
Accelerated Algorithm for Computation of All Prime Patterns in Logical
Analysis of Data
Arthur Chambon
1
, Fr
´
ed
´
eric Lardeux
1
, Fr
´
ed
´
eric Saubion
1
and Tristan Boureau
2
1
LERIA, Universit
´
e d’Angers, Angers, France
2
UMR1345 IRHS, Universit
´
e d’Angers, Angers, France
Keywords:
Logical Analysis of Data, Pattern Generation, Logical Characterization of Data.
Abstract:
The analysis of groups of binary data can be achieved by logical based approaches. These approaches identify
subsets of relevant Boolean variables to characterize observations and may help the user to better understand
their properties. In logical analysis of data, given two groups of data, patterns of Boolean values are used to
discriminate observations in these groups. In this work, our purpose is to highlight that different techniques
may be used to compute these patterns. We present a new approach to compute prime patterns that do not
provide redundant information. Experiments are conducted on real biological data.
1 INTRODUCTION
Context. Logical analysis of data (LAD), introdu-
ced for the first time by Peter Hammer (Hammer,
1986), is based on combinatorial optimization techni-
ques and on the concept of partially defined Boolean
functions. It may be considered as an alternative to
conventional statistical classification methods. LAD
(Crama et al., 1988) can be used in various applica-
tion domains. One of its purpose is the characteriza-
tion of data by means of patterns (and subsequently
by logical formulas). Given two sets of data (groups),
these patterns are indeed subsets of values that are
present in several observations of one set, while not
being present in the other set. Hence, patterns can be
used to identify common characteristics of observati-
ons belonging to the same group. The idea is to focus
on explicit justifications of groups of data, while clas-
sic classification approaches mainly focus on the con-
struction or the identification of these groups. More
precisely, the purposes of LAD are:
• to determine similarities within the same data set,
• to discriminate observations belonging to diffe-
rent data sets.
• to deduce logical rules/formulas that explain data
sets.
As mentioned above, LAD focuses merely on ex-
planation when classification techniques allow the
user to build cluster and to assign groups to incoming
data. Many applications of logical data analysis have
been investigated in medicine (Reddy et al., 2008), in-
dustry (Mortada et al., 2012; Dupuis et al., 2012) or
economy (Hammer et al., 2012).
Example. Let us consider two groups (sets) of ob-
servations P and N (respectively positive and negative
observations) defined over a set A of Boolean varia-
bles. In this example, we consider a set of 8 Bool-
ean variables (labeled from a to h) and 7 observations.
Our purpose is to compute a subset of A that may be
used to explain/justify a priori the membership of ob-
servations to their respective groups.
As mentioned above, contrary to classification ap-
proaches issued from machine learning techniques
(e.g., clustering algorithms), the purpose here is to
provide an explicit justification of the data instead of
an algorithm that assigns groups to data. Note that
we assume that the two groups are built by experts, or
using expert knowledge (this is thus definitely not a
classification nor a clustering problem).
Observ. Groups
Variables
a b c d e f g h
1
P
0 1 0 1 0 1 1 0
2 1 1 0 1 1 0 0 1
3 0 1 1 0 1 0 0 1
4
N
1 0 1 0 1 0 1 1
5 0 0 0 1 1 1 0 0
6 1 1 0 1 0 1 0 1
7 0 0 1 0 1 0 1 0
210
Chambon, A., Lardeux, F., Saubion, F. and Boureau, T.
Accelerated Algorithm for Computation of All Prime Patterns in Logical Analysis of Data.
DOI: 10.5220/0007389702100220
In Proceedings of the 8th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2019), pages 210-220
ISBN: 978-989-758-351-3
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

In LAD methodology, a key concept consists in
identifying patterns of similar values in groups. For
instance, a = 0 and b = 1 is a pattern that is shared
by observations 1 and 3 in P and such that no ob-
servation in N is covered by this pattern. Therefore,
this pattern could be interpreted as a partial explana-
tion of the observations of group P. Among the sets
of patterns, one has to decide which compromise has
to be achieved between their size and their covering
(i.e., the number of observations having the pattern in
group P). Concerning the size of the patterns, some
properties have been exhibited in order to focus on
the most relevant ones. In particular, prime patterns
are patterns whose number of variables cannot be re-
duced unless they are not patterns anymore. Prime
patterns correspond to the simplicity requirement (in
terms of variables), while strong patterns correspond
to an evidential preference where a larger cover is pre-
ferred (we refer the reader to (Chikalov et al., 2013)
for a survey on LAD).
Alternatively, variables f and g can also be used to
generate a Boolean formula φ ≡ ( f ∧ g) ∨ (¬ f ∧ ¬g),
which is true for observations in P (interpreted as
Boolean assignments on variables) and false for ob-
servations in N. Note that the variable b is not suf-
ficient to explain group P since observation 6 in N
has also this variable set to 1. φ is presented here
in disjunctive normal form. Note that such formula
could be convenient for users, either by minimizing
the number of variables (for instance, to simplify their
practical implementation in diagnosis routines) or by
minimizing the size of the formula (for instance, to
improve their readability). Such an approach focu-
ses on minimal characterizations in terms of num-
ber of variables and can be extended to consider se-
veral groups simultaneously (Chhel et al., 2012). In
our work, it is important to consider prime patterns
(without redundant variable) in order to minimize the
number of variables, and to consider as many patterns
as possible in order to have a broader view to mini-
mize the size of the formula.
In our example, we consider only two possible
states for each variables, so we work on binary data.
However, in practice, this is not always the case. In
(Boros et al., 1997), “binarization” of data is introdu-
ced, allowing to transform quantitative data into bi-
nary data. The basic idea of binarization is simple:
each real-valued data is associated to a threshold. The
binary value is 1 (respectively 0) if the real data is
above this threshold (respectively below). Since a sin-
gle threshold is too restrictive, it is important to study
several thresholds combinations. One of the problems
associated with binarization is therefore to find a mi-
nimal number of thresholds that would preserve most
of the information contained in a data set (Hammer
and Bonates, 2006; Boros et al., 1997).
Contributions. In this paper, we present a new al-
gorithm that computes the set of all prime patterns.
Other algorithms can be used to compute prime pat-
terns, in particular the algorithm presented in (Boros
et al., 2000). However our algorithm, based on the de-
tection of solutions in logical characterization of data,
is faster and works on a more important set of varia-
bles. Moreover our algorithm allows us to determine
the coverage of each patterns.
Organization. In Section 2 we recall the concepts
of the logical analysis of data and the computation of
prime patterns. In Section 3 we recall the main con-
cepts of logical characterization of data. In Section 4,
we present our new algorithm. Finally, in Section 5
we compare the performances of our algorithm with
the algorithm of (Boros et al., 2000) on different real
instances (issued from biology) and handmade instan-
ces.
2 LOGICAL ANALYSIS OF DATA
2.1 Terminology and Notation
Let us recall the main concepts of logical analysis of
data (LAD) (Hammer et al., 2004; Hammer and Bo-
nates, 2006; Boros et al., 2011; Chikalov et al., 2013).
LAD is based on the notion of partially defined Bool-
ean functions.
Definition 1. A Boolean function f of n variables,
n ∈ N, is a function f : B
n
→ B, where B is the set
{0,1}.
Definition 2. A vector x ∈ B
n
is a positive vector
(resp. negative vector) of the Boolean function f if
f (x) = 1 (resp f (x) = 0). T ( f ) (resp. F( f )) is the
set of positive vectors (resp. negative vectors) of the
Boolean function f .
In the rest of the paper, Boolean vectors corre-
spond to observations. The set of observations is de-
noted Ω.
Definition 3. A partially defined Boolean function
(pdBf) on B
n
is a pair (P,N) such that P, N ⊆ B
n
and P ∩ N =
/
0.
In a pdBf, we consider two groups of observati-
ons: P (positive group) and N (negative group). A
literal is either a binary variable x
i
or its negation ¯x
i
.
Accelerated Algorithm for Computation of All Prime Patterns in Logical Analysis of Data
211

A term is a pdBf represented by a conjunction of dis-
tinct literals, such that a term does not contain a vari-
able and its negation.
Definition 4. Given: σ
+
,σ
−
⊆ {1,2,...,n}, σ
+
∩
σ
−
=
/
0, a term t
σ
+
,σ
−
is a Boolean function whose
positive set T (t
σ
+
,σ
−
) is of the form:
T (t
σ
+
,σ
−
) = {x ∈ B
n
|x
i
= 1 ∀i ∈ σ
+
and x
j
= 0 ∀ j ∈ σ
−
}
A term t
σ
+
,σ
−
can be represented by an elementary
conjunction, i.e., a Boolean expression of the form:
t
σ
+
,σ
−
(x) = (
^
i∈σ
+
x
i
) ∧ (
^
j∈σ
−
¯x
j
)
We say that a Boolean vector satisfies a term if it
has the same binary values as the term on the variables
of the term. The set of literals of a term t is Lit(t). The
degree of a term t is the number of literals that appear
in this term, ie |Lit(t)|.
Definition 5. A pattern of a pdBf (P,N) is a
term t
σ
+
,σ
−
such that |P ∩ T (t
σ
+
,σ
−
)| > 0 and |N ∩
T (t
σ
+
,σ
−
)| = 0.
A pattern is therefore a term satisfied by at least
one positive vector (from P) and no negative vector
(from N). Of course, patterns are associated to the
positive group P. It is also possible to design patterns
associated to the negative group by considering the
pdBf (N, P) instead of the pdBf (P, N).
Definition 6. The coverage of a pattern p, denoted
Cov(p), is the set Cov(p) = P ∩ T (p).
In other words, the coverage of a pattern p is the
set of positive Boolean vectors satisfying p.
Example 1. Back to introductory example:
Obs Groups
Variables
a b c d e f g h
1
P
0 1 0 1 0 1 1 0
2 1 1 0 1 1 0 0 1
3 0 1 1 0 1 0 0 1
4
N
1 0 1 0 1 0 1 1
5 0 0 0 1 1 1 0 0
6 1 1 0 1 0 1 0 1
7 0 0 1 0 1 0 1 0
p
1
= ¯a ∧ b and p
2
=
¯
f ∧ ¯g are two patterns covering
observations 1 and 3 for p
1
and 2 and 3 for p
2
.
There exist many possible patterns. Using the two
sets Lit(p) and Cov(p), we can define 2 types of par-
tial preorders on patterns: the simplicity preference
and the evidential preference.
Definition 7. The simplicity preference σ, denoted
<
σ
, is a binary relation over a set of patterns P such
that for a couple (p
1
, p
2
) ∈ P
2
, we have p
1
<
σ
p
2
if
and only if Lit(p
1
) ⊆ Lit(p
2
).
Definition 8. The evidential preference E, denoted
<
E
, is a binary relation over a set of patterns P such
that for a couple (p
1
, p
2
) ∈ P
2
, we have p
1
<
E
p
2
if
and only if Cov(p
2
) ⊆ Cov(p
1
).
For a preference <
pi
(π ∈ {σ,E}) and two pat-
terns p
1
and p
2
, we will note the double relation
p
1
<
π
p
2
and p
2
<
π
p
1
by p
1
≈
π
p
2
.
In order to refine the comparison of patterns, we
will consider combinations of preferences.
Definition 9. Let two preferences π and ρ on a set
of patterns P , and let (p
1
, p
2
) ∈ P
2
, the pattern p
1
is preferred to the pattern p
2
with respect to the lexi-
cographic refinement π|ρ, denoted p
1
<
π|ρ
p
2
, if and
only if p
1
<
π
p
2
or p
1
≈
π
p
2
and p
1
<
ρ
p
2
.
Definition 10. Given a preference <
π
on a set of
patterns P , a pattern p
1
∈ P is Pareto-optimal with
respect to π if and only if @p
2
∈ P \{p
1
} such that
p
2
π
p
1
(i.e p
2
<
π
p
1
and p
2
6≈
π
p
1
).
We can thus define types of Pareto-optimal pat-
terns, according to the preferences:
Preference Pareto-optimal pattern
σ Prime pattern
E Strong pattern
E|σ Strong prime pattern
Note the following property demonstrated in
(Hammer et al., 2004).
Property 1. A pattern is pareto-optimal with respect
to E|σ if and only if it is strong and prime.
Example 2 . Let’s take Example 1 to illustrate the
different types of patterns:
• The pattern a ∧ d ∧e is a prime pattern because if
you remove a literal, it is no longer a pattern.
• The e ∧
¯
f ∧ ¯g pattern is a strong pattern because
there is no pattern covering the same observations
plus one.
• The
¯
f ∧ ¯g pattern is both strong and prime. It is
therefore a strong prime pattern.
In (Boros et al., 2000), the notion of support sets
is used. A support set is a subset of variables such
that, by working only on the selected variables, all
positive Boolean vectors are different from negative
Boolean vectors. In other words, we are looking for a
set of variables that discriminate the whole group. A
set support is said to be irredundant if the removal of
any variable gives a set of variables that is no longer
a set support.
2.2 Patterns Generation
The problem of generating optimal patterns can be
solved by a linear program. In (Ryoo and Jang, 2009)
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
212

integer linear programs are proposed, allowing to ge-
nerate different types of patterns, in particular strong
prime patterns. However, if the authors demonstrate
that an optimal solution of their linear program is a
Pareto-optimal pattern, the converse is not true. It is
therefore not possible to generate the set of all prime
patterns (or set of all strong prime patterns) by the li-
near program presented in this article.
If the linear programming approach does not allow
us to generate all the optimal patterns, the algorithm
proposed in (Boros et al., 2000) will generate the set
of all prime patterns. Note also that in (Hammer et al.,
2004) are described algorithms transforming a pattern
into a prime pattern or strong pattern.
2.2.1 Prime Patterns Generation
Concerning the generation of prime patterns, the algo-
rithm in (Boros et al., 2000) proposes the generation
of all prime patterns less than or equal to a degree
D. By choosing the highest degree D
max
of prime pat-
terns, we can generate all of them, but we do not know
how to compute this degree without generating the
set of patterns. We choose an upper bound to D
max
to be certain of our results. By choosing D = n,i.e.
the number of variables, we insure that D > D
max
and
thus, the algorithm will return the set of prime pat-
terns.
Boros’s algorithm first generates prime patterns of
degree 1, then 2 and so on up to degree D. In step
1, the algorithm tests all the terms of degree 1 and
classifies them according to their interest. All terms
that are patterns will be in a set P, and in a set C
1
the
terms of size 1 covering positive and negative obser-
vations. In the following steps (E) we will only con-
sider the terms of the set C
E−1
, terms of degree E − 1,
for which we try to add a literal to transform it into a
term of degree E. If this term is a pattern, it joins the
set P and if it still covers both positive and negative
observations, it joins the set C
E
. Once the step E = D
has been completed, we will have the set P which will
contain all prime patterns of degree less than or equal
to D.
Algorithm 1 presents the pseudo-code of this al-
gorithm.
Since we test all the combinations, we are certain
to generate the set of prime patterns. In addition, to be
generated, a term must be in the set P and thus check
the conditions to be a pattern. As before being in the
set P, the term was in a set C
i
, if we remove a literal,
the term is no longer a pattern. The set P therefore
contains all prime patterns and contains only prime
patterns.
Algorithm 1: Computation of all prime patterns
from (Boros et al., 2000) (PPC 1).
Data: D the maximum degree of patterns that
will be generated.
Result: P
D
the set of prime patterns smaller
than or equal to D.
P
0
=
/
0
//C
i
is the set of terms of degree i that can
become patterns, which cover both positive
and negative observations.
C
0
= {
/
0}
for i = 1 to D do
P
i
= P
i−1
C
i
=
/
0
forall t ∈ C
i−1
do
p=maximum index of variables in t
for s = p + 1 to n do
forall l ∈ {x
s
, ¯x
s
} do
T = t ∧ l
for j = 1 to i − 1 do
t
0
= T with the j-th
variable removed
if t
0
/∈ C
i−1
then
go to ♦
end
end
if T covers a positive vector
but no negative vector then
P
i
= P
i
∪ {T }
end
if T covers both a positive
and a negative vector then
C
i
= C
i
∪ {T }
end
♦
end
end
end
end
return P
D
Note that, for an instance with x observations and n
variables, the complexity is :
O(3
n
× n × (2
n−b
n
3
c
×
n!
b
n
3
c!(n − b
n
3
c)!
+ x))
(3
n
is the number of terms that can be created with
n variables and 2
n−b
n
3
c
×
n!
b
n
3
c!(n−b
n
3
c)!
is the maximal
number of prime patterns).
Accelerated Algorithm for Computation of All Prime Patterns in Logical Analysis of Data
213

3 MULTIPLE
CHARACTERIZATION OF
DATA
Multiple characterization of data is an extension of lo-
gical analysis of data where several groups of data are
considered. The goal is to compute a set of variables,
called solution, sufficient to discriminate each group
of data from the others, simultaneously.
3.1 Presentation
As in LAD, multiple characterization of data (Chhel
et al., 2012) aims to discriminate observations from
different groups. The objective is to determine a set
of variables discriminating all groups simultaneously.
This approach is similar to support sets computation
but the number of groups may be larger than 2.
Example 3. Let us consider again introductory exam-
ple with more than two groups:
Obs Groups
Variables
a b c d e f g h
1
1
0 1 0 1 0 1 1 0
2 1 1 0 1 1 0 0 1
3 0 1 1 0 1 0 0 1
4
2
1 0 1 0 1 0 1 1
5 0 0 0 1 1 1 0 0
6
3
1 1 0 1 0 1 0 1
7 0 0 1 0 1 0 1 0
The variables a and b are not sufficient to discrimi-
nate groups 2 and 3 because the observation 2 is simi-
lar to the observation 6 on these variables. However,
the variables a,b and f discriminate the 3 groups at
the same time because no observations are identical
on these 3 variables.
It is important to note that multiple characterization
of data is different from feature selection since it does
not focus on the most statistically informative vari-
ables, but rather on a combination of variables that
exactly discriminates the groups.
3.2 Terminology and Notations
We use here the notations and formalization propo-
sed in (Chambon et al., 2015). The observations be-
longing to Ω are expressed on Boolean variables be-
longing to the set A. These observations are divided
into several groups belonging to the set of groups G.
These data are represented by a matrix D =.
Definition 11 . An instance of the Multiple Characte-
rization Problem (MCP) is a quadruplet (Ω, A,D,G)
defined by a set of Ω observations whose elements are
expressed on a set of variables A , and are represented
by a matrix of Boolean data D
|Ω|×|A|
and a function
G : Ω → G, such that G(o) is the group to which the
observation o ∈ Ω belongs.
The data matrix is defined as follows:
• The value D[o,a] represents the presence/absence
of the variable a for the observation o.
• A line D[o,.] represents the Boolean vector of pre-
sence/absence of the different variables for the ob-
servation o.
• A column D[.,a] represents the Boolean vector of
presence/absence of the variable a in all observa-
tion.
Thus, two observations o,o
0
∈ Ω can be represen-
ted by the same Boolean vector (so D[o, .] = D[o
0
,.])
and yet be considered as two distinct observations.
In the following we are only interested in satisfia-
ble MCP, i.e. such that D does not contain two iden-
tical observations in two different groups (generaliza-
tion of the notion of support set to multiple groups).
Property 2 . A MCP instance (Ω,A,D,G) is satisfi-
able iff: @(o,o
0
) ∈ Ω
2
such that D[o,.] = D[o
0
,.] and
G(o) 6= G(o
0
)
Definition 12 . Let A ⊂ A, D
A
is the data matrix re-
duced to the subset of variables A.
We now introduce the concept of MCP solution.
Definition 13. Given an instance (Ω,A,D,G), a sub-
set of variables S ⊆ A is a solution if and only if
∀(o,o
0
) ∈ Ω
2
,G(o) 6= G(o
0
) ⇒ D
S
[o,.] 6= D
S
[o
0
,.].
In this case, the matrix D
S
is called a solution matrix.
In other words, S is a solution if two observations,
coming from two different groups, are different on at
least one variable a ∈ S.
Note that in the particular case where we only have
two groups, a MCP solution is a support set (see
Section 2.1).
An instance of the MCP may have several solu-
tions of different sizes. It is therefore important to
define an ordering on solutions in order to compare
and classify them. In particular, for a given solution
S, adding a variable generates a new solution S
0
⊃ S.
In this case we say that S
0
is dominated by S.
Definition 14 . A solution S is non-dominated iff
∀s ∈ S, ∃(o,o
0
) ∈ Ω
2
such that G(o) 6= G(o
0
) and
D
S\{s}
[o,.] = D
S\{s}
[o
0
,.] (i.e. S\{s} is not a solution).
The search for non-dominated solutions thus ma-
kes it possible to avoid searching for redundant infor-
mation while limiting the number of solutions.
Among these solutions, we are interested in com-
puting solutions of minimal size with regards to their
variables.
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
214

Definition 15 . A solution S is minimal iff @S
0
with
|S
0
| < |S| s.t. S
0
is a solution.
According to our notion of dominance between
solutions, a minimal solution in not dominated by
any other solutions. Intuitively, a minimal (non do-
minated) solution cannot be reduced unless two iden-
tical lines appear in two different groups (and conse-
quently the reduced set of variables is not a solution).
As already mentioned, a solution of the MCP is
a generalization of the notion of support set. A non-
dominated solution is thus a generalization of an ir-
redundant support set. Note that the union of irre-
dundant support sets for each couple of groups is not
necessary a non-dominated solution.
3.3 Converting Characterization
Requirements into Constraints
The minimum multiple characterization problem can
be formulated as a linear program (Chhel et al., 2013;
Boros et al., 2000). In fact, finding solutions corre-
spond here to a set covering problem. Given an in-
stance (Ω,A,D,G), let us consider the following 0/1
linear program.
min :
|A|
∑
i=1
y
i
s.t. :
C . Y
t
> 1
t
Y ∈ {0,1}
|A|
,Y = [y
1
,...,y
|A|
]
where Y is a Boolean vector that encodes the pre-
sence/absence of the set of variables in the solution.
C is a matrix that defines the constraints that must be
satisfied in order to insure that Y is a solution. Let
us denote Θ the set of all pairs (o, o
0
) ∈ Ω
2
such that
G(o) 6= G(o
0
). For each pair of observations (o, o
0
)
that do not belong to the same group, defined by an
element of Θ, one must insure that the value of at le-
ast one variable differ from o to o
0
. This will be insu-
red by the inequality constraint involving the 1 vector
(here a vector of dimension |Θ| that contains only 1
values).
More formally, C is a Boolean matrix of size |Θ|×
|A| define as:
• Each line is numbered by a couple of observations
(o,o
0
) ∈ Ω
2
such that G(o) 6= G(o
0
) ((o,o
0
) ∈ Θ).
• Each column represents indeed a variable.
• C[(o, o
0
),a] = 1 if D[o, c] 6= D[o
0
,c], C[(o,o
0
),a] =
0 otherwise.
• We denote C[(o,o
0
),.] the Boolean vector repre-
senting the differences between observations o
and o
0
on each variable. This Boolean vector
is called constraint since one variable a such
C[(o, o
0
),a] = 1 must be selected in order to in-
sure that no identical observations can be found
in different groups .
3.4 Computation of All Non-dominated
Solutions
In (Chambon et al., 2015), Algorithm 2 is presented
in order to compute the set of all non-dominated so-
lutions according to Definition 14.
The idea is thus to select variables a such that
there exists a couple of observations (o,o
0
) ∈ Θ
1
sa-
tisfying C[(o,o
0
),a] = 1 in the constraint matrix C and
C[(o, o
0
),a
0
] = 0 for any variable a
0
6= a.
Algorithm 2: Non-Dominated Solutions: NDS.
Data: C: Constraints matrix of size |Θ|×|A|.
Result: S ol: Non-dominated solutions set.
Sol = {
/
0}
for i = 1 to |Θ| do
//Build a subset of solutions ND
i
ND
i
=
/
0
forall j ∈ A s.t. C[θ
i
, j] = 1 do
forall S ∈ S ol\ND
i
do
if j ∈ S then
ND
i
= ND
i
∪ {S}
end
end
end
//Build a subset of solutions ES
i
ES
i
=
/
0
forall j ∈ A s.t. C[θ
i
, j] = 1 do
forall S ∈ S ol\ND
i
do
if @S
0
∈ ND
i
s.t. S
0
⊆ S ∪ { j} then
ES
i
= ES
i
∪ {S ∪ { j}}
end
end
end
Sol = ND
i
∪ ES
i
end
return Sol;
Algorithm 2 builds incrementally the set S ol of
non-dominated solutions. Each element of Θ =
{θ
1
,θ
2
,...,θ
|Θ|
} is a constraint that must be satisfied.
At each iteration, the solutions are updated in order
to satisfy the constraint θ
i
. The main idea consists in
distinguishing solutions that already satisfy this con-
straint (they are put in a set of non dominated solu-
1
Remind that Θ is a set of couples of observations defi-
ned in 3.3 for indexing lines of the constraint matrix C.
Accelerated Algorithm for Computation of All Prime Patterns in Logical Analysis of Data
215

tions ND
i
) and solutions that need to be modified in
order to satisfy the constraint, (they belong to the set
ES
i
). Note that the modification of these latest soluti-
ons is performed by adding one variable while main-
taining the non-domination property.
This algorithm is related to the Berge’s algorithm
(Berge, 1984) that may be used for computing hitting
sets. Berge’s algorithm consists in incrementing the
solution at each iteration with an element of the con-
straint, and to compare pairs of solutions to remove
dominated solution. Algorithm 2 avoid constructing
dominated solutions.
Note that, as the algorithm builds incrementally
the set of non-dominated solutions. We can use a
maximal bound B for computing only non-dominated
solutions with a number of variables smaller than this
bound. Given a bound B, Algorithm 2 can be modi-
fied when updating ES
i
as ES
i
= ES
i
∪{S ∪{ j}}. This
update can be performed only if ∀s ∈ ES
i
, |s| ≤ B in
order to improve the performance of the algorithm.
4 COMPUTATION OF PRIME
PATTERNS AND GROUP
COVERS
We propose now a new algorithm that uses the com-
putation of all non dominated solutions of the MCP
problem in order to compute prime patterns.
In LAD, the aim is to find a pattern that covers a
maximum number of observations of P, such as no
observation of N contains this pattern. From MCP
point of view, the notion of solution is rather different.
Given a solution S of a MCP instance (Ω,A, D, G) de-
fined as above, the variables of S do not generally cor-
respond to a pattern for the observations in P, unless
all observations are identical on S. In this case a so-
lution of the MCP obviously coincides with a prime
pattern in terms of variables.
In particular, if |P| = 1, the set of all solutions of
the MCP coincides in terms of variables with the set
of all prime patterns that cover the only observation in
P, because in both cases no variable can be removed.
Given a non-dominated solution S of the MCP
(computed by previously mentioned algorithms for
instance), it is easy to transform an observation o of
the group P into prime pattern p. Each variable a of S
appears positively (resp. negatively) in p if D[o,a] = 1
(resp. D[o, a] = 0).
The following simple procedure (algorithm 3)
transforms a non-dominated solution of MCP, consid-
ering only one observation x ∈ P into a prime pattern.
For each observation, we can generate all prime
Algorithm 3: Pattern Transformation.
Data: s: non-dominated solution of the MCP;
x: the only observation in the group P.
Result: p: prime pattern.
pos =
/
0
neg =
/
0
forall a ∈ s do
if x
a
= 1 then
pos = pos ∪ {a}
end
else
neg = neg ∪ {a}
end
end
p = (
V
i∈pos
x
i
) ∧ (
V
j∈neg
¬x
j
)
return p;
patterns that cover this observation. If we generate all
prime patterns for all observations, we generate prime
patterns p, and determine Cov(p) for each one.
Algorithm 4 returns the set Pat of all prime pat-
terns, and the set Cov of coverage of all patterns
p ∈ Pat. Cov is a set of elements V
p
, ∀p ∈ Pat. Each
element V
p
is a set of all observations covered by p.
Note that it is not necessary to compute the set Cov to
generate the set Pat. Hence, each step that involves
the set Cov can be removed.
All algorithms that can compute the set of all
non-dominated solutions (like algorithm NDS) can be
used to determine the set Sol. Note that since we are
working on one group against all the others, we can
also use an algorithm that computes all the irredun-
dant support sets. However, the algorithms presented
in (Boros et al., 2000) only allow to compute a subset
of these irredundant support sets.
Note that Algorithm NDS (Chambon et al., 2015)
can also generate solutions of smaller size than a gi-
ven bound B. Given a bound B, we can only generate
prime patterns with a size inferior to B.
Also note, if we use the algorithm NDS for com-
puting the set Sol, for an instance with x observations
and n variables the complexity is :
O(x
2
× 2
n−b
n
3
c
×
n!
b
n
3
c!(n − b
n
3
c)!
×
n!
b
n
2
c!(n − b
n
2
c)!
)
(
n!
b
n
2
c!(n−b
n
2
c)!
is the maximal number of non-
dominated solutions)
Now, using the set Cov we can run Algorithm 5
to compute only strong prime patterns. From the set
of all covers, we can compute the subset of strong
patterns among prime patterns.
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
216

Algorithm 4: Prime Patterns Computation (PPC 2).
Data: D: matrix of data, with two groups
{P,N}.
Result: Pat: set of all prime patterns
Result: Cov: set of covers of each prime
pattern.
Pat =
/
0
Cov =
/
0
forall o ∈ P do
Generate the constraint matrix C
o
as if o
was the only one observation in P
Sol={set of all non dominated solutions
for C
o
}
forall s ∈ Sol do
p=Transformation Pattern(s,o)
if p /∈ Pat then
Pat = Pat ∪ {p}
//Create a new element V
p
of Cov
which will be a set of
observations covered by p.
V
p
= {o}
Cov = Cov ∪ {V
p
}
end
else
//V
p
is already in Cov; update
V
p
= V
p
∪ {o}
end
end
end
return Pat and Cov;
Algorithm 5: Strong Prime Patterns Computation.
Data: Cov: set of coverage of each prime
pattern.
Pat: set of all prime patterns
Result: SPP: set of all strong prime patterns.
SPP =
/
0
forall p ∈ Pat do
if @p
0
∈ Pat s.t. Cov(p) ⊂ Cov(p
0
) then
SPP = SPP ∪ {p}
end
end
return SPP;
5 EXPERIMENTS
The main purpose of our experiments is to compare
the performance of our new algorithm PPC 2 to the
algorithm PPC 1 for computing sets of prime patterns.
PPC 2 (Algorithm 4) uses the principles presented
in Section 4, encoded in C++ with data structures and
operators from the library boost
2
.
PPC 1 has been recalled in Section 2.2.1. Note that
the source code of this algorithm presented in (Chika-
lov et al., 2013) was not available. It has been im-
plemented in C++ using the same data structures and
operators from the library boost
Experiments have been run on a computer with
Intel Core i7-4910MQ CPU (8×2.90 GHz), 31.3 GB
RAM.
5.1 Data Instances
We consider several sets of observations issued from
different case studies.
• Random is a random instance built with only one
observation in the positive group, and random va-
lue in {0,1}. This instance is used as a basic
test case. The group P is restricted to only 1 va-
lue since otherwise it would have been difficult
to identify common patterns for several randomly
generated observations.
• Instances ra100 phv, ra100 phy, ralsto, ra phv,
ra phy, ra rep1, ra rep2 and rch8 are matrices
built from biological data that correspond to
bacterial strains. Each observation is a bacte-
rial strain and variables are genes (housekeeping
gene, resistance gene or specific effectors). These
bacteria are responsible of serious plant diseases.
Therefore it is important to be able to identify
precisely different groups of bacteria using a re-
stricted set of variables and to identify common
gene profiles. Such identification are very help-
ful for building simple and cheap diagnosis routi-
nes (Boureau et al., 2013). The original files are
available
3
. Initially, several groups are conside-
red in these instances. Therefore, we have con-
sidered the first group of bacteria as the positive
group and the union of the other groups as the ne-
gative group. Note that similar results have been
obtained when considering others groups as posi-
tive group.
• Instances vote r
4
are also binary data used as
benchmarks for classification purpose. Note that
these instances have missing data and have been
completed randomly.
• Instances cr60, os1 and rel1 are datasets corre-
sponding to patients suffering from leukemia. Ob-
servations correspond to specific mutated variants
2
http://www.boost.org/doc/libs/1 36 0/libs/
dynamic bitset/dynamic bitset.html
3
http://www.info.univ-angers.fr/˜gh/Idas/Ccd/ce f.php
4
http://tunedit.org/repo/UCI/vote.arff
Accelerated Algorithm for Computation of All Prime Patterns in Logical Analysis of Data
217
of genes that are suspected to play a role in the
disease. Here the goal is to find genes that could
help to improve prognosis and to select the most
suitable treatments according to the patients pro-
files.
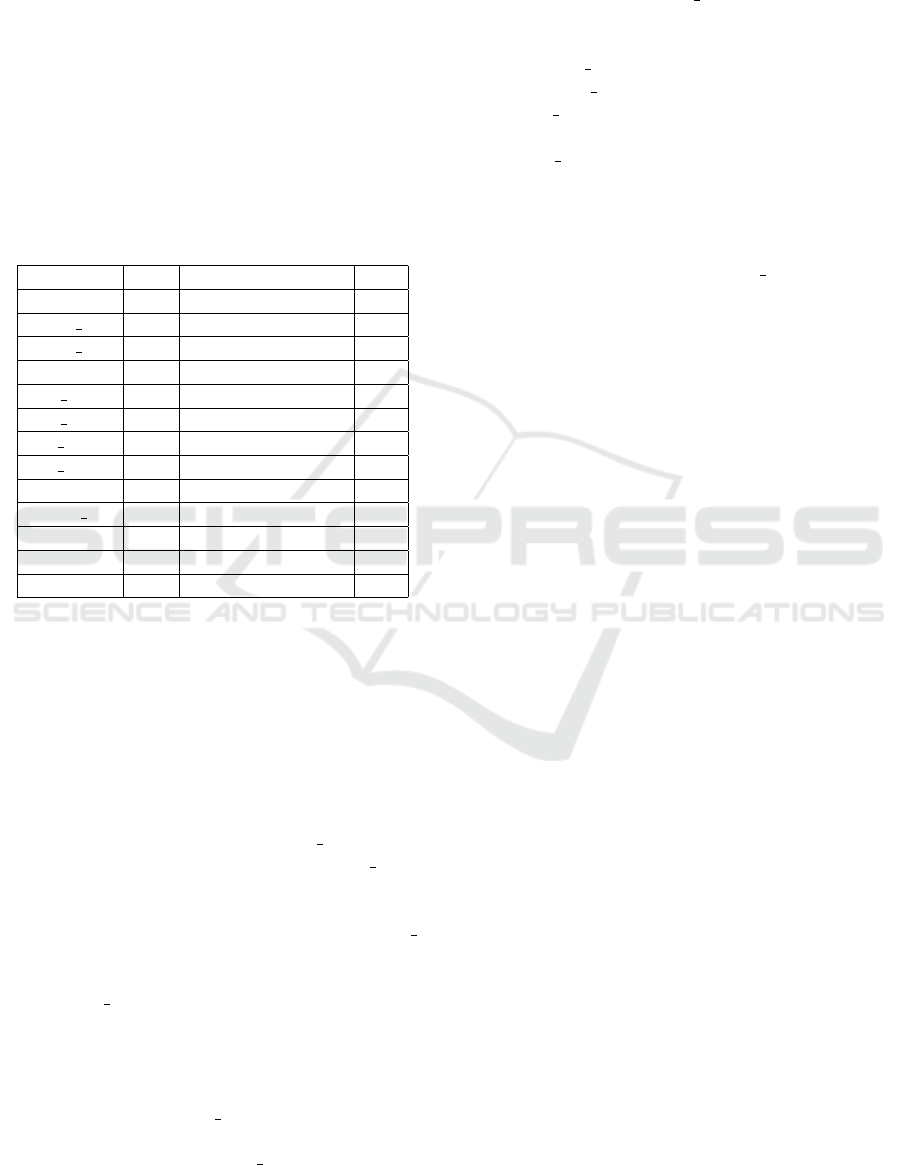
The instances are described in Table 1 with their
number of observations, number of observations in
the positive group (the negative group is of course
the complement) and the maximal number of varia-
bles. For each instance, we consider consider diffe-
rent values x of variables in order to evaluate the per-
formance of the algorithms with regards to this num-
ber of variables.
Table 1: Characteristics of the instances.
Instances Obs Positive group size Var
Random 20 1 35
ra100 phv 100 21 50
ra100 phy 105 31 51
ralsto 73 27 23
ra phv 108 22 70
ra phy 112 31 73
ra rep1 112 38 155
ra rep2 112 37 73
rch8 132 5 37
vote r 435 168 16
cr60 289 58 14
os1 289 224 14
rel1 259 200 14
5.2 Results
Table 2 provides the results obtained on the instances
for computing the set of prime patterns. The first co-
lumn corresponds to the name of the instance, with
the number x of used variables (remind that we consi-
der different sizes for each the instances). The second
column corresponds to the number of prime patterns
for each instance. Next two columns are execution
time (in seconds) for our algorithm PPC 2 (Algorithm
4), and execution time (in seconds) for PPC 1 (Al-
gorithm 1). The last two columns correspond to the
maximal size of the computed patterns (in terms of
number of variables) and the execution time of PPC 1
used with an initial bound equal to this maximal size.
Of course, when using this bound we get the same re-
sults but PPC 1 is faster since it may stop earlier. Note
that in practice the value of the bound is not known
until the set of prime patterns has been computed.
Running time is limited to 24 hours. “-” corresponds
to execution times greater than this limit.
The execution time of PPC 1 increases as the number
of observations increases and especially as the num-
ber of variables increases. PPC 2 is less sensitive to
the number of observations. Nevertheless its compu-
tation time also increases according to the number of
variables.
Let us remark that PPC 1 is able to compute all
prime patterns for instance Random(35) in three days.
Nevertheless, one week is not enough for the instance
rch8(37). PPC 2 is able to compute all prime patterns
for instance ra rep2(65) in a bit more than one hour
while PPC 1 is not able to solve the same instance with
only 20 variables.
In PPC 1, the number of iterations is related to the
number of variables. We observe in the last column
that if a good bound is available (equal to the size
of the largest prime pattern), prime patterns can be
computed more efficiently. Nevertheless, execution
time is still high compared to PPC 2. Moreover, it
requires to know the size of the largest pattern.
6 CONCLUSION
In this paper we have defined a new algorithm to gene-
rate complete sets of prime patterns and strong prime
patterns in LAD context. Compared to the state of
the art algorithms for these problems, our algorithm
is now able to handle larger data sets. The main idea
of its resolution process is to use an extension of the
LAD (multiple characterization of data) in order to
first compute the non dominated solutions and finally
obtain all the prime and strong prime patterns. Expe-
riments show the efficiency of our algorithm in term
of running times and instance sizes.
REFERENCES
Berge, C. (1984). Hypergraphs: combinatorics of finite
sets, volume 45. Elsevier.
Boros, E., Crama, Y., Hammer, P. L., Ibaraki, T., Kogan, A.,
and Makino, K. (2011). Logical analysis of data: clas-
sification with justification. Annals OR, 188(1):33–61.
Boros, E., Hammer, P. L., Ibaraki, T., and Kogan, A. (1997).
Logical analysis of numerical data. Mathematical
Programming, 79(1-3):163–190.
Boros, E., Hammer, P. L., Ibaraki, T., Kogan, A., Mayoraz,
E., and Muchnik, I. (2000). An implementation of
logical analysis of data. IEEE Transactions on Know-
ledge and Data Engineering, 12(2):292–306.
Boureau, T., Kerkoud, M., Chhel, F., Hunault, G., Darrasse,
A., Brin, C., Durand, K., Hajri, A., Poussier, S., Man-
ceau, C., et al. (2013). A multiplex-pcr assay for iden-
tification of the quarantine plant pathogen xanthomo-
nas axonopodis pv. phaseoli. Journal of microbiologi-
cal methods, 92(1):42–50.
Chambon, A., Boureau, T., Lardeux, F., Saubion, F., and
Le Saux, M. (2015). Characterization of multiple
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
218
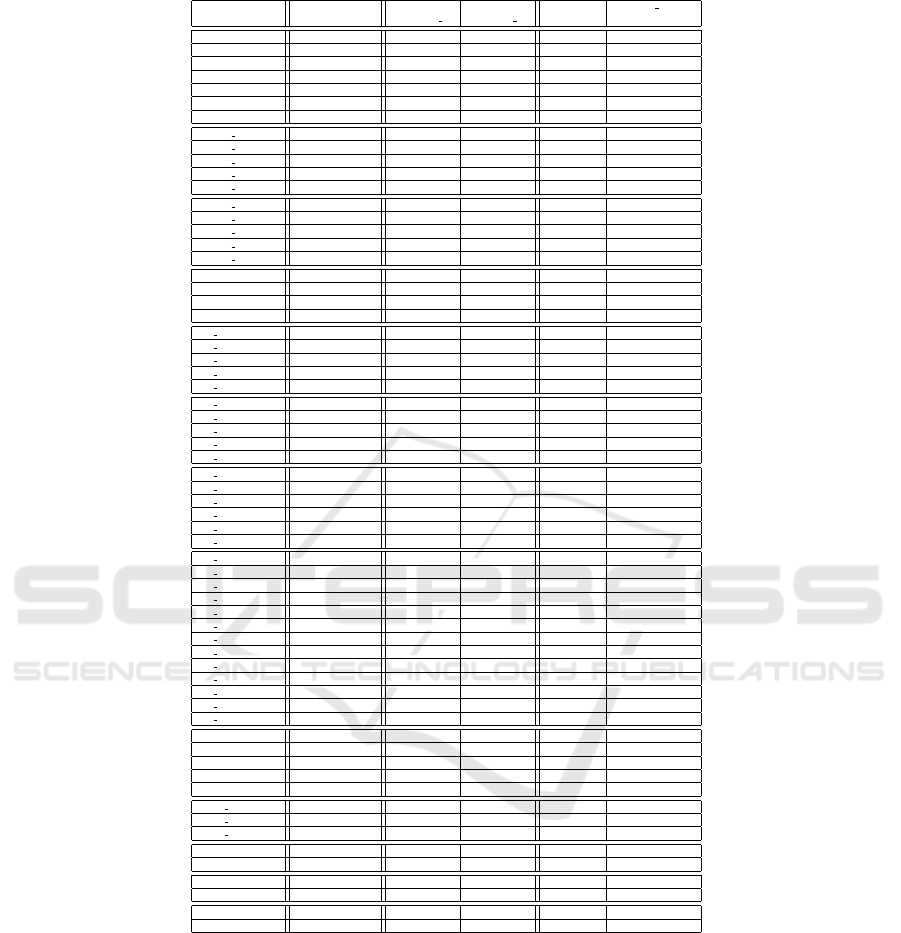
Table 2: Results for prime patterns computation.
Instance Number of Time Time Maximal PPC 1 time
prime patterns PPC 2 PPC 1 size using max size
Random(11) 61 0.003 0.006 4 0.003
Random(15) 170 0.011 0.029 5 0.021
Random(20) 476 0.020 2.306 6 1.185
Random(25) 1143 0.028 113.469 6 16.343
Random(27) 1529 0.052 338.708 6 38.045
Random(30) 2382 0.110 4154.002 6 170.906
Random(35) 4505 0.283 - 6 1668.373
ra100 phv(10) 6 0.001 0.053 3 0.001
ra100 phv(20) 105 0.013 16478.822 5 11.269
ra100 phv(30) 304 0.025 - 6 40646.487
ra100 phv(40) 1227 0.089 - 8 -
ra100 phv(50) 12162 1.381 - 10 -
ra100 phy(10) 5 0.001 0.039 4 0.014
ra100 phy(20) 131 0.015 - 6 555.829
ra100 phy(30) 583 0.037 - 6 85608.555
ra100 phy(40) 1982 0.123 - 8 -
ra100 phy(51) 13112 2.119 - 10 -
ralsto(10) 22 0.003 0.047 5 0.029
ralsto(15) 132 0.008 35.286 6 6.839
ralsto(20) 361 0.013 39725.665 6 436.524
ralsto(23) 1073 0.040 - 8 -
ra phv(10) 6 0.004 0.042 3 0.004
ra phv(25) 194 0.024 - 5 94.069
ra phv(40) 1227 0.101 - 8 -
ra phv(55) 28163 6.061 - 10 -
ra phv(70) 384629 1777.386 - 14 -
ra phy(10) 5 0.002 0.085 4 0.016
ra phy(25) 252 0.016 - 6 8253.219
ra phy(40) 1982 0.121 - 8 -
ra phy(55) 23504 4.914 - 10 -
ra phy(73) 449220 2328.949 - 14 -
ra rep1(10) 4 0.000 0.107 4 0.013
ra rep1(30) 11 0.000 - 4 6.385
ra rep1(60) 415 0.021 - 7 -
ra rep1(90) 9993 1.420 - 10 -
ra rep1(120) 243156 1081.313 - 12 -
ra rep1(155) 2762593 - - 15 -
ra rep2(10) 11 0.002 0.096 4 0.008
ra rep2(15) 46 0.005 133.096 4 0.158
ra rep2(20) 126 0.007 - 6 413.723
ra rep2(25) 303 0.009 - 7 -
ra rep2(30) 745 0.017 - 7 -
ra rep2(35) 2309 0.060 - 9 -
ra rep2(40) 6461 0.403 - 10 -
ra rep2(45) 17048 2.315 - 10 -
ra rep2(50) 43762 14.596 - 10 -
ra rep2(55) 101026 68.378 - 11 -
ra rep2(60) 254042 840.734 - 12 -
ra rep2(65) 720753 4617.556 - 14 -
ra rep2(73) 2474630 60740.333 - 15 -
rch8(15) 1 0.004 5.988 2 0.004
rch8(20) 7 0.009 4261.190 4 0.163
rch8(25) 26 0.012 - 4 0.676
rch8(30) 43 0.016 - 4 2.384
rch8(37) 131 0.021 - 6 77836.071
vote r(10) 169 0.060 0.876 6 0.669
vote r(13) 1047 0.250 52.711 8 48.427
vote r(16) 4454 0.842 4138.240 9 3466.591
cr60(10) 55 0.018 0.923 6 0.585
cr60(14) 196 0.026 350.849 8 219.009
os1(10) 107 0.018 0.958 6 0.571
os1(14) 286 0.035 379.423 7 102.318
rel1(10) 135 0.024 0.976 6 0.604
rel1(14) 388 0.033 376.540 7 107.410
groups of data. In Tools with Artificial Intelligence
(ICTAI), 2015 IEEE 27th International Conference
on, pages 1021–1028. IEEE.
Chhel, F., Lardeux, F., Go
¨
effon, A., and Saubion, F. (2012).
Minimum multiple characterization of biological data
using partially defined boolean formulas. In Procee-
dings of the 27th Annual ACM Symposium on Applied
Computing, pages 1399–1405. ACM.
Chhel, F., Lardeux, F., Saubion, F., and Zanuttini, B. (2013).
Application du probl
`
eme de caract
´
erisation multiple
`
a la conception de tests de diagnostic pour la biolo-
gie v
´
eg
´
etale. Revue des Sciences et Technologies de
l’Information-S
´
erie RIA: Revue d’Intelligence Artifi-
cielle, pages 649–668.
Chikalov, I., Lozin, V., Lozina, I., Moshkov, M., Nguyen,
H. S., Skowron, A., and Zielosko, B. (2013). Logi-
cal analysis of data: theory, methodology and appli-
cations. In Three Approaches to Data Analysis, pages
147–192. Springer.
Crama, Y., Hammer, P. L., and Ibaraki, T. (1988). Cause-
effect relationships and partially defined boolean
functions. Annals of Operations Research, 16(1):299–
325.
Dupuis, C., Gamache, M., and Pag
´
e, J.-F. (2012). Logical
Accelerated Algorithm for Computation of All Prime Patterns in Logical Analysis of Data
219

analysis of data for estimating passenger show rates
at air canada. Journal of Air Transport Management,
18(1):78–81.
Hammer, P. L. (1986). Partially defined boolean functions
and cause-effect relationships. In International confe-
rence on multi-attribute decision making via or-based
expert systems.
Hammer, P. L. and Bonates, T. O. (2006). Logical analy-
sis of data—an overview: from combinatorial optimi-
zation to medical applications. Annals of Operations
Research, 148(1):203–225.
Hammer, P. L., Kogan, A., and Lejeune, M. A. (2012). A lo-
gical analysis of banks’ financial strength ratings. Ex-
pert Systems with Applications, 39(9):7808–7821.
Hammer, P. L., Kogan, A., Simeone, B., and Szedm
´
ak, S.
(2004). Pareto-optimal patterns in logical analysis of
data. Discrete Applied Mathematics, 144(1):79–102.
Mortada, M.-A., Carroll, T., Yacout, S., and Lakis, A.
(2012). Rogue components: their effect and control
using logical analysis of data. Journal of Intelligent
Manufacturing, 23(2):289–302.
Reddy, A., Wang, H., Yu, H., Bonates, T. O., Gulabani, V.,
Azok, J., Hoehn, G., Hammer, P. L., Baird, A. E., and
Li, K. C. (2008). Logical analysis of data (lad) model
for the early diagnosis of acute ischemic stroke. BMC
medical informatics and decision making, 8(1):30.
Ryoo, H. S. and Jang, I.-Y. (2009). Milp approach to pat-
tern generation in logical analysis of data. Discrete
Applied Mathematics, 157(4):749–761.
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
220
