fakhry
et al., 2017), we implemented a residual encoder-
decoder network. The encoding network transforms
an input image into a feature vector space by stack-
ing a series of encoding blocks. The decoding net-
work transforms the feature vector space into a se-
mantic mask by stacking a series of decoding blocks.
The residual connection between the encoding and its
corresponding decoding block give chance to each in-
termediate block to represent the information inde-
pendent of the blocks at any other spatial level. See
(Mehta et al., 2017) for more details about the CNN
network.
We split 58 ROIs into training (30 ROIs) and test
(28 ROIs) sets keeping the distribution of diagnos-
tic categories similar. We used a sliding-window
approach to create samples for training and testing
the CNN architecture. We cropped 256 × 256 pixel
patches at different resolutions, resulting in 5,312
patches from 30 ROIs. We augmented the data using
random rotations (between 5 and 10 degree), horizon-
tal flips, and random crops followed by scaling (i.e.
the crop border was selected randomly between 20
and 50 pixels), resulting in a total of 25,992 patches
that were split into the training and validation sets us-
ing 90:10 split ratio. We trained all of our models end-
to-end using stochastic gradient descent with a fixed
learning rate of 0.0001, momentum of 0.9, weight
decay of 0.0005, and a batch size of 10 on a single
NVIDIA GTX-1080 GPU.
4.2 Tissue Label Frequency and
Co-occurrence Histograms
One of the basic visual differences between diagnos-
tic categories is the existence and amount of different
tissue types. To this end, we calculated frequency his-
tograms for the superpixel labels. However, only the
distribution of the tissue types is not enough to de-
scribe complex spatial relationships. Co-occurrence
histograms, on the other hand, can capture the fre-
quency of the contact between the superpixels with
different tissue labels.
We normalized all histograms to remove the effect
of the size. Since the background was one of the tis-
sue labels, the amount of background affects the his-
togram bins of other tissue types, yet the amount of
background may not be important in diagnostic clas-
Automated Diagnosis of Breast Cancer and Pre-invasive Lesions on Digital Whole Slide Images
63
sification. We created and studied two alternative ver-
sions to all features by removing the background bin,
and then also removing the stroma bin from the his-
tograms before normalization.
4.3 Diagnostic Classification
The diagnostic decision making process is complex.
Pathologists interpret the slides at different resolu-
tions and make decisions about different diagnoses.
For example, the decision to diagnose an invasive car-
cinoma is usually made at a lower resolution, where
a high-level organization of the tissue is available to
the observer. On the other hand, the decision between
atypia and DCIS is made at a higher resolution by ex-
amining structural and cellular changes. Inspired by
this observation, we designed a classification scheme
where a decision is made for one diagnosis at a time.
We designed a set of experiments to test two diag-
nostic classification schemes: (1) A model that classi-
fies an ROI into one of the four diagnostic categories,
and (2) a model that eliminates one diagnosis at a time
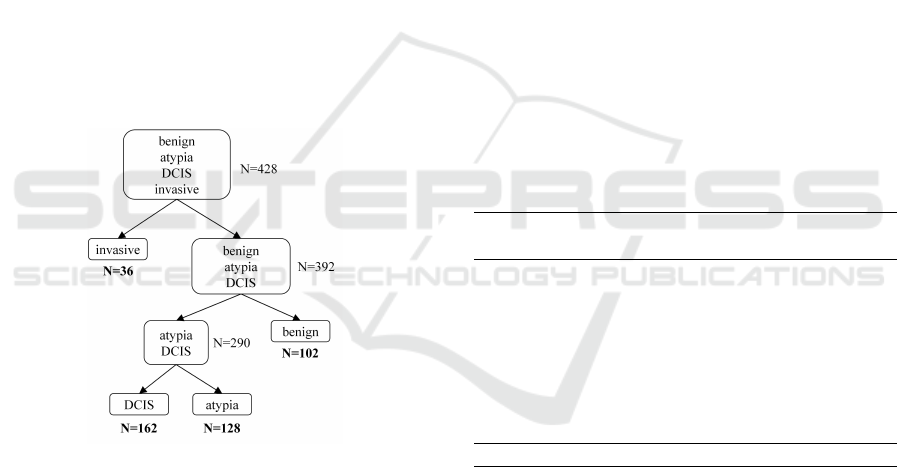
(Figure 4).
Figure 4: One-diagnosis-at-a-time classification.
In diagnostic classification experiments, we used
all expert consensus ROIs (N=428) as described in
Section 3.1. For the 4-diagnosis classification, we
trained an SVM using all samples. For the sec-
ond classification scheme (Figure 4), we trained three
SVMs: 1) invasive vs. not-invasive, using all sam-
ples; 2) atypia and DCIS vs. benign, using benign,
atypia and DCIS samples; 3) DCIS vs. atypia, using
atypia and DCIS samples. When the sample size was
smaller than the number of features, we applied prin-
cipal components analysis (PCA) and used the first
20 principal components to reduce the number of fea-
tures. For all experiments, we trained SVMs in a 10-
fold cross-validation setting using each ROI separa-
tely as a sample. We sub-sampled the training data
to have an equal number of samples for each class.
To remove the effect of sub-sampling, we repeated all
experiments 100 times and reported the average accu-
racies.
5 RESULTS
We evaluated both the tissue label segmentation and
diagnostic classification tasks. Note that any error
produced by the segmentation is propagated to the di-
agnostic classification, since the features used for di-
agnosis are based on tissue labels.
5.1 Tissue Label Segmentation
We evaluated the performances of the SVM method
and the CNN method by comparing the predicted
pixel labels with the ground truth pixel labels pro-
vided by the pathologist. We report precision and re-
call metrics for both models on the test set of 20 ROIs
in Table 2 and confusion matrices in Figure 5.
Table 2: Tissue label segmentation results: Individual label
and average precision and recall values for the SVM-based
and CNN-based supervised segmentations.
Precision Recall
Tissue Label SVM CNN SVM CNN
background .86 .81 .89 .93
benign epi .23 .39 .46 .72
malignant epi .63 .86 .48 .61
normal stoma .68 .28 .24 .88
desm. stroma .63 .72 .61 .20
secretion .01 .32 .24 .49
necrosis .03 .09 .24 .59
blood .35 .52 .46 .83
Average .43 .50 .45 .66
The CNN model performed better than the SVM
method in every label, other than the desmoplastic
stroma label. The CNN method performed especially
well with the rare labels of secretion, blood and necro-
sis with high precision and recall values. However, it
suffers from a low precision, high recall of the nor-
mal stroma label. This may be due to predicting the
majority of the desmoplastic stroma pixels as normal
stroma (See Figure 5).
5.2 Diagnostic Classification
Table 3 shows the average accuracy, (t p +tn)/(t p +
tn + f p + f n), sensitivity, t p/(t p + f n), and speci-
ficity, tn/(tn + f p), values for the three variations of
ICPRAM 2018 - 7th International Conference on Pattern Recognition Applications and Methods
64

background
benign epi
malignant epi
normal
strm
desmopl
strm
secretion
necrosis
blood
Prediction
background
benign epi
malignant epi
normal
strm
desmopl
strm
secretion
necrosis
blood
Prediction
background
Ground Truth
benign epi
malignant epi
normal
strm
desmopl
strm
secretion
necrosis
blood
.89
.01
.01
.00
.02
.07
.01
.00
.01
.46
.36
.01
.08
.03
.04
.00
.02
.12
.48
.00
.22
.15
.00
.00
.04
.01
.06
.24
.42
.18
.02
.03
.04
.03
.17
.03
.61
.11
.01
.00
.13
.02
.07
.18
.16
.24
.08
.13
.12
.04
.13
.00
.26
.20
.24
.01
.03
.01
.05
.13
.23
.05
.04
.46
.93
.01
.01
.03
.01
.01
.00
.00
.01
.72
.16
.09
.01
.01
.00
.00
.06
.09
.61
.13
.07
.02
.03
.00
.03
.02
.01
.88
.05
.00
.00
.01
.06
.02
.05
.66
.20
.00
.00
.01
.15
.06
.05
.07
.02
.49
.09
.07
.10
.01
.11
.01
.01
.15
.59
.01
.01
.01
.01
.10
.04
.00
.00
.83
(a) SVM (b) CNN
Figure 5: Confusion matrices for both SVM-based and
CNN-based models for the eight-label semantic segmenta-
tion task on 20 test ROIs.
background
benign epithelium
malignant epithelium
normal stroma
desmoplastic stroma
secretion
blood necrosis
(a) RGB (b) Ground (c) SVM (d) CNN
Truth
Figure 6: Visualizations of the segmentations produced by
the SVM and CNN using the eight tissue labels: (a) input
image, (b) ground truth labels, (c) the prediction of the SVM
model, (d) the prediction of the CNN model.
tissue label frequency and co-occurrence histograms
with two different segmentation techniques in four
classification tasks, where t p is the number of true
positives, f p is the number of false positives, tn is
the number of true negatives, and f n is the number
of false negatives. Although accuracy is the more
common metric for evaluating classification perfor-
mance, sensitivity and specificity are two metrics that
are widely used in the evaluation of diagnostic tests.
They measure the true positive and true negative rates
of a condition. Sensitivity quantifies the absence of
false negatives, while specificity quantifies the ab-
sence of false positives. Since our dataset is unbal-
anced for different diagnostic classes, we report the
sensitivity and specificity metrics to illustrate the per-
formance of our automated diagnosis experiments.
The four-class classification setting obtains a max-
imum of 0.46 accuracy using all tissue labels with the
CNN-based segmentation. In comparison, one-
diagnosis-at-a-time setting achieves accuracies of
0.94, 0.70 and 0.83 for the differentiation of invasive,
benign and DCIS respectively.
The accuracies are higher for the CNN-based seg-
mentation method as expected, except for the differ-
entiation of atypia and DCIS from benign. However,
the sensitivity and specificity for the experiment with
all tissue labels with SVM-based segmentation are not
as high as the other experiments. The experiment with
no background or stroma with CNN-based segmenta-
tion achieves higher sensitivity (0.94) and specificity
(0.39) despite the low accuracy value (0.60). This is
likely due to the class imbalance between the benign
and non-benign samples.
Removing the background label improves the dif-
ferentiation of invasive from non-invasive lesions;
however, removing the stroma label results in lower
accuracy. The best result for differentiation of inva-
sive is achieved with tissue labels with no background
using CNN-based segmentation. Removing both the
background and stroma labels improves the accuracy
of classification DCIS vs. atypia. Tissue label his-
tograms with no background or stroma with CNN-
based segmentation achieves accuracy of 0.83, sen-
sitivity of 0.88 and specificity of 0.78.
6 DISCUSSION
SVM vs CNN for Segmentation
The CNNs outperformed many of the traditional mod-
els composed of a classifier and hand-crafted image
features in classification, detection and segmentation
tasks. Our experiments showed that it is possible
to obtain a performance boost by using an encoder-
decoder architecture specifically designed for the
breast biopsy images. Although the improvement
seems small, the contribution of CNNs was in the
classes of necrosis, epithelium and stroma, which are
important for distinguishing and classifying DCIS.
The differentiation between necrosis and secretion
might be especially critical in diagnosis.
Furthermore, none of the quantitative measure-
ments evaluated the smooth object boundaries ob-
tained by the CNNs. Because the CNN-based meth-
ods were trained with patches that are 500 times big-
ger than a superpixel, they were able to learn the
structure and segment smooth borders of the objects
as it can be seen in visualizations in Figure 6.
Automated Diagnosis of Breast Cancer and Pre-invasive Lesions on Digital Whole Slide Images
65

Table 3: Comparison of features with different tissue histograms and segmentations for the diagnostic classification tasks.
Accuracy Sensitivity Specificity
Tissue Labels SVM CNN SVM CNN SVM CNN
4-class
All labels .32 .46 - - - -
No background .39 .44 - - - -
No background or stroma .33 .42 - - - -
Invasive vs. Benign-Atypia-DCIS
All labels .48 .82 .13 .24 .97 .95
No background .62 .94 .15 .70 .97 .95
No background or stroma .54 .69 .12 .17 .96 96
Atypia-DCIS vs. Benign
All labels .70 .51 .79 .88 .41 .33
No background .65 .43 .80 .94 .37 .31
No background or stroma .69 .60 .83 .94 .42 .39
DCIS vs. Atypia
All labels .68 .71 .73 .67 .63 .93
No background .65 .78 .62 .75 .92 .85
No background or stroma .72 .83 .70 .88 .76 .78
Importance of Stroma in Diagnosis
When the stroma label was not used in feature calcu-
lations, the accuracy for the classification of invasive
cases dropped indicating the importance of stroma in
differentiation of breast tumors. By encoding two dif-
ferent types of stroma, we incorporated an important
visual cue used by pathologists when diagnosing in-
vasive carcinomas. Our findings are consistent with
the existing literature that showed the importance of
stroma not only in diagnosis, but also in the prediction
of survival time (Beck et al., 2011).
Similarly, removing stroma bins from the his-
tograms did not improve the classification of the
atypia and DCIS cases from the benign proliferations;
however, the difference was not significant in better
performing SVM-segmented features.
Removing stroma improved the classification ac-
curacy between DCIS and atypia, as expected. Since
both lesions are mostly restricted to ductal structures
and the diagnosis is made using cellular features and
the degree of structural changes in the ducts, the most
important feature for this task was the epithelial tis-
sue labels and their frequency and co-occurrence with
other labels. It is likely that removing stroma acted as
a noise filtering (or reduction); thereby learning more
relevant features.
Atypia vs. DCIS
Automated diagnosis of pre-invasive lesions of breast
is an understudied problem mostly due to the lack
of comprehensive datasets and difficulty of the prob-
lem. The differentiation between DCIS, atypical pro-
liferations and benign proliferations is a hard prob-
lem even for humans, yet the distinction between
two categories could alter the treatment of the patient.
Although both diagnoses are associated with higher
risks of developing invasive breast cancer, it is not
uncommon to treat high grade DCIS cases with oral
chemotherapy and even surgery while atypia cases
are usually followed up with additional screenings.
Features based on frequency and co-occurence his-
tograms of tissue labels were able to capture the vi-
sual characteristics of the breast tissue and achieved
good results.
In a previous study, a group of pathologists inter-
preted slides of breast biopsies (Elmore et al., 2015).
The calculated accuracies from the provided confu-
sion matrix are 70%, 98%, 81% and 80% for the tasks
of 4-class, Invasive vs. Benign-Atypia-DCIS, Atypia-
DCIS vs. Benign and DCIS vs. Atypia respectively.
For the same task, our fully automated pipeline accu-
racies are comparable to the actual pathologists. Our
method outperform the pathologists by 3% on the task
of differentiating DCIS from atypia cases.
ICPRAM 2018 - 7th International Conference on Pattern Recognition Applications and Methods
66

Sensitivity and Specificity
The classifier achieves a high accuracy (94%) for the
differentiation of invasive cases from non-invasive le-
sions (benign, atypia and DCIS). For this setting,
relatively low sensitivity (70%) and high specificity
(95%) indicates that the classifier model is produc-
ing more false negatives than false positives. In other
words, the automated system is somewhat under-
diagnosing for the invasive cancer. This might be due
to the limited number of invasive cases in our dataset
in comparison to other diagnostic classes. Further-
more, the invasive samples include some difficult-to-
differentiate cases like micro-invasions. Also, auto-
mated cancer detection is a well-studied problem in
which researchers setup a binary classification prob-
lem with invasive cases and non-invasive cases. Our
contribution in this work is the exploration of pre-
invasive lesions of breast in automated diagnosis set-
ting.
The high sensitivity (79%) and low specificity
(41%) values for the classification of atypia-DCIS and
benign indicates, on the other hand, a clear overdiag-
nosis. This is not as alarming as an underdiagnosis in
the scope of a computer aided diagnosis tool, consid-
ering an overdiagnosed benign case can be caught by
the pathologist and corrected.
Finally, for the DCIS vs. atypia task, our classifier
achieves very good sensitivity and specificity scores,
88% and 78%, respectively.
7 CONCLUSIONS
We aimed to develop image features that can describe
the diagnostically important visual characteristics of
the breast biopsy images. We took an approach that is
motivated by the pathologists’ decision making pro-
cess. We first segment images into eight tissue types
that we determined important for the diagnosis using
two different methods: an SVM-based approach that
uses color and texture features to classify superpix-
els to produce a tissue labeling, and a CNN-based ap-
proach that uses raw images. Then we calculate tissue
label frequency and co-occurrence histograms based
on superpixel segmentation to classify images into
diagnostic categories. In classification, we compare
two schemes: we train an SVM classifier to classify
images into four diagnostic categories and we train
a series of SVM classifiers to classify images one-
diagnosis-at-a-time. Our proposed one-diagnosis-at-
a-time strategy proved to be more accurate, since it
allows classifier to learn different features for differ-
ent diagnostic categories.
Our strategy of diagnosis by elimination is in-
spired by the diagnostic decision making process of
pathologists and it produced comparable accuracies to
humans. Especially at the border of DCIS and atypia,
our features achieve an accuracy of 83% which is 3%
higher than that of pathologists reported on the same
dataset.
We implemented the simple tissue label frequency
and co-occurrence features to demonstrate the power
of semantic segmentation in diagnosing breast can-
cer. Our ongoing work focuses on developing more
sophisticated features based on tissue labels that can
capture the specific structural changes in the breast.
ACKNOWLEDGEMENTS
Research reported in this publication was sup-
ported by the National Cancer Institute awards R01
CA172343, R01 CA140560 and RO1 CA200690.
The content is solely the responsibility of the authors
and does not necessarily represent the views of the
National Cancer Institute or the National Institutes
of Health. We thank Ventana Medical Systems, Inc.
(Tucson, AZ, USA), a member of the Roche Group,
for the use of iScan Coreo Au
TM
whole slide imaging
system, and HD View SL for the source code used to
build our digital viewer. For a full description o f HD
View SL, please see http://hdviewsl.codeplex.com/.
REFERENCES
Achanta, R., Shaji, A., Smith, K., Lucchi, A., Fua, P., and
S
¨
usstrunk, S. (2012). SLIC superpixels compared to
state-of-the-art superpixel methods. IEEE Transac-
tions on Pattern Analysis and Machine Intelligence,
34(11):2274–2281.
Allison, K. H., Reisch, L. M., Carney, P. A., Weaver, D. L.,
Schnitt, S. J., O’Malley, F. P., Geller, B. M., and El-
more, J. G. (2014). Understanding diagnostic variabil-
ity in breast pathology: lessons learned from an expert
consensus review panel. Histopathology, 65(2):240–
251.
Beck, A. H., Sangoi, A. R., Leung, S., Marinelli, R. J.,
Nielsen, T. O., van de Vijver, M. J., West, R. B., van de
Rijn, M., and Koller, D. (2011). Systematic analy-
sis of breast cancer morphology uncovers stromal fea-
tures associated with survival. Science Translational
Medicine, 3(108):108ra113–108ra113.
Chekkoury, A., Khurd, P., Ni, J., Bahlmann, C., Kamen, A.,
Patel, A., Grady, L., Singh, M., Groher, M., Navab,
N., Krupinski, E., Johnson, J., Graham, A., and We-
instein, R. (2012). Automated malignancy detection
in breast histopathological images. In SPIE Medi-
cal Imaging, volume 8315, page 831515. International
Society for Optics and Photonics.
Automated Diagnosis of Breast Cancer and Pre-invasive Lesions on Digital Whole Slide Images
67

Chen, H., Qi, X., Yu, L., Dou, Q., Qin, J., and Heng, P.-
A. (2017). DCAN: Deep contour-aware networks for
object instance segmentation from histology images.
In Medical Image Analysis, volume 36, pages 135–
146.
Dong, F., Irshad, H., Oh, E. Y., Lerwill, M. F., Brach-
tel, E. F., Jones, N. C., Knoblauch, N. W., Montaser-
Kouhsari, L., Johnson, N. B., Rao, L. K. F., Faulkner-
Jones, B., Wilbur, D. C., Schnitt, S. J., and Beck, A. H.
(2014). Computational pathology to discriminate be-
nign from malignant intraductal proliferations of the
breast. PLoS ONE, 9(12):e114885.
Doyle, S., Feldman, M., Tomaszewski, J., and Madabhushi,
A. (2012). A Boosted Bayesian Multiresolution Clas-
sifier for Prostate Cancer Detection From Digitized
Needle Biopsies. IEEE Transactions on Biomedical
Engineering, 59(5):1205–1218.
Elmore, J. G., Longton, G. M., Carney, P. A., Geller, B. M.,
Onega, T., Tosteson, A. N. A., Nelson, H. D., Pepe,
M. S., Allison, K. H., Schnitt, S. J., O’Malley, F. P.,
and Weaver, D. L. (2015). Diagnostic Concordance
Among Pathologists Interpreting Breast Biopsy Spec-
imens. JAMA, 313(11):1122.
Fakhry, A., Zeng, T., and Ji, S. (2017). Residual Deconvolu-
tional Networks for Brain Electron Microscopy Image
Segmentation. IEEE Transactions on Medical Imag-
ing, 36(2):447–456.
He, D. C. and Wang, L. (1990). Texture unit, texture spec-
trum, and texture analysis. IEEE Transactions on
Geoscience and Remote Sensing, 28(4):509–512.
Hou, L., Samaras, D., Kurc, T. M., Gao, Y., Davis, J. E., and
Saltz, J. H. (2016). Patch-based convolutional neural
network for whole slide tissue image classification. In
CVPR.
Kothari, S., Phan, J. H., Young, A. N., and Wang, M. D.
(2011). Histological Image Feature Mining Reveals
Emergent Diagnostic Properties for Renal Cancer. In
2011 IEEE International Conference on Bioinformat-
ics and Biomedicine, pages 422–425.
Mehta, S., Mercan, E., Bartlett, J., Weaver, D., Elmore, J.,
and Shapiro, L. (2017). Learning to Segment Breast
Biopsy Whole Slide Images. ArXiv e-prints.
Oster, N. V., Carney, P. A., Allison, K. H., Weaver, D. L.,
Reisch, L. M., Longton, G., Onega, T., Pepe, M.,
Geller, B. M., Nelson, H. D., Ross, T. R., Tosteson, A.
N. A., and Elmore, J. G. (2013). Development of a di-
agnostic test set to assess agreement in breast pathol-
ogy: practical application of the Guidelines for Re-
porting Reliability and Agreement Studies (GRRAS).
BMC Women’s Health, 13(1):3.
Park, H. L., Chang, J., Lal, G., Lal, K., Ziogas, A., and
Anton-Culver, H. (2017). Trends in treatment pat-
terns and clinical outcomes in young women diag-
nosed with ductal carcinoma in situ. Clinical Breast
Cancer.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-Net:
Convolutional Networks for Biomedical Image Seg-
mentation.
Ruifrok, A. C. and Johnston, D. A. (2001). Quantifica-
tion of histochemical staining by color deconvolution.
Analytical and Quantitative Cytology and Histology,
23(4):291–299.
Sertel, O., Kong, J., Shimada, H., Catalyurek, U., Saltz,
J. H., and Gurcan, M. N. (2008). Computer-aided
prognosis of neuroblastoma: classification of stromal
development on whole-slide images. Pattern Recog-
nition, 6915(6):69150P.
Tabesh, A., Teverovskiy, M., Pang, H.-Y., Kumar, V. P., Ver-
bel, D., Kotsianti, A., and Saidi, O. (2007). Multifea-
ture Prostate Cancer Diagnosis and Gleason Grading
of Histological Images. IEEE Transactions on Medi-
cal Imaging, 26(10):1366–1378.
ICPRAM 2018 - 7th International Conference on Pattern Recognition Applications and Methods
68
