
Circulating Tumor Enumeration using Deep Learning
Stephen Obonyo and Joseph Orero
Faculty of Information Technology, Strathmore University, Ole Sangale Link Road, Nairobi, Kenya
Keywords:
CTC Enumeration, CTC Detection, Artificial Neural Networks, Machine Learning, Deep Learning.
Abstract:
Cancer is the third most killer disease just after infectious and cardiovascular diseases. Existing cancer treat-
ment methods vary among patients based on the type and stage of tumor development. Treatment modalities
such as chemotherapy, surgery and radiation are successful when the disease is detected early and regularly
monitored. Enumeration and detection of Circulating Tumor Cells (CTC’s) is a key monitoring method which
involves identification of cancer related substances known as tumor markers which are excreted by primary
tumors into patient’s blood. The presence, absence or number of CTC’s in blood can be used as treatment
metric indicator. As such, the metric can be used to evaluate patient’s disease progression and determine ef-
fectiveness of a treatment option a patient is subjected to. In this paper, we present a deep learning model
based on Convolutional Neural Network which learns and enumerates CTC’s from stained image samples.
With no human intervention, the model learns the best set of representations to enumerate CTC’s.
1 INTRODUCTION
Cancer is a disease occurring as a result of genetic
mutation or abnormal changes in the genes responsi-
ble for regulating the growth of body cells. The cells
gain ability to keep dividing without control, produ-
cing more cells and forming a tumor (Ferlay et al.,
2010). The abnormal cells infiltrates healthy ones and
spreads throughout the body.
According to Fitzmaurice et al. (2015) cancer cau-
sed more than 8 million deaths globally in the year
2013. The cancer death burden have been recorded in
both first and third world countries though in an une-
qual measure. Cancer cases have been exacerbated by
unique geographical, cultural and demographic fac-
tors such as aging population and increased predispo-
sition to cancer causing conditions such as smoking,
being overweight and physical inactivity.
The death figures in first world countries are re-
latively lower as compared to third world countries.
Developing nations have contributed to 57% of can-
cer total cases. This percentage accounts for approxi-
mately 65% of related deaths worldwide (Torre et al.,
2015). The higher figures can be attributed to inabi-
lity to access proper medication and lack of regular
monitoring.
Today, different types of cancer are treatable if de-
tected early. Standard medical detection modalities
include Radiography, Magnetic Resonance Imaging
(MRI), Computer Tomography (CT) and Ultrasound.
After cancer detection process, a patient is subjected
to appropriate treatment option based on stage and
type of tumor. For a given patient, medical surgery or
radiation would apply, for others chemotherapy, while
hormone therapy for other subjects.
During treatment, a patient must be constantly
monitored to determine effectiveness of treatment
method they are subjected to. This can be achieved
through medical processes such as biopsy or liquid
biopsy. Biopsy is a medical procedure which can be
used to detect tumor related substances known as Ci-
rculating Tumor Cells (CTC’s). The process involves
identification of CTC’s in a raw body tissues. In con-
trast, liquid biopsy involves detection of the CTC’s in
blood after staining the sample.
Tumors excretes CTC’s into patient’s blood. Du-
ring treatment, the presence, absence or the number
CTC’s in blood can be used as patient’s progress me-
tric indicator to evaluate how the subject is responding
to treatment. The metric also determines the effecti-
venesses of treatment option one is enrolled in. Besi-
des monitoring cancer treatment response and deter-
mining the effectiveness of a treatment method, biop-
sies are also key in assessing the cancer recurrence
and progression (Crowley et al., 2013).
In this research work, we present a CTC enumera-
tion model. We show how Convolutional Neural Net-
work (ConvNet) can be used to learn intricate featu-
res for enumerating CTC’s in a given stained sample
image.
Obonyo, S. and Orero, J.
Circulating Tumor Enumeration using Deep Learning.
In Proceedings of the 10th International Joint Conference on Computational Intelligence (IJCCI 2018), pages 297-303
ISBN: 978-989-758-327-8
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
297

An image sample may contain more than one CTC
or none. Based on this, we developed an algorithm
to locate CTC’s and generate training samples from
an input image. We also show that certain models are
incapable of learning abstract representative details as
opposed to the preferred model.
2 RELATED WORK
Application of learning models in cancer manage-
ment have been presented by many researchers. Sim-
ple and classical learning approaches such as decision
trees have been used in detection of common Circula-
ting Tumors variations such as Cytokeratin, Apoptic
and Debris. CTC’s are extracted following standard
procedures such as fictionalized and structured medi-
cal wire, Epithelial Cell Adhesion Molecule, density
gradient centrifugation or membrane filtration. Schol-
tens et al. (2012) used Random Forests and Decision
Trees to classify CTC’s. The target labels were based
on different CTC classes such as Apoptic CTC, CTC
debris, Leukocytes and Debris.
In another study, Svensson et al. (2014) presen-
ted how Naive Bayes classifier and Generative Mix-
ture Model can be used to detect CTC’s in stained
blood sample. Cells were collected using fictionali-
zed medical wire and thereafter stained. The intensity
of RGB (Red, Green, Blue) values of the resulting
sample was then used as the input features to the mo-
del. The classifier learned to discriminate an instance
given the predefined CTC’s class.
Aside from classical learning approaches such as
Decision Trees, Naive Bayes among other algorithms,
deep learning models have also been applied. This le-
arning methodology have led to much better results
in image detection and recognition. According to Le-
Cun et al. (2015) deep learning methods have impro-
ved the state-of-the-art visual object recognition and
detection. This learning approach can automatically
discover best set of features to represent an instance.
This unique characterization has contributed to unpre-
cedented success studies in the field of image recog-
nition.
Mao et al. (2016) proposed use of Deep Convo-
lutional Neural Network, a deep learning model, to
detect Circulating Tumor Cells. The model automati-
cally learned the best set of features used to classify a
sample as either having CTC or not. They demonstra-
ted that automated feature discovery using the lear-
ning model led to much better results than hand craf-
ted. There was a significant variation in performance
when their proposed model was compared to the clas-
sical ones such as Support Vector Machines with ma-
nually engineered input features.
Wang et al. (2016) also employed a similar lear-
ning technique in detection of tumor recurrence yiel-
ding state of the art results. In the work, deep learning
model developed was aimed at predicting the extent
of cancer spread to the other parts of the body.
3 METHODOLOGY
3.1 Dataset
Data used in this research work was secondary data.
It was provided by a PhD candidate in Computer
Science from Missouri University of Science and
Technology. It is the same dataset used in (Mao et al.,
2016). The original work was based on classification
of CTC’s as opposed to enumeration which this pa-
per focuses on. Figure 1 and Figure 2 shows sample
images. Figure 1 is a sample of microscopy image.
Figure 2 on the other hand is a sample fluorescence
image. The latter is used to locate the exact loca-
tion of the CTC’s in the former. Every microscopy
image has a corresponding fluorescence image sho-
wing CTC’s locations.
We developed an algorithm to extract the regions
of interest (ROI) to generate both the positive (with
CTC) and negative (without CTC) training and testing
sets. A total of 1904 training samples were generated.
Out of this, 952 were positive and 952 negative. Every
sample had a corresponding target label equivalent to
the number of CTC’s contained. Positive samples had
N CTC’s and 0 for negative instances. 80% of the
dataset was used for training and the remaining 20%
for testing the model.
3.2 Cropping Algorithm
We developed an algorithm to extract 40 by 40 the
regions of interest (ROI). It accepts x and y coordi-
nates of CTC location in the fluorescence image and
a corresponding microscopy image. Following this,
20-pixel coordinate location to the left of x and to the
right are marked and saved. The same process also
applies for the y coordinate. Random x coordinate
within image width range is generated and another
random y within image height range. These random x,
y values represent the location of new negative sam-
ple instance. If these randomly generated coordinates
range do not overlap with CTC location bounds then
positive and negative sample is cropped out from the
1600x1600 input image and returned. The algorithm
have been summarized by Listing 1.
IJCCI 2018 - 10th International Joint Conference on Computational Intelligence
298

crop_sample(xpixel, ypixel, image)
counter <- 1
initialise xaxes[], yaxes[]
initialise samples[]
xaxes[0] <- xpixel
yaxes[0] <- ypixel
for left_coordinates and
right_coordinates do:
while counter < 21
xaxes[counter+1] = xpixel + counter
yaxes[counter+1] = ypixel + counter
counter <- counter+1
randomx <- random{0,1580}
randomy <- random{0,1280}
if x intersection xaxes is empty and
y intersection yaxes is empty
samples[0] <- crop(xpixel,ypixel)
samples[1] <- crop(x,y)
return samples
Listing 1: The Cropping Algorithm.
Figure 1: 1600x1600 A microscopy image containing
CTC’s.
Figure 2: 1600x1600 Fluorescence image depicting loca-
tion of CTC’s in Figure 1.
3.3 Training Set Generation
Microscopy images contains CTC’s within specific
locations. These images were all converted to one
channel from RGB. Fluorescence images on the other
hand shows the location or coordinates of CTC’s. All
images in this set were converted to gray scale and bi-
nary thresholded. Thresholding helped to accurately
locate the Circulating Tumors Cells.
Binary thresholding involves converting all the
image pixels to either of the two predefined values;
black or white (Gonzalez and Woods, 2002). To
achieve this, pixel intensity values less than 240 were
set to 0 and 255 otherwise for the florescence images.
All fluorescence image samples were thresholded
using similar function and x, y coordinates of white
pixels then mapped to the corresponding one channel
microscopy image. This process was then followed
by cropping out a positive and negative samples of
dimension 40 by 40. The negative set was generated
by randomly generating x, y coordinates, checking for
the overlap with the positive sample bounds and crop-
ping then cropping out. The feature set was created in
this fashion repeating the process over all the images.
The target labels for the negative samples were set 0
and N; number of white pixels for the positive instan-
ces. Resulting feature sets and targets were persis-
tently stored. Table 1 summarizes the dimensions of
training and testing dataset dimensions. Cropped po-
Table 1: A summary of training and testing dataset dimen-
sions.
Dataset Dimension
X train (1523, 40, 40)
y train (1523, 1)
X test (381, 40, 40)
y test (381, 1)
sitive and negative samples exhibit different patterns.
A high-level depiction of this variation is illustrated
by Figure 3 and Figure 4. Figure 3 shows sample posi-
tive while Figure 4 represents negative both randomly
sampled from training dataset.
3.4 Model Architecture
Convolutional Neural Network (ConvNet) model was
used to enumerate the CTC’s given the feature sets
and labels. The model architecture is captured by Fi-
gure 5. The network predicts number of CTC’s given
an image sample thus a regression task. The ConvNet
is composed of Convolution - Max Pooling - Convo-
lution - Max Pooling - Fully Connected Layer (FC1)
- Fully Connected Layer (FC2). The architecture is
shown by Figure 5 During forward propagation 40 by
Circulating Tumor Enumeration using Deep Learning
299

Figure 3: Cropped 40x40 negative sample with 0 CTC’s.
Figure 4: Cropped 40x40 positive sample with given num-
ber of CTC’s.
40 input was fed into the network. A kernel of shape
5*5 was used to convolve the input image with a slide
of 1 resulting in 8 feature maps. The convolution step
was then followed by Rectified Linear Units activa-
tion mathematically formulated by equation 1. The
activation step was then followed by 2*2 max pooling
which samples out set of values based on maximum
value. The second convolution layer resulted in 16 fe-
ature maps which were then flattened and connected
two Fully Connected layers.
f (x) = max(0, x) (1)
The last Fully Connected layer of neurons was con-
nected to one output neuron which produced a conti-
nuous value. This value was then compared to the ac-
tual target label and the difference computed resulting
in an error, E. The value of E is the mean squared er-
ror formulated by equation 3. The error value was
back propagated and used to adjust the network weig-
hts. The model was trained over 5,000 iterations using
backpropagation with batch gradient descent. The
gradient descent update rule for weights W and bias
b is formulated the by equation 2.
w
i
:= w
i
− α
∂
∂w
i
E b
i
:= b
i
− α
∂
∂b
i
E (2)
Besides Convolutional Neural Network, Multi-
layer Neural Network (MLP) and Linear Regression
could have solved the problem. These two models
were also experimented on the same training and test
set. Google cloud platform in relation to software spe-
cifications in table 2 was used for the experimenta-
tion.
Table 2: Development Environment.
Platform Application Version
Python 2.7
Keras 2.1.1
Tensorflow 1.4.0
Numpy 1.14.1
OpenCV 3.3.0
4 RESULTS
Predicting number of CTC’s is a regression task.
Three different models were experimented. The first
model was Convolutional Neural Network (ConvNet)
based on the model architecture represented by Figure
5. The second one was a three layer Multilayer Per-
ceptron (MLP) with 1600 neurons in the two hidden
layers and one neuron in the output layer. Last mo-
del experimented was Linear Regression trained with
stochastic gradient descent over 5000 iterations.
Learning with Multi Layer perceptron is relatively
algorithmically expensive with many layers due to ex-
ponential increase in the number of weights (Nielsen,
2015). Linear Regression on the other hand, though
not algorithmically expensive, does not perform well
as compared to MLP and ConvNet. The results of all
the models based on training and testing errors have
been summarized by Table 3. The variation of loss va-
lue with respect to number of epochs have also been
captured by Figure 6. The ConvNet model was the
best performer on both testing and training feature in-
stances. The performance was benchmarked on root
mean squared error formulated by equation 3. It sums
IJCCI 2018 - 10th International Joint Conference on Computational Intelligence
300
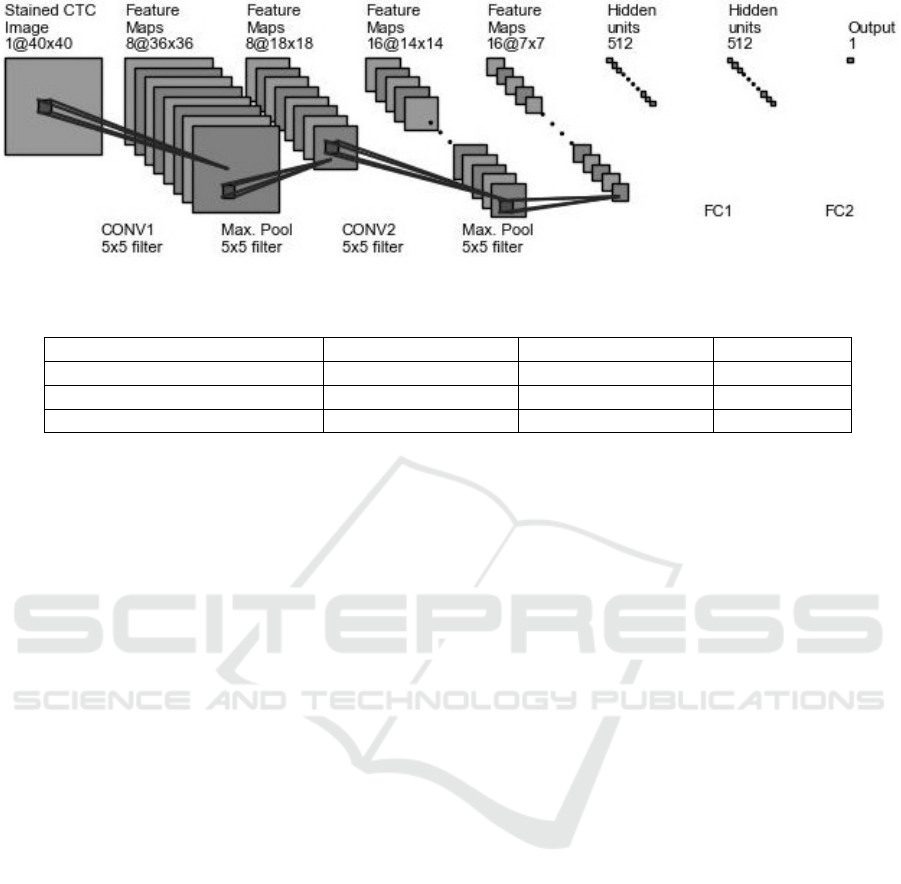
Figure 5: The ConvNet Architecture.
Table 3: Models Results.
Model Train Error Test Error Iterations
Multi Layer Perceptron 0.0364 6.1127 5,000
Convolutional Neural Network 0.0295 4.2822 5,000
Linear Regression 3.475 17.2613 5,000
the difference between predicted ( ˆy
(i)
) and actual va-
lues (y
(i)
).
m
∑
i=1
ˆy
(i)
− y
(i)
(3)
5 DISCUSSIONS
In this research, Convolutional Neural Network out-
performed both MLP and Linear regression with a
Root Mean Squared Error (RMSE) margin of 0.0069
and 3.4455 respectively on training error, and 1.8305
and 12.9791 respectively on test error. This perfor-
mance can be attributed to the fact that deep learning
models are capable of learning the best set of repre-
sentative features automatically. The learned feature
set is weighted and used to enumerate CTC’s given a
new sample instance.
Linear Regression model was trained on all the
pixel intensity values of the input image. It was
the worst performer just after MLP. The Multilayer
layer perceptron model outperformed Linear Regres-
sion with a variation of 3.4386 on train error and
11.146 on the test error. From this performance re-
cord it can be deduced that not all pixel intensity va-
lues are equally informative.
Manually generated features such as RGB histo-
gram or pixel intensity values though can represent
an instance, may require the expert to specify the right
set of features or extensive feature engineering techni-
ques thus expensive. Using all image pixel values is
an assumption that all pixel intensities are representa-
tive of an instance.
In contrary, the feature representations can be le-
arned automatically, on a low dimensional space du-
ring the training process. This is the core functional
and theoretical underpinning of deep learning models
such as Convolutional Neural Network. The deep le-
arning adoption was precipitated by the work done
by Krizhevsky et al. (2012). In the study, the re-
sult obtained halved error value for object recognition
during ImageNet competition. This was unpreceden-
ted performance record as Convolutional Neural Net-
work model outperformed all other classical simple
learning models used before. Following this, other
studies have extended the capability of classical Con-
vNet model leading to state-of-the art results in image
recognition (He et al., 2016), large scale image recog-
nition architectures Szegedy et al. (2015) and unpre-
cedented object detection performance Redmon et al.
(2016); Ren et al. (2015); Liu et al. (2016).
6 CONCLUSIONS
In this paper, we have presented a learning model
which can be used to enumerate Circulating Tumor
Cells (CTC’s) in a stained image sample. An algo-
rithm which extracts regions of interest was develo-
ped and based on the experiments carried out, Con-
volutional Neural Network outperformed both Linear
Regression and classical Multilayer Neural Network
architectures. This performance is attributed to the
fact that the preferred model had the potent to learn
intricate low level representative features on its own.
This is a unique characterization which have been at-
tributed to deep learning models such as ConvNet.
Circulating Tumor Enumeration using Deep Learning
301
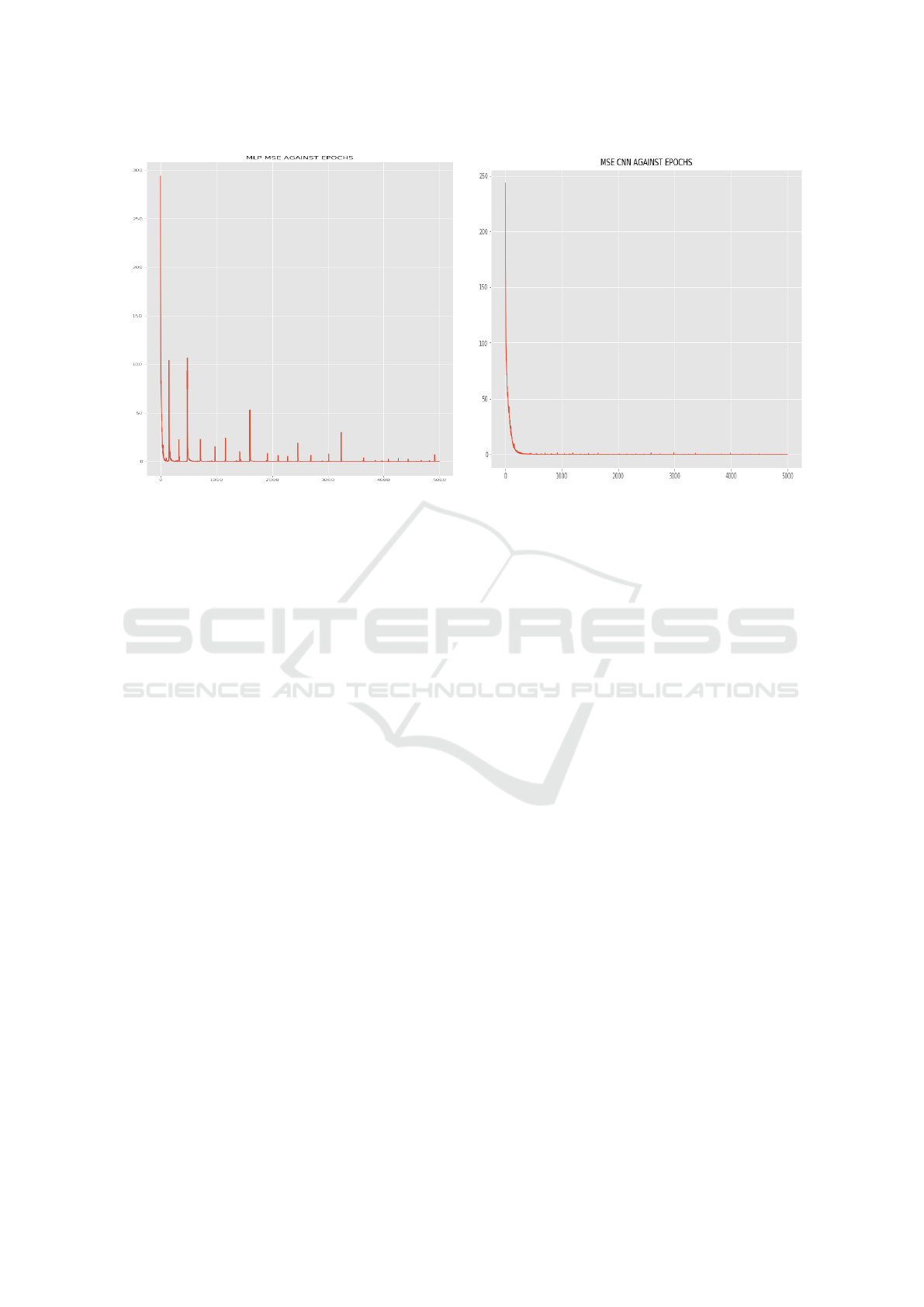
(a) MLPLoss (b) ConvNet Loss
Figure 6: MLP and ConvNet loss variations over epochs during training.
ACKNOWLEDGMENTS
The dataset used in this study was provided by Yunx-
iang Mao. He and others worked on a model which
only classifies CTC’s instead of enumeration which is
presented in this paper.
REFERENCES
Crowley, E., Di Nicolantonio, F., Loupakis, F., and Bar-
delli, A. (2013). Liquid biopsy: monitoring cancer-
genetics in the blood. Nature reviews Clinical onco-
logy, 10(8):472–484.
Ferlay, J., H
´
ery, C., Autier, P., and Sankaranarayanan, R.
(2010). Global burden of breast cancer. In Breast
cancer epidemiology, pages 1–19. Springer.
Fitzmaurice, C., Dicker, D., Pain, A., Hamavid, H., Moradi-
Lakeh, M., MacIntyre, M. F., Allen, C., Hansen, G.,
Woodbrook, R., Wolfe, C., et al. (2015). The global
burden of cancer 2013. JAMA oncology, 1(4):505–
527.
Gonzalez, R. C. and Woods, R. E. (2002). Thresholding.
Digital Image Processing, pages 595–611.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resi-
dual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012).
Imagenet classification with deep convolutional neu-
ral networks. In Advances in neural information pro-
cessing systems, pages 1097–1105.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep lear-
ning. Nature, 521(7553):436–444.
Liu, W., Anguelov, D., Erhan, D., Szegedy, C., Reed, S., Fu,
C.-Y., and Berg, A. C. (2016). Ssd: Single shot mul-
tibox detector. In European conference on computer
vision, pages 21–37. Springer.
Mao, Y., Yin, Z., and Schober, J. (2016). A deep convoluti-
onal neural network trained on representative samples
for circulating tumor cell detection. In Applications of
Computer Vision (WACV), 2016 IEEE Winter Confe-
rence on, pages 1–6. IEEE.
Nielsen, M. A. (2015). Neural networks and deep learning.
Redmon, J., Divvala, S., Girshick, R., and Farhadi, A.
(2016). You only look once: Unified, real-time object
detection. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 779–
788.
Ren, S., He, K., Girshick, R., and Sun, J. (2015). Faster
r-cnn: Towards real-time object detection with region
proposal networks. In Advances in neural information
processing systems, pages 91–99.
Scholtens, T. M., Schreuder, F., Ligthart, S. T., Swennen-
huis, J. F., Greve, J., and Terstappen, L. W. (2012).
Automated identification of circulating tumor cells by
image cytometry. Cytometry Part A, 81(2):138–148.
Svensson, C.-M., Krusekopf, S., L
¨
ucke, J., and Thilo Figge,
M. (2014). Automated detection of circulating tumor
cells with naive bayesian classifiers. Cytometry Part
A, 85(6):501–511.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Angue-
lov, D., Erhan, D., Vanhoucke, V., and Rabinovich, A.
(2015). Going deeper with convolutions. In Procee-
dings of the IEEE conference on computer vision and
pattern recognition, pages 1–9.
IJCCI 2018 - 10th International Joint Conference on Computational Intelligence
302

Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-
Tieulent, J., and Jemal, A. (2015). Global cancer
statistics, 2012. CA: a cancer journal for clinicians,
65(2):87–108.
Wang, D., Khosla, A., Gargeya, R., Irshad, H., and Beck,
A. H. (2016). Deep learning for identifying metastatic
breast cancer. arXiv preprint arXiv:1606.05718.
Circulating Tumor Enumeration using Deep Learning
303
