
Predicting the Early Stages of the Alzheimer’s Disease via Combined
Brain Multi-projections and Small Datasets
Kau
ˆ
e T. N. Duarte, Pedro V. V. de Paiva, Paulo S. Martins and Marco A. G. Carvalho
School of Technology, University of Campinas (UNICAMP), R. Paschoal Marmo, Limeira, Brazil
Keywords:
Classification, Transfer Learning, Mild Cognitive Impairment, Clinical Dementia Rating, Support Vector
Machines.
Abstract:
Alzheimer is a neurodegenerative disease that usually affects the elderly. It compromises a patient’s memory,
his/her cognition, and perception of the environment. Alzheimer’s Disease detection in its initial stage, known
as Mild Cognitive Impairment, attracts special efforts from experts due to the possibility of using drugs to
delay the progression of the disease. This paper aims to provide a method for the detection of this impairment
condition via the classification of brain images using Transfer Learning - Deep Features and Support Vector
Machine. The small number of images used in this work justifies the application of Transfer Learning, which
employs weights from VGG19 initial layers used for ImageNet classification as deep features extractor, and
then applies Support Vector Machines. Majority Voting, False-Positive Priori, and Super Learner were applied
to combine previous classifiers predictions. The final step was a detection to assign a label to the previous
voting outcomes, determining the presence or absence of an Alzheimers pre-condition. The OASIS-1 database
was used with a total of 196 images (axial, coronal, and sagittal). Our method showed a promising performance
in terms of accuracy, recall and specificity.
1 INTRODUCTION
The Alzheimer’s Disease (AD) is a neurodegenera-
tive dementia that affects the human abilities related
to memory, language, perception of the environment
and cognitive skills(Ferreira and Busatto, 2011). Mild
Cognitive Impairment (MCI) is known as a prodromal
stage of AD and corresponds to the range between a
normal aging and dementia. MCI is gaining atten-
tion because by predicting the disease in this stage,
patients are able to find out ways to slow down the di-
sease. Besides, this stage is relevant due to its strong
relationship with the AD progression. As a matter of
fact, 10-12% of the MCI cases convert to AD each
year (Petersen et al., 1999).
The most popular term used to classify specific
stages in the MCI is the Clinical Dementia Rating
(CDR), defined by the following levels: CDR-0 repre-
sents Normal Control (NC) or non-dementia people;
CDR-0.5 corresponds to a very mild dementia; CDR-
1 represents a mild impairment dementia; CDR-2 is
a moderate dementia, whereas CDR-3 indicates se-
vere dementia. These levels allow the medical team to
identify a better prognostic for the AD patient. They
also facilitate the process of classifying images.
Different analysis are currently used to diagnose
or predict AD, such as: (1) Family History; (2) Ima-
ging; (3) Cognitive Tests (Mini-Mental State Exami-
nation); and (4) Neurological Exams.
This work uses imaging to support the prediction
of the Alzheimers disease through the detection of
MCI, which is one indicator of early signs of AD. It is
very difficult to predict AD using only one projection
of the brain (e.g. sagittal). Thus, the combination of
the different planes of the brain, also known as multi-
projection, is used and has reached higher accuracy
than single-projection frameworks (Aderghal et al.,
2017)(Zhou et al., 2017). The results in the litera-
ture achieve higher accuracy also when comparing
some stages of the disease, for instance, when com-
paring CDR-0 and CDR-3 patients (Khedher et al.,
2015)(Suk and Shen, 2016). However, our work fo-
cus on the two initial stages (i.e. CDR-0.5 and CDR-
1) due to our emphasis on predicting the AD dementia
as soon as possible. An example of brain atrophy is
shown in Figure 1.
In this work, we aim at classifying MCI images
using visual information and a small number of sam-
ples. Image data was analyzed using Transfer Lear-
ning with Convolutional Neural Networks (CNN) and
Duarte, K., V. de Paiva, P., Martins, P. and Carvalho, M.
Predicting the Early Stages of the Alzheimer’s Disease via Combined Brain Multi-projections and Small Datasets.
DOI: 10.5220/0007404705530560
In Proceedings of the 14th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2019), pages 553-560
ISBN: 978-989-758-354-4
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
553

(a) (b)
Figure 1: Brain Status (on Aging, 2016): (a) healthy subject
(CDR-0), (b) subject with severe Alzheimer (CDR-3).
Support Vector Machine (SVM).
The main features and contributions of this work
are: (1) The combination of Transfer Learning, SVM,
and Voting for small datasets; (2) The Grouping of
MCI classes to improve the number of images per
class (i.e. thus facilitating the identification of pat-
terns); (3) Application of our approach to the first sta-
ges of MCI, which turns the discrimination of AD a
more challenging task; (4) The availability of the code
and the implementation used to generate this paper.
The remainder of this paper is organized as fol-
lows: In Section 2 we address related work. The pro-
posed method, the fundamentals and details of each
step in the method are addressed in Section 3. The ex-
periments and the evaluation of the method are shown
in Section 4. Finally, in Section 5 we present our con-
clusions.
2 RELATED WORK
Different computational methods using textual data
or visual information have been applied in the me-
dical field. For example, Lebedev (Lebedev et al.,
2014) address the problem of diagnosing AD using
Random Forest (RF) classifiers and different measu-
res obtained from clinical Magnetic Resonance Ima-
ging (MRI) data. The best performance was achieved
with RF and it reached an accuracy over 90% by com-
paring AD with NC images.
The use of a CNN is presented by Wang (Wang
et al., 2017) to automatically recognize MCI in MRI.
The authors addressed the problem of limited training
data using data augmentation and transfer learning to
pre-train the proposed CNN model. Three different
datasets were used for the training and classification
stages: OASIS, LIDC, and ADNI, The authors repor-
ted an accuracy of 90,6% and a F-score of 89,4%.
There is a body of work that combines information
from different modalities or projections. When dea-
ling with the prediction of the AD (Normal, MCI and
AD levels), Fiot (Fiot et al., 2012) employs the use
of Laplacian EigenMaps in order to reduce the data
dimensionality and the K-Nearest Neighbor (KNN)
classifier. They used the MRI, one protein, and six ge-
notype data from the ADNI dataset and reached accu-
racies from 62% up to 83% when comparing Normal
and MCI classes.
Khedher (Khedher et al., 2015) used the data pro-
vided by ADNI dataset to compose a multivariate
method with Partial Least Squares, Principal Compo-
nent Analysis and SVM. Aderghal (Aderghal et al.,
2017) proposed a multi-projection fusion of Axial,
Coronal and Sagittal CNNs using Regions of Interest.
The accuracy reached was 85,94% when comparing
AD and NC. In Suk and Shen (Suk and Shen, 2016),
the authors proposed a framework based on sparse re-
gression as learner representation. Thus, they built a
CNN for clinical decisions. An accuracy of 90,28%
was reached by their method. Using Positron Emis-
sion Tomography (PET) images instead of MRI scans,
Gray (Gray et al., 2011) proposed a region-based ana-
lysis of AD patients, using segmentation of 83 ana-
tomical regions obtained by MRI. The authors used
SVM to classify samples and reached an accuracy of
82% when comparing AD and NC. Cheng and Liu
(Cheng and Liu, 2017) proposed a method based on
2D-CNN to learn features from 3D PET images, and
they achieved an accuracy of 91,40 % .
Some of the literature reviewed in this section rea-
ched a relative high accuracy since they compare NC
and AD images. Clearly, AD images possess more
discriminative features of the disease than MCI ima-
ges. For example, in Khedher (Khedher et al., 2015),
the results obtained by the comparison of NC and
MCI reached an accuracy of 80,27%. On the other
hand, when comparing only the NC and AD classes,
the accuracy was increased to 88,49%. Our work defi-
nes a scenario with a focus on MCI, which represents
the initial levels of disease.
In essence, our work differs from previous works
by the following features: (1) the use of Transfer Le-
arning (TL) instead of fully-CNNs. The TL-CNNs
have not been previously applied within the same con-
text of this work; (2) the use of the first two classes
of MCI (i.e. CDR-0.5 and CDR-1). The most chal-
lenging prediction of the early stages of the Alzhei-
mer disease via classification lies within the first clas-
ses because these images are quite similar to those
from healthy individuals; (3) the proposed method re-
ached satisfactory results even when using a small set
of images.
VISAPP 2019 - 14th International Conference on Computer Vision Theory and Applications
554

3 PROPOSED METHOD
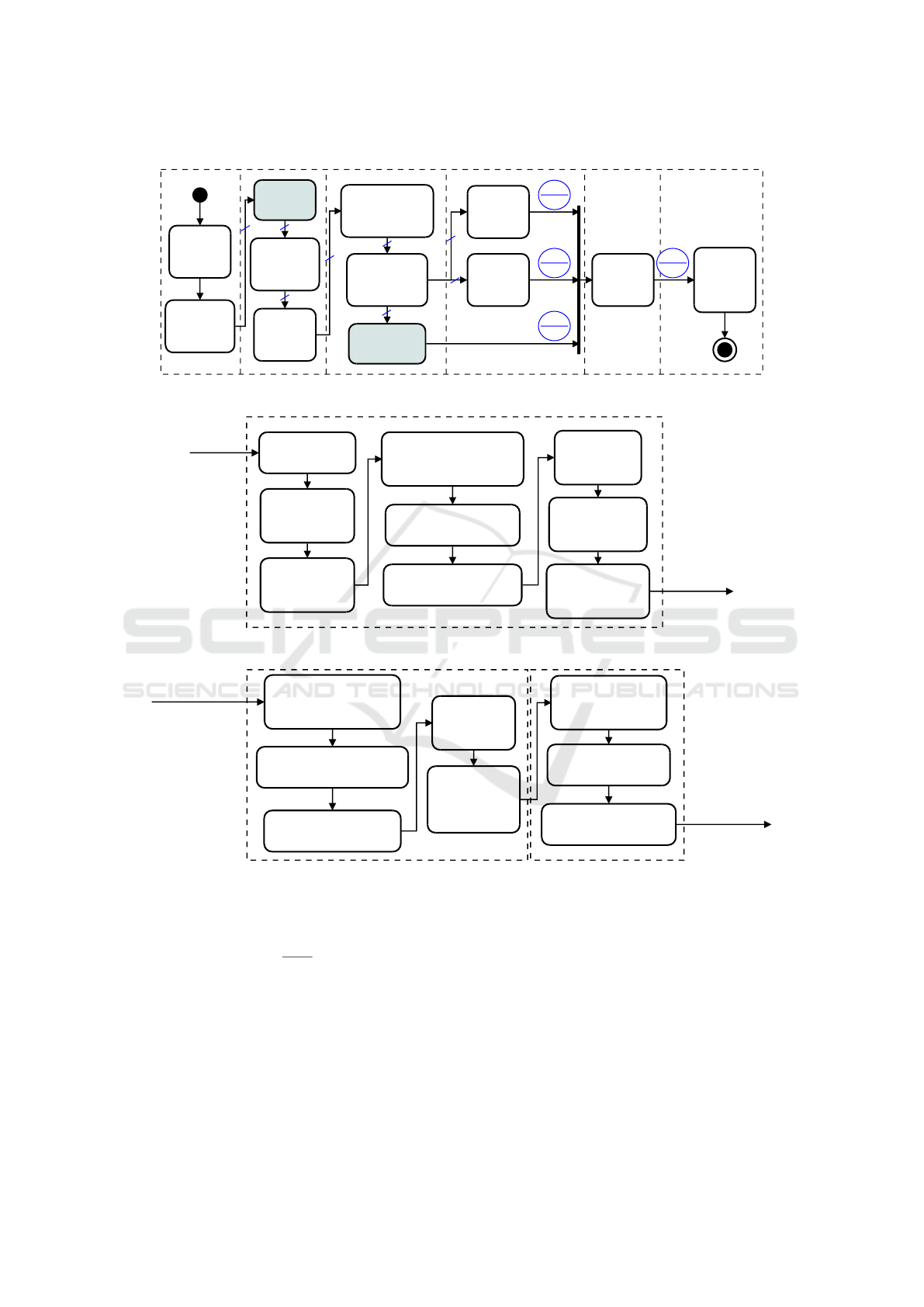
We arranged the method in six major steps (Figure
2): (1) Input data: including acquisition and cate-
gorization; (2) Preprocessing: it involves skull strip-
ping, image enhancement, and data normalization;
(3) Classification: it combines Transfer Learning and
SVM. Super Learner was also applied to the SVM
outputs; (4) Voting: the method applied Majority Vo-
ting and False-Positive Priori to the outputs of SVM;
(5) Detection: it also applies Majority Voting to the
inputs and it labels the samples combining the previ-
ous procedures; (6) Output: generate a label defining
either the presence or the absence of MCI.
Our method (Figure 2(a)) starts out by extracting
each slice of a 3D-brain projection. The MCI classes
are grouped using Spectral Clustering (Figure 2(b)).
Thus, the skull is removed and the intensity adjus-
ted. For the sake of classification, Transfer Learning
is applied in each representation to generate deep fea-
tures, which are further used in SVMs to classify ima-
ges. Methods such as Majority Voting, FP Priori, and
Super Learner are used to combine the results of the
projections. Finally, Majority Voting is carried out to
group the results of each method. The final response
is the detection whether a subject has MCI or not.
In Figure 2(a), each crossed blue line with identi-
fier three (3) informs the number of projections in the
flow. Also, in each blue circle, the upper part means
an example of response for each projection (i.e. axial,
coronal, and sagittal), whereas the lower part shows
the result after voting or classification. We now dis-
cuss each step of the method in more detail.
3.1 Input Data
In this work, the Open Access Series of Imaging Stu-
dies (OASIS)-1: Cross-sectional MRI Data in Young,
Middle Aged, Nondemented and Demented Older
Adults was used. The CSV file dataset is formed by
436 subjects data (135 NC, 70 CDR-0.5, 28 CDR-
1, 2 CDR-2, and 201 unlabeled) with age between
18 and 96 years. For each patient, the following in-
formation is given: (1) Demographic: Age, gender,
education, Socioeconomic Status (SES); (2) Clini-
cal: Mini-Mental State Examination (MMSE), Cli-
nical Dementia Rating; (3) Derivated anatomic vo-
lumes: estimated Total Intracranical Volume (eTIV),
Atlas Scaling Factor (ASF), normalized Whole Brain
Volume (nWBV); and (4) Imaging: Magnetic Reso-
nance Imaging. The above related text data were or-
ganized in a table in order to facilitate the data mining
interpretation.
Each subject is associated with a 208x176x176
pixel image. Three slices from different views of the
brain were extracted: (1) The Axial MRI, which is
composed of 208 x 176 pixels (slice #90); (2) Coro-
nal MRI, consisting of 176x176 pixels (slice #110),
and (3) Sagittal MRI with 208x176 pixels (slice #95).
The extraction of each brain projection is showed
in the Figure 3.
3.2 Preprocessing
Preprocessing plays a key role in every classifier’s
performance. In this work, the same preprocessing
flow was applied to each individual brain projection,
which were programmatically extracted from the ori-
ginal 3D brain representation.
3.2.1 Grouping of the MCI Classes
This step employed information provided by the OA-
SIS dataset, which consists of clinical, demographic,
and functional information of subjects. The algorithm
used for grouping the MCI-classes (in step 5) was
Spectral clustering. It required four preparation steps,
i.e. steps 1-4 for data cleaning and transformation:
1. Removal by age: Guerreiro (Guerreiro and Bras,
2015) indicated that the brain has a normal
atrophy due to aging. Thus, a healthy 96 year-old
brain shows discriminative values in relation to an
18-year-old brain. The authors also point out that
AD commonly occurs at age 65 or older. In order
to avoid misclassification, 238 subjects under age
60 were removed from the original dataset with
436, thus remaining 198 subjects.
2. Removal by classes: Two subjects classified as
CDR-2 were also removed, thus remaining 196
subjects. The goal was to avoid mislabeling due
to the small number of samples;
3. Data filling: 19 subjects had no SES informa-
tion. Instead of removing these data, this condi-
tion was mitigated by adding values using Pro-
gressive Sampling and Linear Regression;
4. Data transformation: The goal is to fit the data
for the Spectral Clustering inputs using the follo-
wing steps: (a) Scale adjustment: The eTIV and
nWBV attributes were originally specified in dif-
ferent scales. Thus, if the data were below a cer-
tain threshold (10 in our case), they were multip-
lied by 10
3
; (b) Attribute binarization: The Gen-
der attribute originally had (M/F) categorical va-
lues. They were binarized by setting one (1) for
M and zero (0) for F; (c) Normalization: In order
to assign the same weight to each type of attri-
bute, the data were normalized using the Standard
Score (z-score) (Equation (1)):
Predicting the Early Stages of the Alzheimer’s Disease via Combined Brain Multi-projections and Small Datasets
555
Acquire
3D Brain
Image
Extract 2D
slice of each
projection
Group AD
classes
Extract non-
cerebral
tissue
Enhance
the image
intensity
Generate deep
features using
Transfer Learning
Apply
Support Vector
Machine
Apply
the
FP Priori
Apply the
Majority
Voting
Apply the
Super Learner
1. Input 2. Preprocessing 3. Classification 4. Voting 6. Output
Generate
a Label
(has or
not MCI)
5. Detection
Arbitration
(Decision
Making)
3
3
3
3
3
3
1,0,0
0
1,0,0
1
1,0,0
1
1,1,0
1
3
3
(a)
Group AD classes
Acquire datasheet
in OASIS-1
Remove patients
in 18-59 years
old range
Remove patients
classified as
CDR-2
Update SES information
using Progressive Sampling-
Linear Regression
Normalize eTIV and nWBV
to a same scale
Convert gender from
categorical to binary values
Normalize the
datasheet using
Standard Score
Apply Spectral
Clustering to
find binary classes
Update the classes
according to
above results
from input
block
to Extract
non-cerebral
tissue
(b)
Apply the Super Learner
Split dataset into Training
and Test set using
10-fold cross validation (1)
Divide each training fold using
10-fold cross validation (2)
Generate a classifier using
the new training set (2)
Apply the new
classifier
in Test set (2)
Concatenate the
results of each
classifier into
a vector
using each projection separately
Concatenate all the
classifiers responses
(vectors) into a matrix
Generate a model using
RF in the responses
After applying TL, perform
Super Learner in
using all projections together
from Apply Support
Vector Machine
to Arbitration
(Decision Making)
(c)
Figure 2: Overview of the proposed method for MCI recognition. (a) Entire model for recognition; (b) Grouping of AD
Classes step; (c) Super Learner approach.
~z =
~x − ¯x
σ
, (1)
where ~x represents the data points, ¯x is the mean
value of an attribute, and σ is the standard devia-
tion.
5. Spectral Clustering: The two initial CDR-classes
with dementia (i.e. CDR-0.5 and CDR-1) were
grouped using spectral clustering to create the
group of non-healthy subjects. This technique is
based on a similarity matrix and eigenvalues and
the goal is to perform a reduction in the dimen-
sionality of the data. The outcome is the assign-
ment of the CDR-0 class to one group and both
the CDR-0.5 and CDR-1 classes to another.
3.2.2 Skull Stripping
Also known as Brain Extraction, this step deals with
the removal of non-cerebral tissue from an MRI scan.
The OASIS-1 dataset provides brain images that are
VISAPP 2019 - 14th International Conference on Computer Vision Theory and Applications
556

(a) (b) (c) (d)
Figure 3: 3D representation of the Brain using OASIS-1: (a) Head illustration (adapted from (Lordkaniche, 2011)), (b) Axial
Plane, (c) Coronal Plane, (d) Sagittal Plane.
already extracted. However, brain extraction may be
independently accomplished my means of the Brain
Extraction Tool (Jenkinson et al., 2005).
3.2.3 Intensity Enhancement
The pixels in magnetic-resonance images have dis-
tinct intensities, which requires that the contrast must
be adjusted so that they all fall within the same inter-
val. Thus, the pixels were normalized using a 256-bin
MinMax through Equation (2):
I
IE
= low
o
+ (high
o
− low
o
) ×
I − low
i
high
i
− low
i
(2)
where I
IE
is the enhanced image, I represents the
original image, high represents the highest-intensity
pixel value, low is the lowest-intensity pixel value, i
and o are reference indexes to input and output data
respectively.
3.3 Classification
Classification is the problem of identifying to which
of a set of categories (sub-populations) a new obser-
vation (data) belongs. It adds a label to a data sample,
thus assigning it to a specific set. In this work, the la-
bels are binary as they specify either the presence or
the absence of MCI (i.e. CDR-0.5 and CDR-1).
The two types of classifiers used were Transfer
Learning and SVM. SVM’s are used to divide a da-
taset with binary classes in a hyperplane representa-
tion. In this work, we set the SVM using the follo-
wing attributes: C=1.0, kernel = radial basis function.
Since SVM’s are quite well documented in the li-
terature(Hearst et al., 1998)(Cristianini and Shawe-
Taylor, 2000), in this section we focus on Transfer
Learning.
CNN is a specific type of neural network that uses
grid-topologies as input data, such as images. Un-
like Artificial Neural Networks, CNNs have Convo-
lutional Layers for the processing of linear functions
and Pooling Layers for the non-linear ones. However,
CNNs have some restrictions: (1) They can only be
performed with a large number of images (i.e. thou-
sands). Depending on the model, the number of weig-
hts can reach millions for each convolutional layer
and their processing demands a relatively high com-
putational effort; (2) In the medical field there are few
domains that have large (i.e. enough) amount of avai-
lable data.
TL-CNN is a methodology that transfers pre-
trained network information to another network,
whether it belongs to the same domain or not. It is
an alternative to the application of CNNs when the
datasets are relatively small.
In this work, we used TL-CNN as follows: (1) A
VGG19 pre-trained network using the ImageNet da-
taset was loaded; (2) As the images inside ImageNet
have no relation to our images, only the six first layers
were maintained (the first layers only extract generic
features); (3) All the layers from the seventh up to the
last one were excluded; (4) Once the images are input
to the neural network, the outputs of the sixth layer
are L matrices with M × N dimension, where L repre-
sents the number of kernels and M × N represents the
image size; (5) Finally, we reshaped the output to one
row per image (also known as deep features).
3.4 Voting
Each method is pooled before a final classification de-
cision is carried out. In this work, two voting appro-
aches were used: (1) FP Priori: The decision of the
ensemble is TRUE if at least one of the classifiers re-
sults in a false-positive (FP). (2) Majority Voting: In-
dividual classifiers are combined by taking a simple
majority vote of their decisions. For any given in-
stance, the class chosen by most number of classifiers
is the ensemble decision.
Predicting the Early Stages of the Alzheimer’s Disease via Combined Brain Multi-projections and Small Datasets
557

3.5 Detection and Output
The procedure for detection is quite simple: the out-
put of the Majority Voting, the Super Learner and the
FP Priory are submitted to another Majority Voting to
generate the final label. The final output is a boolean
flag (or label) indicating whether a subject has MCI
(labeled as 1) or not (labeled as 0).
3.6 Software Implementation
The following tools were used for the implemen-
tation of the method: (1) In the Input step, the
208x176x176-pixel images were sliced using the
MATLAB’s r2015b Image Processing toolbox; (2)
The grouping of MCI classes was implemented early
in the preprocessing step using Python 3.4 Jupyter
Notebook, with the Scikit-learn package. Some steps
were manually coded (e.g. Progressive Sampling).
The image enhancement and skull stripping were ge-
nerated using MATLAB; (3) The Classification step
was all implemented using Python 3.4 Jupyter Note-
book. The extraction of deep features were obtained
using the Keras toolbox (with TensorFlow backend).
Once the CNN features were extracted, the SVM was
applied using the Scikit-learn package. The Super Le-
arner was manually implemented; (4) In the Voting
step, both the techniques (FP Priori and Majority Vo-
ting) were manually implemented using Python 3.4
Jupyter Notebook. (5) Detection was implemented
using Python 3.4 Jupyter Notebook; (6) Finally, the
metrics were generated using Python 3.4 Jupyter No-
tebook with Scikit-learn package.
All images and codes implemented in this work
are available in the GitHub platform(Duarte and
Paiva, 2018).
4 EXPERIMENTS AND RESULTS
Five metrics were adopted in this work: (1) Precision
- P; (2) Recall - R; (3) F-score - F; (4) Accuracy -
A; (5) Specificity - S. They are obtained by combi-
ning the true-positive (TP), true-negative (TN), false-
positive (FP), and false-negative (FN) values. These
evaluation metrics are obtained as follows:
P =
T P
T P + FP
, R =
T P
T P + FN
, F = 2 ×
P ∗ R
P + R
(3)
A =
T P + T N
T P + T N + FP + FN
, S =
T N
FP + T N
(4)
The data were first divided into a training and test
set by applying a 10-fold cross-validation. After the
results have been obtained, it is performed the average
between metrics using all folds. Thus, reducing the
significance of random factors. Regarding the TL-
CNN methodology, the training set was also divided
in 10% for validation and 90% remaining kept trai-
ning set.
To obtain a reasonable accuracy in the voting step,
it is significant to ensure that there is a small correla-
tion among the classifier outputs. The correlation ma-
trix for these outputs is illustrated in Table 1. The ana-
lysis using the Pearson Correlation coefficient shows
that the only correlations that are “moderate” are the
Axial and the Coronal (0.72). The correlation bet-
ween the other attributes is considered “weak”. Even
though the correlation between Axial and Coronal is
moderate, we opted to keep the three projections since
it is not possible carry out any decision with only two
projections, i.e. a third projection is always needed
for tie-breaking.
Table 1: Correlation Matrix from classifiers’ predictions.
Axial Coronal Sagittal
Axial 1 0.72 0.42
Coronal 0.72 1 0.27
Sagittal 0.42 0.27 1
Table 2 presents the results of our method and
the literature (NC vs MCI) regarding the five metrics.
When comparing with classifiers from literature, the
best ensemble evaluated regarding the specificity va-
lue was FP Priori (0.754).
The dataset was reduced to prevent any bias in the
classification step. For example, the subjects under
60 years were all removed due to their high discrimi-
native patterns in regard to subjects above 60 years.
The CDR-2 samples were also removed due to their
insufficient number (i.e. two), which would cause a
mislabeling at the time the CNN was trained. In the
end, 53% of the samples were removed.
The results are shown in Figure 4. Our method
ranked 5th (FP Priori) for Recall; 4th (Arbitrary) for
Accuracy, and 3rd (FP Priori) for Specificity. Nevert-
heless, our best result was achieved with the Arbitrary
voting (precision = 0.72, F-Score = 0.716, and accu-
racy = 0.726). It is important to notice that the num-
ber of samples in our case was lower than the others
in the literature (196 x 287 Kheder). Unlike Khed-
her who used ADNI, we used the OASIS dataset. We
have also not extended the data. Among the work re-
viewed in this section, the only that employed multi-
modality was Khedher (Khedher et al., 2015), which
from the literature was the best work evaluated. Ho-
wever, the authors do not specify the CDR classes that
were considered.
VISAPP 2019 - 14th International Conference on Computer Vision Theory and Applications
558
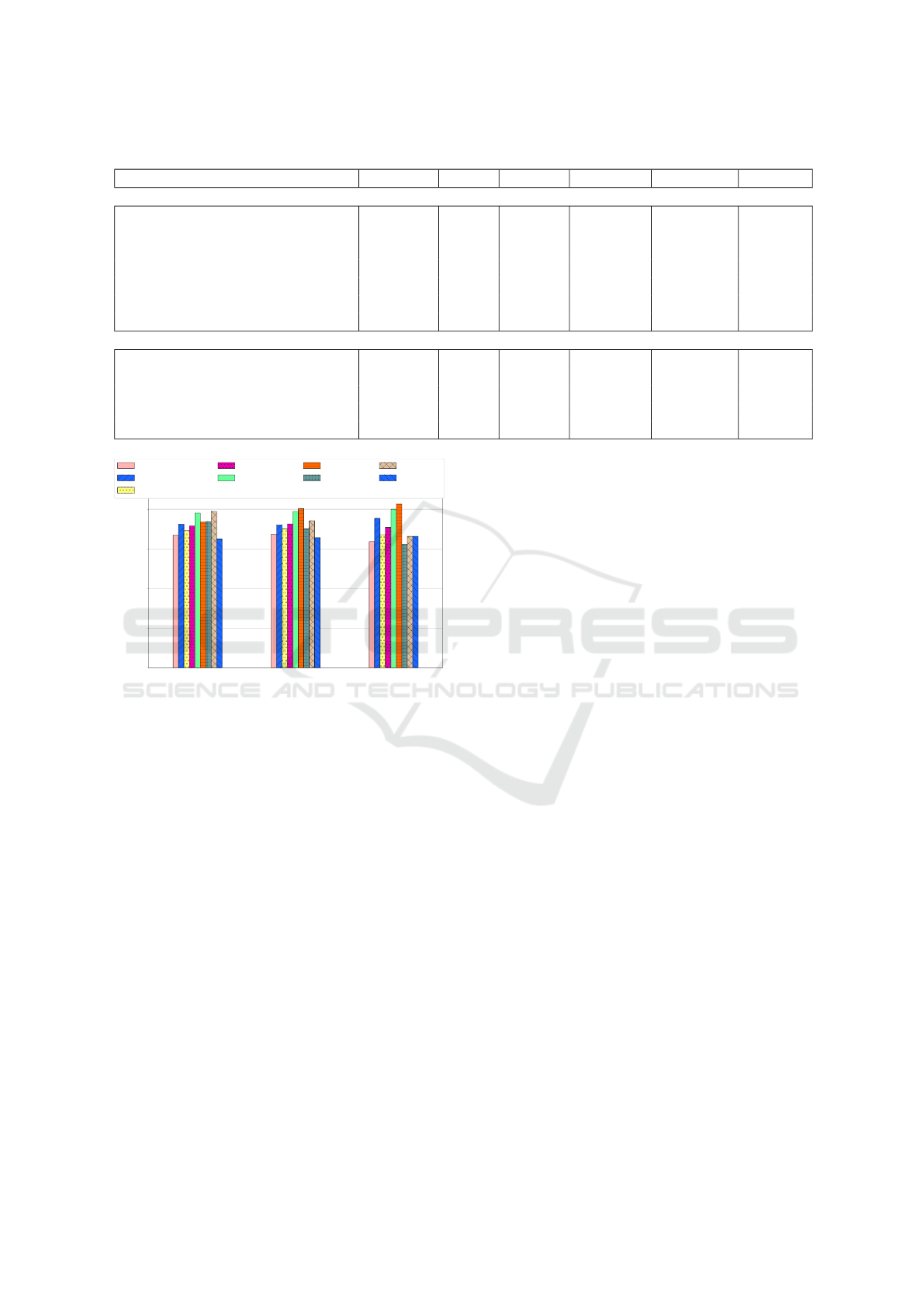
Table 2: Evaluation of Classifiers.
Classifier/Voting Precision Recall F-Score Accuracy Specificity Samples
Our Method
CNN Axial 0.706 0.701 0.698 0.711 0.683
CNN Coronal 0.638 0.633 0.627 0.637 0.597
CNN Sagittal 0.694 0.688 0.674 0.679 0.626
Majority Voting 0.676 0.669 0.665 0.674 0.637 196
FP Priori 0.710 0.725 0.702 0.721 0.754
Super Learner 0.696 0.692 0.688 0.700 0.671
Arbitrary (Decision Making) 0.720 0.716 0.714 0.726 0.709
Literature
Chen and Liu(Cheng and Liu, 2017) - 0,781 - 0,789 0,800 246
Khedher (Khedher et al., 2015) - 0,735 - 0,803 0,827 287
Gray (Gray et al., 2011) - 0,738 - 0,702 0,623 609
Suk and Shen(Suk and Shen, 2016) - 0,789 - 0,742 0,663 805
Aderghal (Aderghal et al., 2017) - 0,650 - 0,656 0,663 1020
Recall Accuracy Specificity
0.0
0.2
0.4
0.6
0.8
(%) of each metric
Maj Voting
FP Priori
Super Learner
Arbitration
Chen
Khedher
Gray
Suk
Aderghal
Figure 4: Evaluation of classifiers regarding Recall (Sensi-
tivity), Accuracy and Specificity metrics.
5 SUMMARY AND CONCLUSION
Alzheimer’s disease is currently ranked as the sixth
leading cause of death in the United States. Howe-
ver, recent estimates point out that the disorder may
be the third cause of death for older people, just be-
hind heart disease and cancer. Thus, computational
tools, methods, initiatives and efforts that support the
combat of Alzheimer or MCI are welcome.
In this work, we have presented a multi-projection
method based on a combined TL-CNN and SVM ap-
plication using Axial, Coronal, and Sagittal planes,
focused on to classify the first classes of the Alzhei-
mer’s Disease. Our process was defined by six major
steps: (1) input data, which is consisted of visual data
(MRI scans); (2) preprocessing, which is composed of
class grouping, skull stripping, and intensity enhance-
ment; (3) classification, using Transfer Learning and
Support Vector Machine to find AD-classes. The final
steps were to aggregate the voting (4) and classifier
methods to obtain the detection (5) and output label
(6) for a subject.
The focus of this work was on predicting the Alz-
heimer’s disease by detecting the MCI classes of AD
using Transfer Learning and SVM combined from
different brain projections. The method was applied
to a small dataset covering the prodromal stages of the
disease, where it is more challenging to find differen-
ces between patterns on the first MCI classes. We ar-
gue that this may be regarded as the worst-case input
scenario for the proposed method. For future work,
we suggest data augmentation as a preprocessing step
in order to improve accuracy, due to its capability of
generating different and additional patterns from in-
put data.
ACKNOWLEDGEMENTS
The authors thank CAPES (Brazilian Coordination of
Superior Level Staff Improvement), the reviews of
prof. Vitor R. Coluci, Guilherme P. Coelho, Ana E.
A. Silva, and Jo
˜
ao R. Bertini. Also, we thank CE-
PID FAPESP BRAINN (Brazilian Institute of Neu-
roscience and Neurotechnology). Data were provi-
ded by OASIS: Cross-Sectional: Principal Investiga-
tors: D. Marcus, R, Buckner, J, Csernansky J. Morris;
P50 AG05681, P01 AG03991, P01 AG026276, R01
AG021910, P20 MH071616, U24 RR021382.
REFERENCES
Aderghal, K., Benois-Pineau, J., Afdel, K., and Catheline,
G. (2017). FuseMe: Classification of sMRI images
by fusion of Deep CNNs in 2D+e projections. In
Content-based Multimedia Indexing , Florence, Italy.
Predicting the Early Stages of the Alzheimer’s Disease via Combined Brain Multi-projections and Small Datasets
559

Cheng, D. and Liu, M. (2017). Combining convolutional
and recurrent neural networks for alzheimer’s disease
diagnosis using pet images. In 2017 IEEE Internati-
onal Conference on Imaging Systems and Techniques,
pages 1–5.
Cristianini, N. and Shawe-Taylor, J. (2000). An Introduction
to Support Vector Machines and Other Kernel-based
Learning Methods. Cambridge University Press.
Duarte, K. T. N. and Paiva, P. V. V. (2018). Combining
tl, svm, and voting for alzheimer’s detection. https:
//github.com/enemy537/Multi-ModalCNN.
Ferreira, L. K. and Busatto, G. F. (2011). Neuroimaging
in alzheimers disease: current role in clinical practice
and potential future applications. Clinics, 66:19–24.
Fiot, J.-B., Fripp, J., and Cohen, L. D. (2012). Combi-
ning imaging and clinical data in manifold learning:
Distance-based and graph-based extensions of lapla-
cian eigenmaps. In 2012 9th IEEE International Sym-
posium on Biomedical Imaging. IEEE.
Gray, K. R., Wolz, R., Keihaninejad, S., Heckemann, R. A.,
Aljabar, P., Hammers, A., and Rueckert, D. (2011).
Regional analysis of fdg-pet for use in the classifica-
tion of alzheimer’s disease. In IEEE International
Symposium on Biomedical Imaging: From Nano to
Macro, pages 1082–1085.
Guerreiro, R. and Bras, J. (2015). The age factor in alzhei-
mer’s disease. Genome Medicine, 7(1).
Hearst, M., Dumais, S., Osuna, E., Platt, J., and Scholkopf,
B. (1998). Support vector machines. IEEE Intelligent
Systems and their Applications, 13(4):18–28.
Jenkinson, M., Pechaud, M., and Smith, S. (2005). BET2:
MR-based estimation of brain, skull and scalp surfa-
ces. In Eleventh Annual Meeting of the Organization
for Human Brain Mapping.
Khedher, L., Ramrez, J., Grriz, J., Brahim, A., and Segovia,
F. (2015). Early diagnosis of alzheimers disease based
on partial least squares, principal component analysis
and support vector machine using segmented mri ima-
ges. Neurocomputing, 151:139 – 150.
Lebedev, A., Westman, E., Westen, G. V., Kramberger,
M., Lundervold, A., Aarsland, D., Soininen, H.,
Kłoszewska, I., Mecocci, P., Tsolaki, M., Vellas, B.,
Lovestone, S., and Simmons, A. (2014). Random fo-
rest ensembles for detection and prediction of alzhei-
mers disease with a good between-cohort robustness.
NeuroImage: Clinical, 6:115–125.
Lordkaniche (2011). Share-cg face 01 - 3d mo-
del. http://www.sharecg.com/v/55159/view/5/
3D-Model/Face-01.
on Aging, N. I. (2016). Alzheimer’s disease fact
sheet. https://www.nia.nih.gov/health/
alzheimers-disease-fact-sheet.
Petersen, R. C., Smith, G. E., Waring, S. C., Ivnik, R. J.,
Tangalos, E. G., and Kokmen, E. (1999). Mild cog-
nitive impairment: clinical characterization and out-
come. Arch. Neurol., 56:303–308.
Suk, H.-I. and Shen, D. (2016). Deep ensemble sparse re-
gression network for alzheimer’s disease diagnosis. In
Wang, L., Adeli, E., Wang, Q., Shi, Y., and Suk, H.-I.,
editors, Machine Learning in Medical Imaging, pages
113–121, Cham. Springer International.
Wang, S., Shen, Y., Chen, W., Xiao, T., and Hu, J. (2017).
Automatic recognition of mild cognitive impairment
from mri images using expedited convolutional neu-
ral networks. In Lintas, A., Rovetta, S., Verschure,
P. F., and Villa, A. E., editors, Artificial Neural Net-
works and Machine Learning – ICANN, pages 373–
380. Springer.
Zhou, T., Thung, K.-H., Zhu, X., and Shen, D. (2017). Fe-
ature learning and fusion of multimodality neuroima-
ging and genetic data for multi-status dementia diag-
nosis. In Wang, Q., Shi, Y., Suk, H.-I., and Suzuki, K.,
editors, Machine Learning in Medical Imaging, pages
132–140, Cham. Springer International Publishing.
VISAPP 2019 - 14th International Conference on Computer Vision Theory and Applications
560
