
Quisper Ontology Learning from Personalized Dietary Web Services
Tome Eftimov
1
, Gordana Ispirova
1, 2
, Paul Finglas
3
, Peter Koro
ˇ
sec
1, 4
and Barbara Korou
ˇ
si
´
c Seljak
1
1
Computer Systems Department, Jo
ˇ
zef Stefan Institute, Jamova cesta 39, 1000 Ljubljana, Slovenia
2
Jo
ˇ
zef Stefan International Postgraduate School, Jamova cesta 39, 1000 Ljubljana, Slovenia
3
Quadram Institute Bioscience, Norwich research Park, Norwich, Norfolk, NR4 7UA, U.K.
4
Faculty of Mathematics, Natural Sciences and Information Technologies, Glagolja
ˇ
ska ulica 8, 6000 Koper, Slovenia
Keywords:
E-health, Ontology Learning, Part of Speech Tagging, Personalized Dietary Web Service, Semantic Web,
String Similarity.
Abstract:
Unhealthy diet can lead to diseases such as diabetes, allergies, and some types of cancer, among other health-
related problems. In order to help users and clinical dietitians access the relevant knowledge about food and
nutrition data in e-health systems that use different data sources, ontologies about food and related domains,
such as clinical medicine, individual user profile, etc., are very important in providing successful and smart e-
health systems. In this paper we present an ontology-learning process using personalized dietary web services
that are dealing with food-related data and knowledge rules. The result of the ontology-learning process
is an OWL ontology that is developed in a semi-automatic way and can be used for the harmonization of
personalized dietary web services and will enable researchers to share information in this domain. In addition,
it can also use aggregated data from different sources to provide new knowledge and help people live healthier
lives.
1 INTRODUCTION
There is clear evidence that eating a healthy diet can
prevent diet-related chronic diseases and can increase
the quality of life. Generalized dietary guidelines and
recommendations can help with following a healthy
diet and they are available in books, magazines, tele-
vision programs and the Internet. For example, Di-
etary Reference Intakes (DRIs) are reference values
that are quantitative estimates of nutrient intakes and
can be used for planning and assessing the diet of a
healthy person (Hellwig et al., 2006). In addition,
there are food-composition studies, which are car-
ried out in order to determine the chemical nature
of the principle components in food that affect hu-
man health (Greenfield and Southgate, 2003). Nowa-
days, there are several websites that contain informa-
tion about nutritional principles or provide person-
alized dietary services. For example, the QuaLiFY
project presents the QuaLiFY Server Platform (Quis-
per), which provides access to scientifically validated
data and knowledge rules relevant to personalized
nutritional products and services (http://quisper.eu/).
Quisper is a technology-based platform that aims to
facilitate data exchange and collection, and allows
users to connect to it through an API for the pur-
poses of research, applications or services. The archi-
tecture of Quisper is shown on Figure 1. If Qusiper
web services and other services that provide informa-
tion about nutritional principles share and publish the
same ontology for the terms that they all use, then
information systems can use the extracted and ag-
gregated information from these web services to an-
swer questions or as the input data for other applica-
tions. The main question is how to extract the knowl-
edge from the data sources that use different ways of
describing and classifying the data. In order to do
this there is a need of an ontology for researchers
who need to share information in this domain. There
are a lot of approaches that deal with the ontology-
learning process, which is a process of extracting con-
ceptual knowledge from an input and building an on-
tology from it (Biemann, 2005; Hazman et al., 2011).
Ontology-learning systems, according to the type of
the data from which they are learned, can be catego-
rized as unstructured, semi-structured and structured.
In this paper we present an ontology-learning pro-
cess that uses personalized dietary web services. In
Section II we present the problem in depth. Section
III describes the ontology-learning process in general,
and in Section IV the Quisper ontology, which is the
result of the ontology-learning process, is presented.
Eftimov, T., Ispirova, G., Finglas, P., Korošec, P. and Seljak, B.
Quisper Ontology Learning from Personalized Dietary Web Services.
DOI: 10.5220/0006951302790286
In Proceedings of the 10th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2018) - Volume 2: KEOD, pages 279-286
ISBN: 978-989-758-330-8
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
279

Figure 1: Quisper architecture.
Section V gives some examples and possible uses of
the Quisper ontology. In Section VI we compare the
Quisper ontology with existing food ontologies, and
in Section VII we conclude the paper by discussing
the importance of this ontology.
2 PROBLEM DEFINITION
There are several questions to take into consideration
when we want to learn an ontology using personalized
dietary web services. These web services use input
and output variables and they are explained with text
descriptions by the programmer who implemented the
web service. The first issue is that people use human
languages and write the descriptions of the used terms
in an unstructured form. For example, in different
web services we can find different descriptions for the
same term and different codes that are used, (”Vita-
min B12”, ”Vitamin B-12”), (”carotene, alpha -”, ”al-
pha carotene”). From this we can conclude that there
is a lack of a structured method for representation, and
this happens because of the different ways that peo-
ple express themselves. Another challenge is to cap-
ture terms that are Single Nucleotide Polymorphism
(SNP) data, because in most cases they are used by the
names provided by SNPedia (”rs1205”, ”rs174546”)
(http://www.snpedia.com/), and a description of these
terms is missing; it is provided by a reference (website
or paper). We need to organize the extracted terms in
some taxonomy, and in the end, using the information
about the taxonomy and the relations between some
variables used in the web services, we need to define
the classes of the ontology, the attributes of the classes
called properties, and the restrictions of the proper-
ties. On Figure 2, the problem definition is presented.
Figure 2: Problem definition.
3 ONTOLOGY LEARNING
3.1 POS Tagging-probability Weighted
Method
In order to find the similar terms that are used in
different web services we propose the POS tagging-
probability weighted method. Let D
i
be the set of de-
scriptions of terms that are used in one web service,
with the web service consisting of n terms (they can
be input or output variables).
D
i
= ∪
n
j=1
d
i, j
, (1)
where d
i, j
is a description of the term, and j = 1, .., n.
Because we are working with terms that are related to
chemical names, on each description we used part of
speech tagging to extract nouns, adjectives and num-
bers (Schmid, 1994; Tian and Lo, 2015; Voutilainen,
2003). The nouns carry the most information about
the term’s description, the adjectives give details and
explain the term and the numbers are in most cases
related to the chemical nomenclature.
Let us define
NN
i, j
= {nouns extracted f rom d
i, j
},
JJ
i, j
= {ad jectives extracted f rom d
i, j
},
CD
i, j
= {numbers extracted f rom d
i, j
}. (2)
To find the similarity between two descriptions of the
terms, d
i
1
, j
1
and d
i
2
, j
2
, we define an event X as a prod-
uct of three other events.
X = NN · JJ ·CD, (3)
where NN is the similarity between the nouns ex-
tracted in NN
i
1
, j
1
and NN
i
2
, j
2
, JJ is the similarity be-
tween the adjectives extracted in JJ
i
1
, j
1
and JJ
i
2
, j
2
,
and CD is the similarity between the numbers ex-
tracted in CD
i
1
, j
1
and CD
i
2
, j
2
.
Because all these events are independent, the proba-
bility of the event X can be found with
P(X) = P(NN) · P(JJ) · P(CD). (4)
Now we need to define the probabilities of each of the
events NN, JJ, and CD. Because we need to find the
similarity between two sets, it is logical to use the Jac-
card index, J, which is used in statistics for compar-
ing the similarity and diversity of sample sets (Real
and Vargas, 1996). For this purpose we use a modi-
fication of the Jaccard index in combination with the
Laplace probability estimate. We do this so even if
KEOD 2018 - 10th International Conference on Knowledge Engineering and Ontology Development
280

the description of some term does not contain adjec-
tives or nummbers the relevant match can be found,
thus we will always have non-zero probabilities. The
probabilities of the events can be calculated with
P(Y ) =
|Y
i
1
, j
1
∩Y
i
2
, j
2
| + 1
|Y
i
1
, j
1
∪Y
i
2
, j
2
| + 2
=
J(Y
i
1
, j
1
, Y
i
2
, j
2
) +
1
|Y
i
1
, j
1
∪Y
i
2
, j
2
|
1 +
2
|Y
i
1
, j
1
∪Y
i
2
, j
2
|
, (5)
where Y ∈ {NN, JJ, CD}.
The probability of the event X is obtained by substi-
tuting the relations (5) into the relation (4), which is
the weight assigned to each matching pair, and at the
end for each term the pair with the greatest weight is
the most relevant match found.
To find the similar terms that are used in different web
services, we rewrite the method presented above in a
matrix representation. Instead of a set of descriptions
of the terms D
i
, we can rewrite D
i
to be a n ×1 vector,
whose elements are descriptions of the terms.
The result of the POS tagging on this vector is three
vectors, NN
i
, JJ
i
and CD
i
, which are also n × 1 vec-
tors. The elements of the NN
i
, JJ
i
, and CD
i
vectors
are NN
i, j
, JJ
i, j
, and CD
i, j
, which are sets of extracted
nouns, adjectives, and numbers, respectively, where
j = 1, .., n.
Let us suppose that we have two web services and the
descriptions of the terms that they use are represented
in the vectors D
1
and D
2
, which are n × 1 and m × 1,
respectively. After using POS tagging we obtained
the vectors NN
1
, JJ
1
, CD
1
, NN
2
, JJ
2
, and CD
2
. In or-
der to find the similar terms, we define the following
operations between the vectors
Y
intersect
= Y
1
∩ Y
>
2
Y
union
= Y
1
∪ Y
>
2
, (6)
where Y ∈ {NN, JJ, CD}.
Y
intersect
and Y
union
are n × m matrices, where the el-
ements are defined as a matrix multiplication, but in-
stead of multiplication, the elements are defined as
the cardinality of the intersection between the two sets
and the cardinality of the union between the two sets,
respectively.
[Y
intersect
]
i, j
= |Y
1,i
∩Y
2, j
|,
[Y
union
]
i, j
= |Y
1,i
∪Y
2, j
|, (7)
where i = 1, .., n and j = 1, .., m. To use the Jaccard
index in combination with the Laplace probability es-
timate, we need to add 1 and 2 to each element of
Y
intersect
and Y
union
, respectively.
Y
interesect
= Y
intersect
. + 1,
Y
union
= Y
union
. + 2, (8)
where .+ means that we need to add 1 or 2 to each
element in the matrix.
At the end, we calculate
P(Y ) =
Y
interesect
Y
union
, (9)
where the division is made element by element.
Using the relation (9) separately for NN, JJ and CD
and substituting them into the relation (4), which in
this case will be a multiplication element by element,
we obtained the probability matrix, in which the rows
are descriptions of the terms used from the first web
service and the columns are descriptions of the terms
used from the second web service. To find the most
relevant match for each term used in the first web ser-
vice, we need to find the maximum element in each
row, or if we want to find the most relevant match for
each term from the second web service we need to
find the maximum element in each column.
After obtaining the matching pairs, we need to define
which of them are true matches. To do this, we define
one value as a threshold (it can be defined according
to experimental results), then we compare the proba-
bility of each matching pair with the threshold value,
and if the probability is greater than or equal to the
threshold, then the pair is a true match.
3.2 Taxonomy of the Extracted Terms
3.2.1 Initial Taxonomy
Using the POS tagging-probability weighted method,
we are able to capture the similar terms that are used
in the web services and also the terms that are used
only in one of them. At the end, we organized the
extracted terms in a taxonomy, which is an XML
document, whose structure is presented with the ex-
ample in Section IV.B. The initial taxonomy is the
one we obtained by matching two web services. The
root element consists of one element WSProviders
and more elements of the type Matching. The ele-
ment WSProviders consists of elements of the type
WSProvider, which has two attributes, name and ver-
sion, which are the name and the version of the web
service. The Matching element has an attribute class,
which is the class where this matching belongs and
two children elements, Descriptions and MatchScore.
The Descriptions element has one or two elements
of the type Description. When it has only one ele-
ment of the type Descripiton, this means that the term
Quisper Ontology Learning from Personalized Dietary Web Services
281

is typical only for one web service and the attributes
source, description, code are the name of the web ser-
vice where this term is used, the description of the
term and the code of the term, respectively. In this
case, the element MatchScore has an element Match-
Pair, which has two elements of the type WSProvider,
which are the names of the web services that are
matched and the element matchScore has the value 0,
which means that this term is typical only for one web
service and there is no matching of this term between
the two web services. In the case when the Descrip-
tions element has two elements of the type Descrip-
tion, then the matching between these two web ser-
vices is found and it is the same term that is used in
both web services, but maybe with different descrip-
tions or codes. In this case, the MatchScore element
has the element MatchPair, which has two elements
of the type WSProvider, which are the names of the
web services that are matched and the element match-
Score has a value that is obtained by the POS tagging-
probability weighted method.
3.2.2 Updated Taxonomy
After the initial taxonomy is created, the next task is
to match it with the new web service. To match them
we use the POS tagging-probability weighted method,
where D
1
is the vector from descriptions used in the
new web service and D
2
is the vector from the de-
scriptions used in the initial taxonomy. At the end,
the new taxonomy consists of the extracted terms,
where some Matching elements have three elements
of the type Description in the Descripitons element.
This is the case when all the web services use the
same term, but maybe with different descriptions and
codes, and the MatchScore element consists of three
elements of type MatchPair, one per each pair of web
services, where the matchScore elements have val-
ues that are obtained by the POS tagging-probability
weighted method. So the created taxonomy can have
Matching elements that have one, two or three De-
scripiton elements in the Descripitons element.
We used this approach for each new web service, and
at the end we have the taxonomy that consists of sim-
ilar terms that are used between the web services and
the terms that are typical only for one web service.
3.3 Ontology Learning Process
The ontology-learning process has three steps (Figure
3). In the first step, the initial taxonomy is created.
The second step is related to updating the initial tax-
onomy. At the end, we use the created taxonomy and
the information presented in the web services to build
an OWL ontology from scratch using Prot
´
eg
´
e.
Figure 3: Ontology-learning process.
4 QUISPER ONTOLOGY
4.1 Data
The data we used for developing the ontology
is from Quisper - Quality information services
for personalized nutrition (http://quisper.eu/).
For our purpose we used three web services
that are part of Quisper - Quality informa-
tion services for personalized nutrition, Eu-
roFIR Food Transport (http://www.eurofir.org/),
Food4me (http://www.food4me.org/), and SafeCape
(http://www.safecape.gr/).
The EuroFIR Food Transport provides an interface
for the food-composition-information data inter-
change, from which we extracted the names of
the entities used in the EuroFIR food-composition
database, and the names of all the possible compo-
nents that can be found in food.
From Food4me we used the Personalized Dietary
Advice Service (PDAS), which uses food intake,
biomarkers from blood analysis, SNP data and other
body metrics (weight, BMI, etc.) as inputs and
provides decision trees that lead to personal dietary
advice, from which we extracted the input variables.
The third web service is SafeCape, which contains
scientifically validated genotype-phenotype-nutrition
associations and can be used to generate personalized
nutrition and lifestyle recommendations for individ-
uals. For our task we used the descriptions of the
input and output variables that are used from this web
service.
Each piece of data is described with a name (origi-
nal, English, scientific), description, value(s), unit(s),
reference (documentation) and quality index. In our
case we are dealing with the following data formats:
food composition (food name - component name -
KEOD 2018 - 10th International Conference on Knowledge Engineering and Ontology Development
282

Table 1: Number of descriptions used from each web ser-
vice.
Web Service Number of descriptions
EuroFIR Food
Transport 924
Food4me 48
SafeCape 100
value(s) - unit(s) - reference(s)), dietary reference in-
takes (age group - sex - component name - value(s) -
unit(s) - reference(s)), biomarker analysis (age group
- sex - biomarker name - value(s) - unit(s) - DRV(s) -
reference(s)) and knowledge rules (54 years - Male
- rs5128 - GG - rs698 - AG - VitA goal - 2700
IU/810µg - PhysicalActivity goal - 45 min/day).
The first step before we started with the ontology-
learning process was to collect the data that is used
by each web service. After the data was collected we
had several discussions in order to realize what is im-
portant in terms of analysis. Then, we looked at the
nature of the data in order to define what kind of pre-
processing is required. At the end, we preprocessed
the data by removing punctuation marks, converted
each description to lower-case letters, and introduced
whitespace and number tokenization. It is important
to note here that this step takes the most time in the
ontology-learning process.
4.2 Quisper Ontology-learning Process
Following the steps presented in Figure 3, we started
with the matching between the web services Food4me
and EuroFIR Food Transport. Using the descriptions
of the terms used in Food4me and EuroFIR Food
Transport, we applied the POS tagging-probability
method to find the similar terms, and we used the
threshold 0.1 to find the true matchings. Some true
matchings that are the result of the matching be-
tween these two web services are presented in Ta-
ble 2. We found 27 true matchings between Food4me
and EuroFIR Food Transport, or having in mind that
in Food4me we have 48 terms, 27 of them are also
used in EuroFIR Food Transport. We need to men-
tion here that we also found 3 matchings that have a
value greater than 0.1, but they are not true match-
ings, and this happened because in the descriptions of
the terms we did not find nouns, which carry most of
the information, so we manually removed them. Af-
ter finding similar terms between the web services, we
went again through all of them in order to find if there
are some matchings of terms that are SNP. We did this
because, if the web service has terms that are SNP, in-
stead of using descriptions of the terms, they use the
names from SNPedia (rs4880, rs174546), which is a
Table 2: Some true matchings between Food4me and Eu-
roFIR Food Transport.
Food4me EuroFIR Food Transport weight
Alpha-carotene carotene, alpha- 0.187
Carotenoids carotenoids 0.167
Iron iron, total 0.125
Vitamin B12 vitamin b-12 0.250
Vitamin B6 vitamin b-6, total 0.200
wiki investigating human genetics. We catch them
using a regular expression, because all of them start
with rs, followed by a number. Because there are no
SNP terms in EuroFIR Food Transport, we did not
find matchings for these kinds of terms between these
two web services. At the end, we added the extracted
similar terms in the taxonomy, and after this we added
the terms from the two web services for which we did
not find a match. The result of the matching between
these two web services is our initial taxonomy.
After creating the initial taxonomy, we wanted
to add the terms that are used in the SafeCape web
service, which is our new web service. As explained
above, instead of matching the SafeCape web service
separately with the Food4me and EuroFIR Food
Transport web services, we continued with matching
the SafeCape with the initial taxonomy. First, we
performed the POS tagging-probability weighted
method to match the similar terms between it and
the initial taxonomy, after that we found the similar
SNP terms that are used, and at the end we updated
the initial taxonomy with the extracted similar terms
and also we added the terms that are typical only for
the SafeCape web service. It was obvious that if a
term from SafeCape has a match with a term from
Food4me, and this term from Food4me has a match
with a term from EuroFIR Food Transport, then the
term from SafeCape also has a match with the term
from EuroFIR Food Transport, because the relation
of similarity is transitive. The similarity relation is
a relation of equivalence, because it is transitive,
symmetric and reflexive. In the case of matching
SNP terms, the matchScore element in the MatchPair
element between SafeCape and Food4me is set to
1, because the matching is between these two web
services and we do not have the value that is obtained
using the POS tagging-probability weighted method.
Having the taxonomy of the matchings between
the three web services, the next step was to develop an
ontology that includes the defining classes in the on-
tology, arranging the classes in a taxonomic hierarchy,
defining slots and describing the allowed values for
these slots and filling in the values of the slots for in-
stances. An ontology with a set of instances of classes
constitutes a knowledge base. We started developing
Quisper Ontology Learning from Personalized Dietary Web Services
283
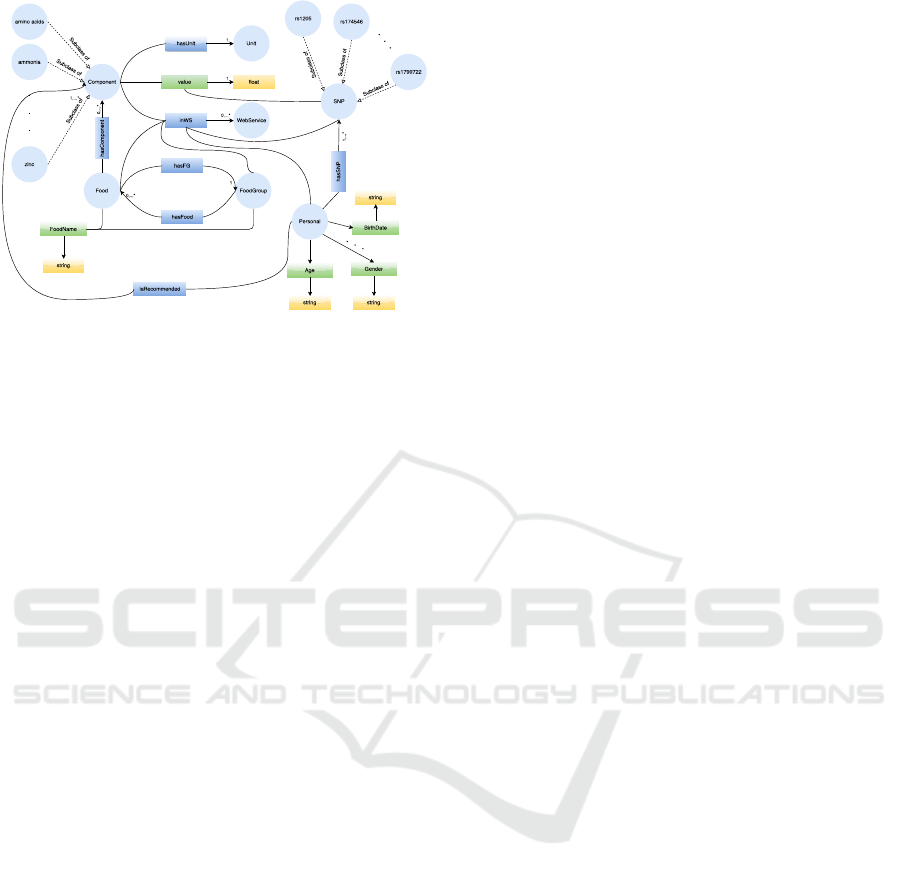
Figure 4: Graphical representation of the ontology.
< Tax o n om y >
< WS p r ov i de r s >
< WS p r ov i d er name = " Eu r o F I RF o od T r a n s p or t "
ver s i on = " 1.0.4 " / >
< WS p r ov i d er name = " F ood 4 m e " ve r s ion = " 1. 0 . 0 "/ >
< WS p r ov i d er name = " S af e C ape " v e rsi o n =" 1 . 0 . 0 " / >
</ W S pr o vi d e rs >
< Mat c h in g class = " C o mpo n e nt " >
< De s cr i p ti o ns >
< De s c ri p ti o n s o u r ce = " E u r o FIR "
de s c ri p ti o n =" alpha - carotene , carotene , alpha -"
code = " C ARTA " / >
< De s c ri p ti o n s o u r ce = " F o o d 4me "
de s c ri p ti o n =" Alpha - c a r ot e n e "
code = " 413 4 8 30 0 1 "/ >
</ D e sc r ip t i o n s >
< Ma t c hS c o re >
< Ma t c hPa i r >
< WS p r ov i d er name = " Eu r o F I RF o od T r a n s p or t "/ >
< WS p r ov i d er name = " F ood 4 m e " / >
< ma t c hS c o re > 0 .187 5 </ m a tc h S co r e >
</ M a tch P air >
</ M a tc h S co r e >
</ M a t ch i n g >
.
.
.
</ T a x on o m y >
the ontology from scratch, beginning with the ques-
tions about the domain of the ontology, for what we
are going to use this ontology and for what types of
questions the information in the ontology should pro-
vide answers (Noy et al., 2001).
Considering the extracted terms presented in the tax-
onomy and the fact that all the terms in the web ser-
vices are classified into 5 groups, Component, Food,
FoodGroup, Perosnal and SNP, and the relations be-
tween the terms in the XML document of the Eu-
roFIR Food Transport web service, we needed to de-
cide which of them will be concepts, properties or
instances. By discussing this with human experts,
our proposed ontology has 7 classes, which are the
most general concepts and they are Component, Food,
FoodGroup, Personal, SNP, Unit, and WebService.
For the class Component, we define two object prop-
erties hasUnit and inWS, which both have the domain
Component and the ranges are Unit and WebService,
respectively. The hasUnit object property has a car-
dinality of exactly 1, and the inWS object property
has a minimum cardinality of 1, while the maximum
cardinality is unrestricted. Also, we define one data
property, which is value and it can be a float value.
The value data property denotes the value of the com-
ponent that has a unit defined by the hasUnit object
property and the inWS object property denotes where
this kind of information can be found. We added all
the extracted concepts in the taxonomy, which are
classified as Component, as subclasses of the class
Component. For the class Food, we define three ob-
ject properties and one data property. The object
properties are inWS, which is the same as that used for
the Component, hasComponent, which denotes which
components the food consists of and the minimum
cardinality is 0 and the maximum cardinality is un-
restricted, and hasFG, which denotes the food group
to which this food belongs; it has cardinality of ex-
actly 1. The data property is FoodName and denotes
the name of the food and it is a string value. For class
FoodGroup, we define two object properties and one
data property. One of the object properties is inWS,
and the other is hasFood, which denotes the food that
belongs to this food group, and it has minimum cardi-
nality 0, and the maximum cardinality is unrestricted.
The data property is FoodName, which is the name
of the food group and it is a string value. The ex-
tracted concepts in the taxonomy that are classified
in these classes are added as instances. Both classes
Personal and SNP have an object property isWS. and
the class SNP has a data property value. The SNP
class has subclasses, which are the extracted concepts
in the taxonomy, and one data property value. The
class Personal has several data properties, which are
the concepts extracted in the taxonomy (related with
the individual user profile) and two object properties,
isRecommended and hasSNP, in order to describe the
dietary reference intakes and the biomarkers’ analy-
ses, respectively. The proposed ontology is presented
on Figure 4 and can be requested from authors for fu-
ture applications.
KEOD 2018 - 10th International Conference on Knowledge Engineering and Ontology Development
284

5 EXAMPLES AND USES OF THE
QUSIPER ONTOLOGY
5.1 Example
The first purpose for which this ontology was cre-
ated is to enable the harmonization between the per-
sonalized web services that are part of the Qualify
project. Having in mind that they are all dealing
with food-related data and different ways of describ-
ing and classifying data, the question was how to ex-
tract knowledge from them. For this we developed
a RESTful web service called ”HarmonizedQuisper”
that is a part of the QuaLiFY Server Platform (Quis-
per). The web service provides a single point of ac-
cess to the QuaLiFY Server Platform web services.
Requests use the purpose-designed Data Query Lan-
guage (DQL). The DQL is used for defining the in-
formation content and the search options in the re-
quests. The language is similar to SQL, but it is not
connected with a data model in any specific database;
it is based on a data model, which is the Quisper
ontology. The reserved words with which we de-
fine the query sentence structure are the concepts pre-
sented in the Quisper ontology. The responses de-
pend on which web service is called. The main task
where this ontology is used is to find the appropri-
ate web service to which we need to address our
input, and using it together with the created taxon-
omy to translate the input query to the appropriate
query for the web service that needs to be requested.
For example, if we have the data query ”SELECT
ComponentValue WHERE ComponentName=’alpha-
carotene’ ORDER BY FoodName DESC”, the Har-
monizedQuisper using the Qusiper ontology and tax-
onomy knows that this question is addressed to the
EuroFIR Food Transport and translates the reserved
words to the words that are used by the EuroFIR Food
Transport.
5.2 Uses of the Quisper Ontology
Another advantage is that the researchers who are
dealing with this kind of data can annotate the data
using this ontology and this will enable us to share in-
formation in this domain and to use in an easy way the
data from different sources to provide new knowledge
in this domain.
6 COMPARISON WITH
EXISTING FOOD
ONTOLOGIES
In (Boulos et al., 2015), the authors give a review
of the existing food ontologies that can be used with
some modifications and together with some relevant
non-food ontologies. This is in order to enable users
to make the correct, healthy food-and-drink choices
that are personalized for their particular health condi-
tion, age, body weight, lifestyle and preferences, and
also give their coverage gaps, the incompleteness or
limited scope, which are challenges for developing
a universal, comprehensive food ontology. They re-
view the following food ontologies: FoodWiki (C¸ elik,
2015), AGROVOC (Caracciolo et al., 2012), Open
Food Facts (https://world.openfoodfacts.org/who-we-
are), Food Product Ontology (Kolchin and Zamula,
2013), and FOODS (Diabetics Edition) (Snae and
Br
¨
uckner, 2008). FoodWiki’s aim is to represent an
abstract model of different types of foods available
to the users, together with their nutritional informa-
tion, including the type and the amount of nutrients,
and the recommended daily intake. It consists of four
main concepts: Person, Diseases, Product and Food
Ingredients/Compounds. AGROVOC is a large and
mature multilingual thesaurus, which includes termi-
nology widely used in practice for subject fields in
agriculture, fisheries, forestry, food and related do-
mains. Open Food Facts is a global food database
based on contributions from individuals around the
world, it allows learning about the nutritional infor-
mation of a specific food, and compares products
from around world. It is also beneficial for the food
industry to track, monitor, and strategically plan its
food production. The Food Product Ontology de-
scribes food products with common representation,
vocabulary and language for the food-product do-
main. It is an extended version of a widely used
standardized ontology for product, price, store and
company data. FOODS is a food-oriented ontology-
driven system that uses food ontology to deliver a
web-based food-menu recommender system for pa-
tients with diabetes in Thailand. Compared to all
these five ontologies, the Quisper ontology covers a
wider domain. It is not focused only on food-related
data, but also includes information for each individual
from his/her user profile, biomarker analysis, dietary
reference intakes and recommendations. It is an OWL
ontology and it can be easily integrated and combined
with some of these five food ontologies. Also, it can
be easily extended using the proposed POS tagging-
probability weighted method, in the case when a new
personalized web service will appear.
Quisper Ontology Learning from Personalized Dietary Web Services
285

7 CONCLUSIONS
This paper presents an ontology-learning process
which uses personalized dietary web services in or-
der to develop an ontology that can be used for the
harmonization of personalized dietary web services
and will enable researchers to share information in
this domain. Having this kind of ontology will en-
able researchers to use aggregated data and informa-
tion from different sources to provide new knowledge,
new protocols and help people live healthier lives.
ACKNOWLEDGEMENTS
This work was supported by the project QuaLiFY,
which received funding from the European Union’s
Seventh Framework Programme for research, techno-
logical development and demonstration under grant
agreement no 613783. This work was supported
by the project ISO-FOOD, which received fund-
ing from the European Union’s Seventh Frame-
work Programme for research, technological devel-
opment and demonstration under grant agreement no
621329 (2014-2019). This work was conducted us-
ing the Prot
´
eg
´
e resource, which is supported by grant
GM10331601 from the National Institute of General
Medical Sciences of the United States National In-
stitutes of Health. We would like to thank the Co-
founder of SafeCape Software Solutions Ltd, Agge-
los Androulidakis, for helping us with the SafeCape
web service, the Hyve and the TNO for helping us
with the Food4Me web service. The Hyve is a SME
working on development and professional support for
open source software used in life science research
that created the IT-part and the TNO is an indepen-
dent research organisation that adapted the input of
Food4Me to fit both the web-service.
REFERENCES
Biemann, C. (2005). Ontology learning from text: A survey
of methods. In LDV forum, volume 20, pages 75–93.
Boulos, M. N. K., Yassine, A., Shirmohammadi, S., Nama-
hoot, C. S., and Br
¨
uckner, M. (2015). Towards an
“internet of food”: Food ontologies for the internet of
things. Future Internet, 7(4):372–392.
Caracciolo, C., Stellato, A., Rajbahndari, S., Morshed,
A., Johannsen, G., Jaques, Y., and Keizer, J. (2012).
Thesaurus maintenance, alignment and publication as
linked data: the agrovoc use case. International Jour-
nal of Metadata, Semantics and Ontologies, 7(1):65–
75.
C¸ elik, D. (2015). Foodwiki: Ontology-driven mobile safe
food consumption system. The Scientific World Jour-
nal, 2015.
Eftimov, T. and Seljak, B. K. (2015). Pos tagging-
probability weighted method for matching the inter-
net recipe ingredients with food composition data. In
Knowledge Discovery, Knowledge Engineering and
Knowledge Management (IC3K), 2015 7th Interna-
tional Joint Conference on, volume 1, pages 330–336.
IEEE.
Greenfield, H. and Southgate, D. A. (2003). Food composi-
tion data: production, management, and use. Food &
Agriculture Org.
Hazman, M., El-Beltagy, S. R., and Rafea, A. (2011). A sur-
vey of ontology learning approaches. database, 7:6.
Hellwig, J. P., Otten, J. J., Meyers, L. D., et al. (2006). Di-
etary Reference Intakes:: The Essential Guide to Nu-
trient Requirements. National Academies Press.
Kolchin, M. and Zamula, D. (2013). Food product ontol-
ogy: Initial implementation of a vocabulary for de-
scribing food products. In Proceeding of the 14th
Conference of Open Innovations Association FRUCT,
Helsinki, Finland, pages 11–15.
Noy, N. F., McGuinness, D. L., et al. (2001). Ontology
development 101: A guide to creating your first ontol-
ogy.
Real, R. and Vargas, J. M. (1996). The probabilistic basis
of jaccard’s index of similarity. Systematic biology,
pages 380–385.
Schmid, H. (1994). Probabilistic part-of-speech tagging us-
ing decision trees. In Proceedings of the international
conference on new methods in language processing,
volume 12, pages 44–49. Citeseer.
Snae, C. and Br
¨
uckner, M. (2008). Foods: a food-oriented
ontology-driven system. In Digital Ecosystems and
Technologies, 2008. DEST 2008. 2nd IEEE Interna-
tional Conference on, pages 168–176. IEEE.
Tian, Y. and Lo, D. (2015). A comparative study on the
effectiveness of part-of-speech tagging techniques on
bug reports. In Software Analysis, Evolution and
Reengineering (SANER), 2015 IEEE 22nd Interna-
tional Conference on, pages 570–574. IEEE.
Voutilainen, A. (2003). Part-of-speech tagging. The Oxford
handbook of computational linguistics, pages 219–
232.
KEOD 2018 - 10th International Conference on Knowledge Engineering and Ontology Development
286
