
Rule-based and Machine Learning Hybrid System for Patient Cohort
Selection
Rui Antunes
a
, João Figueira Silva
b
, Arnaldo Pereira
c
and Sérgio Matos
d
DETI/IEETA, University of Aveiro, Campus Universitário de Santiago, Aveiro, Portugal
Keywords:
Electronic Health Record, Patient Cohort Selection, Machine Learning, Rule-based.
Abstract:
Clinical trials play a critical role in medical studies. However, identifying and selecting cohorts for such
trials can be a troublesome task since patients must match a set of complex pre-determined criteria. Patient
selection requires a manual analysis of clinical narratives in patients’ records, which is a time-consuming task
for medical researchers. In this work, natural language processing (NLP) techniques were used to perform
automatic patient cohort selection. The approach herein presented was developed and tested on the 2018 n2c2
Track 1 Shared-Task dataset where each patient record is annotated with 13 selection criteria. The resulting
hybrid approach is based on heuristics and machine learning and attained a micro-average and macro-average
F1-score of 0.8844 and 0.7271, respectively, in the n2c2 test set. Part of the source code resultant from this
work is available at https://github.com/ruiantunes/2018-n2c2-track-1/.
1 INTRODUCTION
Clinical trials are a vital process in medical research
as they enable the analysis of cause-effect relations.
It is through such analysis that it is possible to as-
sess how efficient are the drugs or therapies in test
(Mann, 2003). Despite its utmost importance, clinical
trials require the selection of patient cohorts which is
a process performed by medical researchers, known
to be both burdensome and time-consuming, where
researchers might have to sift through various data
sources whilst considering how to correctly combine
and apply complex selection criteria.
To simplify this selection process, attempts have
been sought to automate cohort selection by perform-
ing patient phenotyping with informatics techniques,
and this has in fact been demonstrated to be possible
for some studies by the eMERGE consortium, which
showed that algorithms can be used with effectiveness
for phenotyping purposes (Pathak et al., 2013).
While automating cohort selection is certainly of
great interest, it faces major challenges namely how
to define inclusion and exclusion criteria such that an
algorithm can automatically and efficiently select pa-
a
https://orcid.org/0000-0003-3533-8872
b
https://orcid.org/0000-0001-5535-754X
c
https://orcid.org/0000-0003-3361-6269
d
https://orcid.org/0000-0003-1941-3983
tients in a dataset, or even how to integrate data from
various sources (Pathak et al., 2013), such as omics
and EHR (Electronic Health Record) data. EHR data
is of particular interest as it can contain textual infor-
mation stored in a structured form (data inserted in
strict form fields), or in clinical narratives where text
data is stored in an unstructured format (e.g. free text
report in a discharge record). Unstructured data has
been getting increased attention since fusing informa-
tion extracted from structured and unstructured data,
instead of single-handedly resorting to the structured
variant, can lead to significant performance improve-
ments in a system (Ludvigsson et al., 2013).
Extracting proper information from unstructured
data such that it can be represented in a structured
counterpart is a very difficult task. However, the ca-
pability to efficiently perform such extraction is of
paramount importance, as automatic patient cohort
selection systems can greatly benefit from it (Shiv-
ade et al., 2014). It is due to this widely recognized
potential that much research has focused on levering
unstructured data from EHRs, using for that purpose
techniques such as NLP (Natural Language Process-
ing) to process unstructured text and extract meaning-
ful content (Pathak et al., 2013).
The present work focuses on exploring NLP tech-
niques to extract information from unstructured data
in EHRs, and also on assessing whether a system
based on heuristics (rules) and machine learning algo-
Antunes, R., Silva, J., Pereira, A. and Matos, S.
Rule-based and Machine Learning Hybrid System for Patient Cohort Selection.
DOI: 10.5220/0007349300590067
In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), pages 59-67
ISBN: 978-989-758-353-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
59

rithms can efficiently select patient cohorts for clini-
cal trials using for that purpose medical records from
“Track 1: Cohort selection for clinical trials” from
the 2018 National NLP Clinical Challenges (n2c2)
Shared-Task
1
. This paper is structured as follows: in
Section 2 we explain all data resources used for this
work, Section 3 covers the methodology developed to
solve the n2c2 task, obtained results are presented and
discussed in Section 4, and finally Section 5 presents
some conclusions.
2 DATA
2.1 Dataset
The dataset used for this work was provided by the
2018 n2c2 (Track 1 Shared-Task) organization, and
is split into a training and a test set containing 202
and 86 samples, respectively. Each sample comprises
between 2 to 5 dated records of a single patient, the
records are de-identified and the dates are modified to
protect the identities of the participants. Nevertheless,
the relative time intervals between patient records are
kept to allow a timeline interpretation of these.
Table 1: Dataset selection criteria (based on the provided
guidelines).
Tag Criteria
ABDOMINAL
Intra abdominal surgery, intestine resection,
bowel obstruction
ADVANCED-CAD
Having at least two conditions about
cardiovascular diseases (taking medications,
myocardial infarction, angina, ischemia)
ALCOHOL-ABUSE Current alcohol abuse
ASP-FOR-MI Use of aspirin to prevent myocardial infarction
CREATININE Serum creatinine larger than the limit of normal
DIETSUPP-2MOS Taken a dietary supplement in the past 2 months
DRUG-ABUSE Drug abuse
ENGLISH Patient must speak English
HBA1C HbA1c value between 6.5% and 9.5%
KETO-1YR Diagnosis of ketoacidosis in the past year
MAJOR-DIABETES Major diabetes-related complication
MAKES-DECISIONS Patient must make their own medical decisions
MI-6MOS Myocardial infarction in the past 6 months
1
https://n2c2.dbmi.hms.harvard.edu/track1
Table 2: Class distribution in the provided dataset.
Tag
Training set Test set
Met Not met Met Not met
ABDOMINAL 76 126 30 56
ADVANCED-CAD 125 77 45 41
ALCOHOL-ABUSE 7 195 3 83
ASP-FOR-MI 163 39 68 18
CREATININE 82 120 24 62
DIETSUPP-2MOS 106 96 44 42
DRUG-ABUSE 10 192 3 83
ENGLISH 192 10 73 13
HBA1C 67 135 35 51
KETO-1YR 0 202 0 86
MAJOR-DIABETES 113 89 43 43
MAKES-DECISIONS 194 8 83 3
MI-6MOS 18 184 8 78
Each sample of the dataset has a list of 13 bi-
nary selection criteria that were manually annotated
by medical professionals. Moreover, each criterion
can be classified with a value of ‘met’ or ‘not met’ in-
dicating if a patient does or not meet the pre-defined
requirements of the criterion. Table 1 is based on the
guidelines provided by the n2c2 organizers and shows
a summary of the 13 selection criteria where each cri-
terion was attributed a unique tag for identification
purposes. From now on, we will refer to selection
criteria as tags. Each tag is a criterion that represents
a single binary classification problem.
Table 2 shows the dataset distribution where one
can see that certain tags are highly imbalanced. There
are tags where the ‘met’ class is much more frequent
(e.g. ENGLISH), but the opposite is also verified with
the ‘not met’ class prevailing (e.g. DRUG-ABUSE). It
is also relevant to note that the tag KETO-1YR does
only contain ‘not met’ labels, making machine learn-
ing models unable to learn this criterion.
2.2 External Resources
To expand the provided dataset, we used as external
resource the MIMIC-III critical care database (John-
son et al., 2016), which is a large and freely-available
database containing medications, laboratory measure-
ments, imaging reports and other clinical data from
around 40 thousand adult patients. In this work, we
used around 2 million clinical reports (1) to create
word embeddings to be used in deep learning algo-
rithms, (2) to be priorly selected, pseudo-labeled and
used as additional training data in a semi-supervised
setting, and (3) to find text patterns to help in the de-
HEALTHINF 2019 - 12th International Conference on Health Informatics
60

Table 3: ICD-9 medical codes related with some of the se-
lection criteria.
Tag ICD-9 diagnosis and procedure codes
ABDOMINAL
536.3, 536.4, 537.2, 537.3, 537.5, 539, 555.0,
555.2, 560, 564.4, 569.6, 751.1, 863, 864, 865,
866, 868, 996.81, 996.82, 996.86, 996.87,
E879.5, 42, 43, 44, 45, 45.4, 45.7, 47, 50, 51, 52
ALCOHOL-ABUSE 303, 305.0, 980, V11.3
ASP-FOR-MI E935.3
DIETSUPP-2MOS V65.3, 280, 264, 265, 266, 267, 269
DRUG-ABUSE
304, 305.2, 305.3, 305.4, 305.5, 305.6, 305.7,
305.8, 305.9
MAJOR-DIABETES
249, 249.4, 249.5, 249.6, 249.7, 249.8, 250,
250.4, 250.5, 250.6, 250.7, 337.1, 357.2, 362.0,
588.1, 997.6, E878.5, 84.0, 84.1, 84.3, 84.91
MI-6MOS 410, 412
velopment of hand-crafted rules.
Since the clinical reports in the MIMIC-III
database possess ICD-9 diagnosis and procedure
codes
2,3
, we decided to explore those ICD-9 codes
for the selection of relevant clinical reports from
the MIMIC-III database. To do that, we manually
mapped each tag into a list of possible ICD-9 codes
(the resulting mapping is presented in Table 3), and
used the mapped codes to select relevant records from
the database. The filtered list of clinical reports was
then classified using a machine learning approach and
reports with higher confidence were selected to be
used as additional positive (‘met’) training samples.
3 METHODOLOGY
The objective of this work was to explore NLP tech-
niques to solve the problem of automatic patient co-
hort selection. The problem in question consists in
classifying 13 binary criteria for each patient given
their clinical textual records. Classifying each tag
as ‘met’ or ‘not met’ was considered a single binary
problem, where machine learning models were tested
separately and rule-based methods were developed in-
dividually for each criterion. Our final system was
a combination of both, where some tags were bet-
ter solved using heuristics and others using machine
learning algorithms.
2
http://www.icd9data.com
3
The ICD-9 codes are generated during patient admis-
sion for billing purposes.
In this work, we used five classical machine learn-
ing classifiers from the scikit-learn and xgboost li-
braries (Pedregosa et al., 2011; Chen and Guestrin,
2016), and built two deep learning models using the
Keras library (Chollet et al., 2015). These are pre-
sented in more detail in the Subsections 3.3 and 3.4.
3.1 Timeline Restrictions
For a majority of tags, all the clinical records of each
patient were concatenated resulting in a unique tex-
tual document per patient, and for simplicity pur-
poses we ignored date information in clinical records.
However, for tags KETO-1YR and MI-6MOS only the
records from the past 1 year and past 6 months, re-
spectively, were considered since these criteria have
time restrictions. Despite criteria DIETSUPP-2MOS
restricting intake of dietary supplements in the past
2 months, older records were also considered since
these could indicate past supplements still being in-
gested.
3.2 Rule-based Methods
From inspecting the training dataset, its statistics and
understanding the selection criteria, we perceived that
developing hand-crafted rules to find text patterns
would be the most effective solution for certain tags.
For instance, this behaviour applied to tags CRE-
ATININE and HBA1C where float values had to be
found in the text near “creatinine” and “HbA1c” men-
tions, being an information that is not considered in
the supervised learning approach (only in heuristics).
Moreover, certain tags had one of the classes with
very small support, and in those cases we expected
that machine learning classifiers could not correctly
learn due to the lack of training samples, whereas
rule-based methods were expected to have better pre-
diction capability. With this in mind, rules were im-
plemented for all tags with the exception of the AB-
DOMINAL and MAJOR-DIABETES tags.
We developed two rule-based classifiers: one for
submitting the results to the n2c2 Shared-Task, and
another (after submission) by improving some of the
old rules by doing a more exhaustive error anal-
ysis on the training set. However, we acknowl-
edged that this manual modification of the rules be-
ing evaluated in the training set could lead to over-
fitting. The rules were altered for the following
9 tags: ADVANCED-CAD, ALCOHOL-ABUSE, ASP-
FOR-MI, CREATININE, DRUG-ABUSE, ENGLISH,
HBA1C, MAKES-DECISIONS, and MI-6MOS.
Both of the developed rule-based classifiers re-
ceive as input the raw text of the concatenated
Rule-based and Machine Learning Hybrid System for Patient Cohort Selection
61

records. The rules implemented in both classifiers
not only try to identify keywords specific to the
criterion of interest using regular expressions, but
also make complex decisions using if-else conditions.
Rules for catching negation cases were also taken
into account. Reports from the MIMIC-III database
were also consulted to expand the rules, namely for
the criteria ALCOHOL-ABUSE, DRUG-ABUSE, EN-
GLISH, and MAKES-DECISIONS. Additionally, the
DrugBank database (Wishart et al., 2018) was used
for compiling a list of supplements for the criteria
DIETSUPP-2MOS.
3.3 Classical Machine Learning
To feed the classical machine learning classi-
fiers, documents were firstly vectorized using a
bag-of-words (BoW) approach. In the tokenization
step, all the words were converted to lowercase ex-
cept for those with all uppercase letters as they could
represent acronyms, and stopwords were discarded.
Preliminary results showed that feeding the classifiers
with bigrams and trigrams in addition to unigrams
did not result in significant improvements, thus in this
work we only considered the use of unigrams.
The scikit-learn and xgboost APIs were used to
explore the following classical machine learning clas-
sifiers: AdaBoostClassifier, BaggingClassifier, De-
cisionTreeClassifier, GradientBoostingClassifier, and
XGBClassifier. All classifiers were used with their
respective default hyperparameter settings.
3.4 Deep Learning
In this work we tested two deep learning classifiers:
an artificial neural network (NN) and a convolutional
neural network (CNN). Both models were imple-
mented with the Keras API and Table 4 presents a
detailed structure of each model.
Each document was represented by the concate-
nation of its words using word embeddings, with a
fixed length of 5000 words. The word embeddings
were created from around 2 million MIMIC-III clini-
cal reports, using the word2vec architecture (Mikolov
et al., 2013) from the gensim library (
ˇ
Reh˚u
ˇ
rek and
Sojka, 2010). The final word embedding model con-
tained around 100 thousand distinct words.
From preliminary experiments we decided to use
word embeddings generated with the skip-gram ar-
chitecture, a feature size of 50, a window of 5, and
with all the words converted to lowercase. Further-
more, the models were trained using a batch size of
256 samples for a period of 30 epochs.
Table 4: The structure of the deep learning models.
Model Structure
NN
Embedding layer
Flatten layer
Dense Layer with 128 units
ReLU activation
Dense layer with 128 units
ReLU activation
Dropout with rate 0.2
Single unit with sigmoid activation
CNN
Embedding layer
Conv1D layer with 128 filters
ReLU activation
Global max pooling operation
Dense layer with 128 units
ReLU activation
Dropout with rate 0.2
Single unit with sigmoid activation
3.5 Overall System
The system herein described is composed of heuris-
tics and machine learning based methods, with clas-
sical machine learning and deep learning models
having been tested. Our approach consisted in
selecting the methods which achieved better re-
sults in the training set and transposing them to
the test set. The source code of our work de-
veloped for sections 3.2 and 3.3 is available at
https://github.com/ruiantunes/2018-n2c2-track-1/.
The rule-based methods take as input raw text,
while the classical machine learning classifiers use
BoW unigrams, and the deep learning models use
word embeddings.
In the supervised learning approaches, pre-se-
lected MIMIC-III clinical reports (using the ICD-9
codes) were priorly classified considering the prob-
ability output by an ensemble classifier pre-trained
with the training set
4
. We tested this setup in 7 tags as
shown in Table 3.
Additionally, an optional pre-processing step was
developed for the removal of tabular information from
text with the aim of limiting document content to nat-
ural text.
At the final stage of the pipeline, the pre-process-
ing style, classifier (heuristics or machine learning)
and data (training with/without additional MIMIC-III
reports) are chosen so that the best combination can
4
The ensemble provides the average of the probabilities
obtained from five different classical machine learning clas-
sifiers.
HEALTHINF 2019 - 12th International Conference on Health Informatics
62
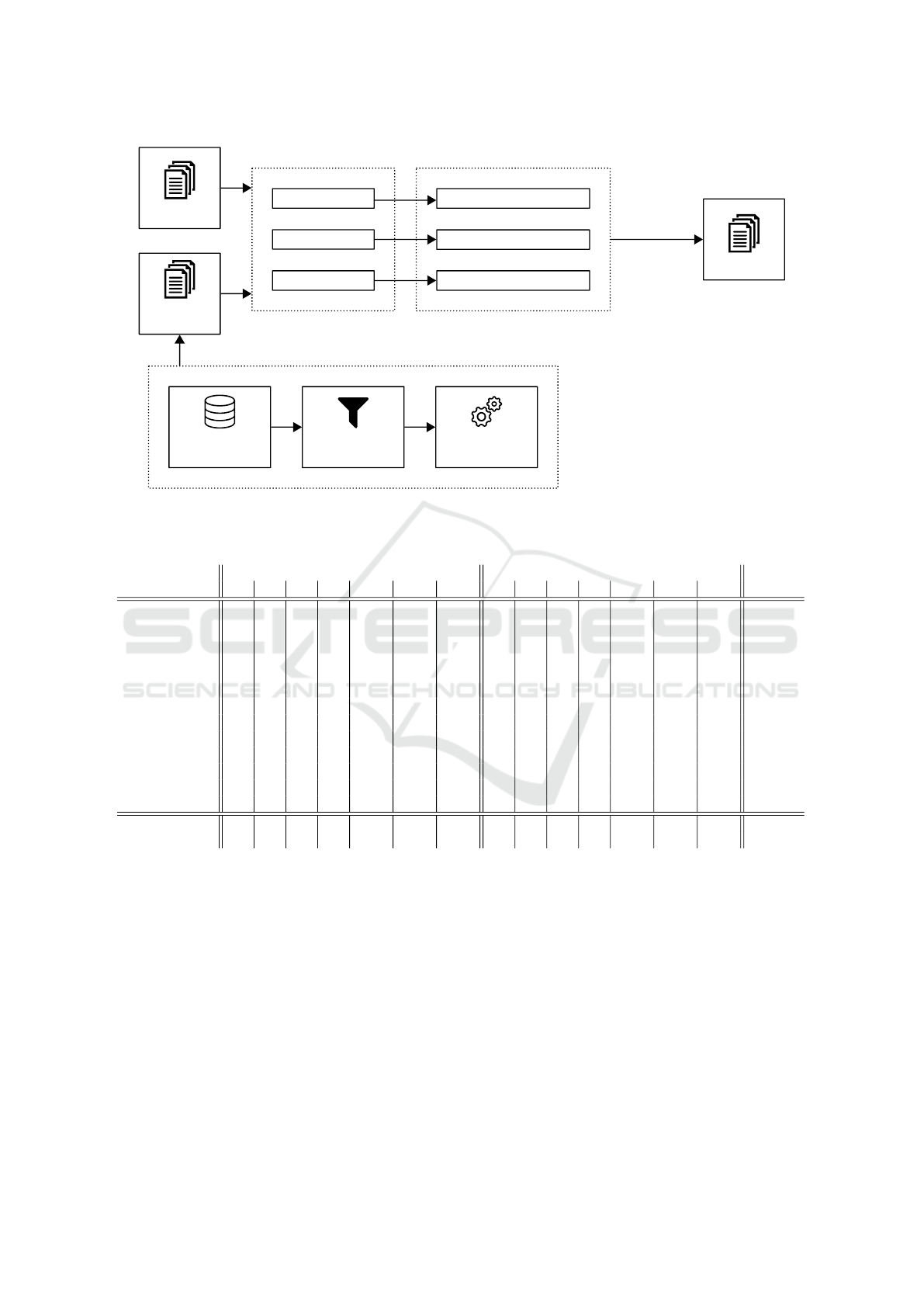
Pre-processing
raw texts
BoW unigrams
word embeddings
rule-based
classical machine learning
Classification
deep learning
training set
additional
MIMIC set
test set
MIMIC-III
clinical reports
Filtered by
ICD-9 codes
Pseudo-labeled by
machine learning
select best
configuration
Figure 1: Final system architecture.
Table 5: Detailed results with a baseline classifier applied to the test set. TP: true positive. TN: true negative. FP: false
positive. FN: false negative. P: precision. R: recall. F1: F1-score.
Tag
Met Not met
Overall F1
TP TN FP FN P R F1 TP TN FP FN P R F1
ABDOMINAL 0 56 0 30 0.0000 0.0000 0.0000 56 0 30 0 0.6512 1.0000 0.7887 0.3944
ADVANCED-CAD 45 0 41 0 0.5233 1.0000 0.6870 0 45 0 41 0.0000 0.0000 0.0000 0.3435
ALCOHOL-ABUSE 0 83 0 3 0.0000 0.0000 0.0000 83 0 3 0 0.9651 1.0000 0.9822 0.4911
ASP-FOR-MI 68 0 18 0 0.7907 1.0000 0.8831 0 68 0 18 0.0000 0.0000 0.0000 0.4416
CREATININE 0 62 0 24 0.0000 0.0000 0.0000 62 0 24 0 0.7209 1.0000 0.8378 0.4189
DIETSUPP-2MOS 0 42 0 44 0.0000 0.0000 0.0000 42 0 44 0 0.4884 1.0000 0.6562 0.3281
DRUG-ABUSE 0 83 0 3 0.0000 0.0000 0.0000 83 0 3 0 0.9651 1.0000 0.9822 0.4911
ENGLISH 73 0 13 0 0.8488 1.0000 0.9182 0 73 0 13 0.0000 0.0000 0.0000 0.4591
HBA1C 0 51 0 35 0.0000 0.0000 0.0000 51 0 35 0 0.5930 1.0000 0.7445 0.3723
KETO-1YR 0 86 0 0 0.0000 0.0000 0.0000 86 0 0 0 1.0000 1.0000 1.0000 0.5000
MAJOR-DIABETES 43 0 43 0 0.5000 1.0000 0.6667 0 43 0 43 0.0000 0.0000 0.0000 0.3333
MAKES-DECISIONS 83 0 3 0 0.9651 1.0000 0.9822 0 83 0 3 0.0000 0.0000 0.0000 0.4911
MI-6MOS 0 78 0 8 0.0000 0.0000 0.0000 78 0 8 0 0.9070 1.0000 0.9512 0.4756
micro-averaged 312 541 118 147 0.7256 0.6797 0.7019 541 312 147 118 0.7863 0.8209 0.8033 0.7526
macro-averaged 0.2791 0.3846 0.3183 0.4839 0.6154 0.5341 0.4262
be applied in the test set. Figure 1 shows the final
overall system architecture.
Note that, for the KETO-1YR tag, the machine
learning models were not trained (due to the lack of
training samples) being the output pre-defined to al-
ways be ‘not met’ in this case.
4 RESULTS AND DISCUSSION
In this section we present several results obtained by
applying different methods in the training and test
sets. The performance of the machine learning algo-
rithms was evaluated using 3-fold cross-validation in
the training set.
We used two evaluation metrics proposed by the
n2c2 organizers which take into account the dataset
imbalance: overall micro and macro F1-scores. This
overall score is the average of the two F1-scores of
the ‘met’ and ‘not met’ classes. The evaluation met-
rics were calculated for each tag, thus enabling the
analysis of each criterion separately.
For a clear understanding and detailed exposi-
tion of all the calculated metrics, Table 5 presents
the results from a baseline classifier which simply
attributed the most frequent label in the training set
to all test samples. This baseline classifier attained
a micro-F1 of 0.7526 and a macro-F1 of 0.4262 on
Rule-based and Machine Learning Hybrid System for Patient Cohort Selection
63
Table 6: Overall averaged F1-scores in the training and test sets. Ada: AdaBoostClassifier. Bag: BaggingClassifier. DT:
DecisionTreeClassifier. GB: GradientBoostingClassifier. XGB: XGBClassifier. NN: neural network. CNN: convolutional
neural network. RB: rule-based classifier. MRB: modified rule-based classifier.
3-fold CV on the training set
Evaluation on the test set
Tag
Classical Machine Learning Deep Learning Rule-based Classical Machine Learning Deep Learning Rule-based
Ada Bag DT GB XGB NN CNN RB MRB Ada Bag DT GB XGB NN CNN RB MRB
ABDOMINAL 0.6071 0.5024 0.5281 0.5868 0.5654 0.5717 0.4257 0.7574 0.5590 0.7079 0.7807 0.6334 0.4658 0.3944
ADVANCED-CAD 0.6780 0.6525 0.6034 0.7208 0.7611 0.4951 0.4977 0.8251 0.8251 0.7977 0.7889 0.6178 0.8227 0.8114 0.4113 0.6673 0.7832 0.8089
ALCOHOL-ABUSE 0.4899 0.4912 0.4807 0.4847 0.4912 0.4912 0.4912 0.8598 1.0000 0.5896 0.4911 0.4850 0.5896 0.4911 0.4911 0.4911 0.4850 0.4850
ASP-FOR-MI 0.5144 0.4843 0.4946 0.5025 0.4603 0.4466 0.4466 0.7916 0.8625 0.4847 0.4401 0.5271 0.5469 0.4908 0.4416 0.4416 0.7095 0.7426
CREATININE 0.7760 0.7959 0.7258 0.7723 0.8042 0.4189 0.5846 0.8895 0.9118 0.7329 0.7219 0.5933 0.7219 0.7110 0.4948 0.5411 0.8295 0.7862
DIETSUPP-2MOS 0.6526 0.6432 0.5937 0.6926 0.7126 0.6539 0.5308 0.7975 0.6728 0.5597 0.5930 0.6510 0.6162 0.5390 0.5083 0.7943
DRUG-ABUSE 0.7123 0.5795 0.6802 0.7370 0.4873 0.4873 0.4873 0.7020 1.0000 0.4850 0.4911 0.6815 0.6601 0.4911 0.4911 0.4911 0.7312 0.9255
ENGLISH 0.7780 0.7780 0.8837 0.8837 0.5795 0.4873 0.4873 0.9172 1.0000 0.7559 0.7929 0.7915 0.7929 0.5983 0.4591 0.4591 0.6554 0.6554
HBA1C 0.5429 0.5139 0.5588 0.5279 0.5702 0.4568 0.4006 0.9374 0.9601 0.6098 0.6048 0.6210 0.5773 0.5773 0.3676 0.3723 0.9382 0.8439
KETO-1YR 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.4971
MAJOR-DIABETES 0.7375 0.6929 0.6554 0.7483 0.7429 0.5656 0.5473 0.7902 0.8023 0.6975 0.8721 0.8023 0.6044 0.5966
MAKES-DECISIONS 0.4873 0.4899 0.6192 0.5706 0.4899 0.4899 0.4899 0.8256 1.0000 0.4911 0.4881 0.6277 0.4850 0.7440 0.4911 0.4911 0.6067 0.4911
MI-6MOS 0.4753 0.5306 0.5936 0.5097 0.5730 0.4767 0.4767 0.8026 0.8778 0.4724 0.4756 0.4625 0.4724 0.4756 0.4756 0.4756 0.7281 0.8102
micro-averaged 0.8198 0.8108 0.7682 0.8222 0.8355 0.7813 0.7858 0.8331 0.8134 0.7775 0.8356 0.8258 0.7566 0.7676
macro-averaged 0.6117 0.5888 0.6090 0.6336 0.5952 0.5031 0.4897 0.6261 0.5935 0.6081 0.6517 0.6110 0.4794 0.4946
Table 7: Overall averaged F1-scores with classifiers trained with 100 additional ‘met’ training MIMIC-III reports. Ada:
AdaBoostClassifier. Bag: BaggingClassifier. DT: DecisionTreeClassifier. GB: GradientBoostingClassifier. XGB: XGBClas-
sifier.
Tag
3-fold CV on the training set Evaluation on the test set
Ada Bag DT GB XGB Ada Bag DT GB XGB
ABDOMINAL 0.6669 0.5890 0.6075 0.6655 0.6581 0.7511 0.7617 0.6595 0.7352 0.7507
ALCOHOL-ABUSE 0.6729 0.7949 0.7040 0.7040 0.7159 0.5753 0.4911 0.4819 0.4911 0.4911
ASP-FOR-MI 0.5848 0.5365 0.4976 0.4951 0.4873 0.4673 0.5232 0.5784 0.4788 0.4379
DIETSUPP-2MOS 0.6977 0.6781 0.6219 0.7335 0.7106 0.6012 0.6007 0.5697 0.6510 0.6977
DRUG-ABUSE 0.7228 0.7093 0.6456 0.6962 0.7093 0.7440 0.6910 0.6601 0.6815 0.7440
MAJOR-DIABETES 0.7335 0.7214 0.6231 0.7537 0.7489 0.7790 0.6512 0.7089 0.8372 0.8140
MI-6MOS 0.6930 0.7023 0.7020 0.6842 0.6842 0.4724 0.4724 0.4625 0.4724 0.4724
the test set. To produce simpler presentations of the
results, from now on we will only present the over-
all F1-scores. However, detailed results containing
true/false positive/negative counts, as presented in Ta-
ble 5, were examined during the refinement of our ap-
proach.
Table 6 shows the results of rule-based and ma-
chine learning methods evaluated on the training and
test sets. Looking only at the evaluation in the train-
ing set, one can see that for each tag where rules were
implemented, the rule-based method was the best per-
forming classifier. On the other hand, deep learning
models produced the worst results.
The AdaBoostClassifier and the GradientBoost-
ingClassifier achieved the two highest macro-aver-
age F1-scores both in training and test sets. For the
tags where the modified rule-based classifier was im-
plemented, this classifier achieved the best results
in the training set, but the same was not verified
for the test set which shows that certain rules were
overfit to the training set. For the tags ABDOMI-
NAL, ADVANCED-CAD, and MAJOR-DIABETES, the
results obtained with classical machine learning sig-
nificantly improved in the test set proving that train-
ing with more data helped.
Table 7 shows the results when 100 additional
‘met’ training MIMIC-III reports are used for train-
ing. Because these additional reports were classified
with pre-trained classifiers on the full training dataset,
these results are indirectly biased. This explains why
the results in the training set had significant improve-
ments, whereas for the test set obtained improvements
were less significant.
Table 8 presents the results obtained when apply-
ing classical machine learning with tabulated infor-
mation removed from the raw texts. Significant dif-
ferences were not found when compared to the results
presented in Table 6.
HEALTHINF 2019 - 12th International Conference on Health Informatics
64
Table 8: Overall averaged F1-scores with tabulated information removed from the raw texts. Ada: AdaBoostClassifier. Bag:
BaggingClassifier. DT: DecisionTreeClassifier. GB: GradientBoostingClassifier. XGB: XGBClassifier.
Tag
3-fold CV on the training set Evaluation on the test set
Ada Bag DT GB XGB Ada Bag DT GB XGB
ABDOMINAL 0.6325 0.5466 0.5676 0.5957 0.5733 0.7399 0.5419 0.6765 0.8294 0.6052
ADVANCED-CAD 0.6668 0.6952 0.5761 0.7405 0.7405 0.8089 0.7531 0.6549 0.8350 0.8350
ALCOHOL-ABUSE 0.4899 0.4912 0.4794 0.4847 0.4912 0.5753 0.4911 0.5753 0.5753 0.4911
ASP-FOR-MI 0.5064 0.5076 0.5190 0.5084 0.4603 0.4908 0.4454 0.5947 0.5086 0.4342
CREATININE 0.7726 0.7726 0.6966 0.7864 0.7978 0.6718 0.6946 0.6142 0.7141 0.7329
DIETSUPP-2MOS 0.6261 0.6574 0.6081 0.6631 0.6830 0.7089 0.6073 0.5216 0.6158 0.6728
DRUG-ABUSE 0.7123 0.4873 0.5825 0.7093 0.4873 0.4819 0.4911 0.6601 0.6601 0.4911
ENGLISH 0.7780 0.9079 0.8422 0.8837 0.5795 0.7559 0.7929 0.7737 0.7929 0.5983
HBA1C 0.6087 0.5568 0.5462 0.5216 0.5816 0.5951 0.4574 0.5232 0.5681 0.5577
KETO-1YR 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000 0.5000
MAJOR-DIABETES 0.7637 0.6859 0.6354 0.7725 0.7094 0.7906 0.7673 0.6510 0.8604 0.8136
MAKES-DECISIONS 0.4873 0.4899 0.5629 0.4886 0.4899 0.4881 0.4911 0.6546 0.4850 0.7440
MI-6MOS 0.4753 0.5306 0.5601 0.5097 0.5730 0.4724 0.4756 0.4658 0.4724 0.4756
micro-averaged 0.8270 0.8185 0.7657 0.8263 0.8306 0.8298 0.8031 0.7675 0.8336 0.8304
macro-averaged 0.6169 0.6022 0.5905 0.6280 0.5897 0.6215 0.5776 0.6051 0.6475 0.6117
Table 9: Overall averaged F1-scores with the best combination of methods selected by inspecting the evaluation in the training
set. The methods that provided the best results in the training set were chosen.
Selected method Tag
Evaluation on
Training Test
AdaBoostClassifier with 100 additional training MIMIC reports ABDOMINAL 0.6669 0.7511
Rule-based classifier ADVANCED-CAD 0.8251 0.8089
Modified rule-based classifier ALCOHOL-ABUSE 1.0000 0.4850
Modified rule-based classifier ASP-FOR-MI 0.8625 0.7426
Modified rule-based classifier CREATININE 0.9118 0.7862
Rule-based classifier DIETSUPP-2MOS 0.7975 0.7943
Modified rule-based classifier DRUG-ABUSE 1.0000 0.9255
Modified rule-based classifier ENGLISH 1.0000 0.6554
Modified rule-based classifier HBA1C 0.9601 0.8439
Rule-based classifier KETO-1YR 0.5000 0.4971
GradientBoostingClassifier with tabulated information discarded MAJOR-DIABETES 0.7725 0.8604
Modified rule-based classifier MAKES-DECISIONS 1.0000 0.4911
Modified rule-based classifier MI-6MOS 0.8778 0.8102
micro-averaged 0.9143 0.8844
macro-averaged 0.8596 0.7271
Rule-based and Machine Learning Hybrid System for Patient Cohort Selection
65

Finally, Table 9 shows the final results when the
best combination was selected by inspecting the re-
sults in the training set. The best combination in
the training set achieved a micro-F1 of 0.9143 and a
macro-F1 of 0.8596 whereas in the test set it attained
a micro-F1 of 0.8844 and a macro-F1 of 0.7271. Re-
sults show that there is a clear overfitting to the train-
ing set because the macro-F1 on the test set is around
13 percentage points smaller. In this optimal configu-
ration, rule-based methods were mostly selected.
5 CONCLUSIONS
In this paper we proposed a system for the automatic
classification of 13 binary selection criteria given only
patient clinical records. The development of systems
as the one herein described is vital for helping physi-
cians in the selection of patient cohorts for clinical tri-
als, which is a task known to be both time-consuming
and complex.
Our system contains rule-based methods and ma-
chine learning algorithms that are accordingly se-
lected to better classify each criterion. In this work,
we developed hand-crafted rules for almost all the cri-
teria. However, the process of creating adequate rules
is hard and cumbersome since it requires an analysis
of the data, not excluding the medical expertise that
is oftentimes required. Moreover, while rule-based
methods achieved good results, these require the de-
velopment of a distinct algorithm for each criterion
while machine learning classifiers do not face this
problem, being easier to re-use.
In this task, classical machine learning classifiers
worked much better when compared to deep learning
classifiers. In most cases, deep learning models pre-
dicted the same label every time, behaving similarly
to a baseline classifier and proving that the dataset had
a reduced size. Our results do also show that machine
learning classifiers provided better results for crite-
ria with balanced labels, evidencing that other criteria
lack training data.
As future work, a better pre-processing step can
be followed and the developed rules can be improved
with the help of medical expertise. Furthermore, an-
other possible way of improving the performance of
the system in some criteria consists in the implemen-
tation of different techniques for augmenting training
data with data from external resources. Finally, other
techniques for using distributed word representations
could be considered, and optimization of the classi-
fier hyperparameters could be performed through grid
search.
ACKNOWLEDGEMENTS
This project was partially funded by the In-
tegrated Programme of SR&TD “SOCA” (Ref.
CENTRO-01-0145-FEDER-000010) and “MMIR”
(Ref. PTDC/EEI-ESS/6815/2014), co-funded by
Centro 2020 program, Portugal 2020, European
Union, through the European Regional Development
Fund.
Rui Antunes is supported by the Fundação
para a Ciência e a Tecnologia (PhD Grant
SFRH/BD/137000/2018). João Figueira Silva is
supported by the Fundação para a Ciência e a Tec-
nologia (PhD Grant PD/BD/142878/2018). Arnaldo
Pereira is supported by the Fundação para a Ciência
e a Tecnologia (PhD Grant PD/BD/142877/2018).
REFERENCES
Chen, T. and Guestrin, C. (2016). XGBoost: a scalable
tree boosting system. In Proceedings of the 22nd
ACM SIGKDD International Conference on Knowl-
edge Discovery and Data Mining, pages 785–794, San
Francisco, California, USA. ACM.
Chollet, F. et al. (2015). Keras. https://keras.io.
ˇ
Reh˚u
ˇ
rek, R. and Sojka, P. (2010). Software framework for
topic modelling with large corpora. In Proceedings of
the LREC 2010 Workshop on NewChallenges for NLP
Frameworks, pages 45–50, Valletta, Malta. ELRA.
Johnson, A. E. W., Pollard, T. J., Shen, L., Lehman, L.-
w. H., Feng, M., Ghassemi, M., Moody, B., Szolovits,
P., Anthony Celi, L., and Mark, R. G. (2016). MIMIC-
III, a freely accessible critical care database. Scientific
Data, 3.
Ludvigsson, J. F., Pathak, J., Murphy, S., Durski, M.,
Kirsch, P. S., Chute, C. G., Ryu, E., and Murray, J. A.
(2013). Use of computerized algorithm to identify in-
dividuals in need of testing for celiac disease. Jour-
nal of the American Medical Informatics Association,
20(e2):e306–e310.
Mann, C. J. (2003). Observational research methods.
Research design II: cohort, cross sectional, and
case-control studies. Emergency Medicine Journal,
20(1):54–60.
Mikolov, T., Chen, K., Corrado, G., and Dean, J. (2013).
Efficient estimation of word representations in vector
space. arXiv e-print.
Pathak, J., Kho, A. N., and Denny, J. C. (2013). Electronic
health records-driven phenotyping: challenges, recent
advances, and perspectives. Journal of the American
Medical Informatics Association, 20(e2):e206–e211.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer,
P., Weiss, R., Dubourg, V., Vanderplas, J., Passos,
A., Cournapeau, D., Brucher, M., Perrot, M., and
Duchesnay, E. (2011). Scikit-learn: machine learning
HEALTHINF 2019 - 12th International Conference on Health Informatics
66

in Python. Journal of Machine Learning Research,
12:2825–2830.
Shivade, C., Raghavan, P., Fosler-Lussier, E., Embi, P. J.,
Elhadad, N., Johnson, S. B., and Lai, A. M. (2014).
A review of approaches to identifying patient pheno-
type cohorts using electronic health records. Jour-
nal of the American Medical Informatics Association,
21(2):221–230.
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J.,
Marcu, A., Grant, J. R., Sajed, T., Johnson, D., Li,
C., Sayeeda, Z., Assempour, N., Iynkkaran, I., Liu,
Y., Maciejewski, A., Gale, N., Wilson, A., Chin, L.,
Cummings, R., Le, D., Pon, A., Knox, C., and Wil-
son, M. (2018). DrugBank 5.0: a major update to the
DrugBank database for 2018. Nucleic Acids Research,
46(D1):D1074–D1082.
Rule-based and Machine Learning Hybrid System for Patient Cohort Selection
67
