
Strategies to Access Patient Clinical Data from Distributed Databases
Jo
˜
ao Rafael Almeida
1,2
, Olga Fajarda
1
, Arnaldo Pereira
1
and Jos
´
e Lu
´
ıs Oliveira
1
1
Institute of Electronics and Informatics Engineering of Aveiro (IEETA), University of Aveiro, Aveiro, Portugal
2
Department of Computation, Computer Science Faculty, University of A Coru
˜
na, A Coru
˜
na, Spain
Keywords:
Clinical Research, Electronic Health Records, Observational Studies, Common Data Model, Semantic Web.
Abstract:
Over the last twenty years, the use of electronic health record systems has become widespread worldwide,
leading to the creation of an extensive collection of health databases. These databases can be used to speed
up and reduce the cost of health research studies, which are essential for the advance of health science and
the improvement of health services. However, despite the recognised gain of data sharing, database owners
remain reluctant to grant access to the contents of their databases because of privacy and security issues, and
because of the lack of a common strategy for data sharing. Two main approaches have been used to perform
distributed queries while maintaining all data control in the hands of the data custodians: applying a common
data model, or using Semantic Web principles. This paper presents a comparison of these two approaches by
evaluating them according to parameters relevant to data integration, such as cost, data quality, interoperability,
extendibility, consistency, and efficiency.
1 INTRODUCTION
Health research studies are determinant for the ad-
vance of health science and the improvement of health
services. Pharmaceutical and public health surveil-
lance, the development of new treatments, the ex-
pansion of knowledge about diseases and monitor-
ing health crises are essentially done using health re-
search studies (Nass et al., 2009). This type of work
involves several time-consuming and expensive steps,
namely, the identification and recruitment of consent-
ing subjects, and the gathering of the data, which in
some cases means following the recruited subjects
over a long period. However, health research studies
can be speed up and much cheaper, if they are done
using data collected for other purposes, like data from
health-related registry systems or data collected from
previous studies (Cheng and Phillips, 2014).
Nowadays, due to the worldwide generalisation of
electronic health record (EHR) systems and the digi-
tisation of health-related information, a vast number
of electronic health databases, containing diversified
clinical digital data, exists (Geissbuhler et al., 2013).
Besides turning the research more efficient, by saving
time and money, the use of these databases for health
research studies has the advantage of increasing the
quality of the research, especially when combining
data from several databases (Piwowar and Chapman,
2010). Furthermore, the use of existing databases pre-
vents the collection of duplicate data and gives the
researcher access to a larger, more diverse popula-
tion, as well as to certain groups of people, which,
for example, do not participate in clinical trials, such
as children and older people (Schneeweiss and Avorn,
2005). Moreover, every clinical trial puts the research
subjects through some risk and, therefore, the sub-
stitution of a clinical trial by the secondary use of
clinical digital data prevents unnecessary risk (Doolan
et al., 2017). Even when clinical trials are necessary,
e.g. for the development of new therapies, existing
health care data can be used to identify clinical trial
participants, and, consequently, accelerate this com-
plex process (Ohmann and Kuchinke, 2007; Pakho-
mov et al., 2007). Drug safety surveillance is, essen-
tially, done using EHRs, because some adverse drug
events are only observed after the release of the drug
to a larger, diversified population (Trifir
`
o et al., 2014).
Retrospective cohort studies and case-control stud-
ies are other kinds of health research studies that can
be done using existing health databases (Ganz et al.,
2014; Reisner et al., 2015).
However, despite the recognition of the ines-
timable value of the secondary use of existing digi-
tal clinical data, and the importance of the open data
movement and the FAIR Data principles (Wilkinson
et al., 2016), health database owners remain reluctant
466
Almeida, J., Fajarda, O., Pereira, A. and Oliveira, J.
Strategies to Access Patient Clinical Data from Distributed Databases.
DOI: 10.5220/0007576104660473
In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), pages 466-473
ISBN: 978-989-758-353-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

in sharing the content of their databases (Pisani and
AbouZahr, 2010). Even the data obtained through
public research funding projects are not shared with
the research community (Lopes et al., 2015). The re-
luctance of the health database owners to share their
data is due to several reasons. The main reasons con-
cerns data ownership, intellectual property rights and
the lack of a common strategy for data sharing.
Two main approaches are used to enable the ac-
cess to clinical data from distributed databases, with-
out losing patient data privacy: (i) applying a common
data model or (ii) using Semantic Web (SW) princi-
ples. In this paper, we compare these two approaches
according to parameters relevant to data integration,
such as cost, data quality, interoperability, extendibil-
ity, consistency, and efficiency.
The rest of the paper is organized as follows: in
Section 2 we present an overview of existing solu-
tions, in Section 3 we describe the CDM, and the SW
approaches, in Section 4 we discuss and compare both
approaches, and finally in Section 5 we conclude the
paper.
2 RELATED WORK
Several solutions have been developed for the se-
cure sharing of patient clinical data from distributed
databases. CALIBER
1
, for instance, is a research
platform consisting of a combination of highly trained
staff, tools and data resources, “research ready”
variables extracted from linked electronic health
records coming from England’s hospital records, pri-
mary care, social deprivation information, and cause-
specific mortality data. The resources available con-
sists of data up to 2016 including more than 10 mil-
lion people with approximately 400 million person-
years of follow-up. The main purpose of CALIBER is
to promote an open community developing methods
and tools to accelerate replicable science across all
clinical and scientific disciplines spanning the trans-
lational cycle (from drug discovery through to public
health). However, the process to gain access to CAL-
IBER resources is very slow and bureaucratic.
PopMedNet
2
is a scalable and extensible open-
source platform to simplify the implementation
and operation of distributed health data query-
ing networks. This platform was developed by
HMORN (Health Maintenance Organisation Re-
search Network), a consortium of 19 U.S. regional
healthcare delivery organisations. Through a set of
1
http://www.ucl.ac.uk/health-informatics/caliber
2
http://www.popmednet.org/
web-based services and tools, PopMedNet enables
the creation and use of distributed data networks. It
supports both menus driven queries and distributed
analyses using complex, single-use or multi-use pro-
grams and returns aggregated counts of eligible study
cohorts (Brown et al., 2012).
OHDSI (Observational Health Data Sciences and
Informatics)
3
is an international, interdisciplinary and
multi-stakeholder project with the aim to develop ap-
plications to access and analyse large-scale observa-
tional health data. This collaborative was initiated at
the end of the Observational Medical Outcomes Part-
nership (OMOP) project, in order to continue the re-
search started. The OMOP was a public-private US
project with the objective to develop solutions to per-
form medical product safety surveillance using obser-
vational healthcare databases (Hripcsak et al., 2015).
The main outcome of the OMOP consortium was the
creation of the OMOP Common Data Model (CDM),
which standardises the content, structure and conven-
tion of healthcare databases (Overhage et al., 2011).
The OMOP CDM is considered to be the most com-
plete an efficient common data model available (Kahn
et al., 2012; Ogunyemi et al., 2013; Ross et al., 2014).
Over the last five years, besides continuing to improve
the OMOP CDM, the OHDSI community developed
several analytic tools, namely, Achilles, HERMES,
HERACLES, and CIRCE. In 2016, this community
released a web-based platform, called ATLAS
4
, that
integrates features form the previously mentioned ap-
plications. This web-based platform provides tools to
browse standardised vocabularies, explore databases,
define cohorts, and make a population-level analysis
of observational data converted to the OMOP CDM.
The European Medical Information Frame-
work (EMIF)
5
is a European project, launched in
2013, with the purpose of improving the access of
researchers to patient-level data from distinct health
data repositories across Europe. The EMIF Platform
is an integrated system where researchers can browse
three different levels of information: metadata, aggre-
gated data, and raw data. Every Data Custodian con-
trols to whom and the level of information that can be
shared (Trifan et al., 2018). Several solutions have
been developed to simplifying the access to health
data, in order to meet the needs of the Data Custodi-
ans involved in the project. EMIF has adopted OMOP
CDM for EHR data harmonisation, as also the use
of solutions to infer knowledge through query federa-
tion.
Applying the idea of having a common data
3
http://www.ohdsi.org/
4
http://www.ohdsi.org/web/atlas/
5
http://www.emif.eu
Strategies to Access Patient Clinical Data from Distributed Databases
467

model, there are some methodologies and tools with
a similar goal, such as the Semantic Web frame-
works (Berners-Lee et al., 2012). The principles of
SW and Linked Data (LD) (Speicher et al., 2015) can
be used to solve data integration and interoperability
problems. One of the pillars for the realisation of the
SW is the way data is represented. The Resource De-
scription Framework (RDF) covers this important is-
sue, with the data model proposed by the World Wide
Web Consortium (W3C) in a suite of normative spec-
ifications (Schreiber and Raimond, 2015).
Nowadays, semantic technologies are at the core
of many systems that support data-intensive research
areas, as is the case with system biology, inte-
grative neuroscience, bio-pharmaceutics and transla-
tional medicine, just to mention a few cases (Chen
et al., 2013). In addition, numerous repositories are
using the SW data model that can be accessed over the
Internet (Zaveri and Ertaylan, 2017), due to the exis-
tence of stable standards and best practice guidelines.
Bringing together people and machines, the semantic
technologies offer the ability to describe data better
and to map and link distributed datasets. In this way,
an information network is created that can be used by
searching the information from a single entry point.
The literature reports the use of various SW so-
lutions to integrate data from EHR systems. To in-
crease the usability of EHR systems, (Lasierra et al.,
2017) described a method to model patient-centered
clinical EHR workflows. To allow interoperable shar-
ing of patient data between healthcare organisations,
(Alamri, 2018) proposes a semantic-mediation archi-
tecture to support semantic interoperability. By us-
ing this intermediate layer, the clinical information is
exploited using richer ontological representations to
create a “model of meaning” for enabling semantic
mediation. For the facilitation of RDF data manage-
ment and query federation across several repositories,
(Sernadela et al., 2017) developed SCALEUS
6
, a se-
mantic web migration tool that can be deployed on
top of traditional systems to bring knowledge, infer-
ence rules, and query federation to the existent data.
In a single package, it includes a triplestore support-
ing multiple independent datasets, simplified API and
services for data integration and management, and
a SPARQL query engine, supporting real-time infer-
ence mechanisms and optimised text searches over the
knowledge base. This platform was used to facilitate
RDF data management and query federation across
the several tools of the RD-Connect initiative, an EU
FP7 project which aimed to create an integrated plat-
form connecting databases, registries, biobanks and
clinical bioinformatics for rare disease research.
6
http://bioinformatics-ua.github.io/scaleus/
3 QUERYING METHODOLOGIES
3.1 Querying Pipeline
Common technical and governance solutions must be
developed to simplify the access to health data. An
approach to do so is using the methodology presented
by (Fajarda et al., 2018), where a pipeline is used to
achieve the querying process, as shown in Figure 1.
An implementation of this kind of solution was done
in the EMIF project. The pipeline considers three
main roles:
• the Researcher, someone who needs to query sev-
eral databases, to which he has no direct access,
to conduct research;
• Data Custodians, individuals responsible for ad-
ministering their databases;
• the Study Manager, the person responsible for
managing the research study and act as an inter-
mediary between the Researcher and Data Custo-
dians.
The study starts with a researcher who wants to
query some databases. This person creates a study
request, writing his/her question in the EMIF Cat-
alogue
7
, a platform that allows researchers to find
databases which fulfil their particular research study
requirements. This platform, also, allows the research
to select the desired databases that s/he would like to
query (Silva et al., 2018).
After receiving the study request, the Study Man-
ager starts a workflow using TASKA
8
, a work man-
agement system (Almeida et al., 2018) that will sup-
port the whole study orchestration.
The first task of the workflow consists of the co-
hort/query definition, which results in a script. Us-
ing TASKA, this script can be sent to all the selected
Data Custodians at once. Every Data Custodian ex-
ecutes the script in their database, and the results of
the querying are, then, exported to the Study Man-
ager. After receiving all the results, the Study Man-
ager compiles them to answer the Research’s request.
Finally, the Research receives the response to his/her
request, and the pipeline ends.
The Study Manager has an important role in this
pipeline since s/he has direct access to the Data Custo-
dians and must have the knowledge necessary to work
with the query definition tools. Consequently, a per-
son not familiar with these tools can easily query sev-
eral databases of her/his choice and to which s/he has
no direct access.
7
https://emif-catalogue.eu/
8
https://bioinformatics.ua.pt/taska
HEALTHINF 2019 - 12th International Conference on Health Informatics
468
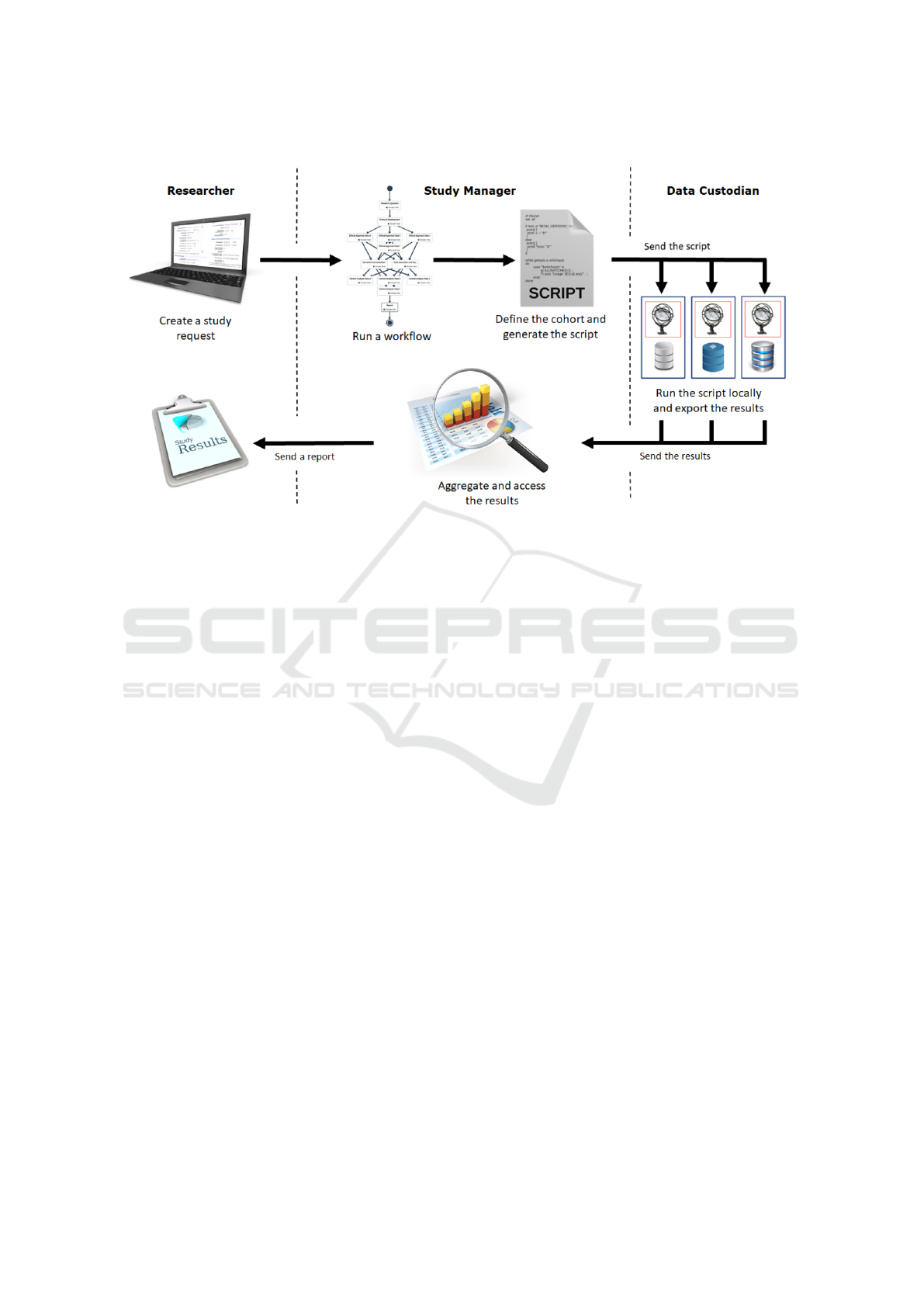
Figure 1: Workflow of the querying process (Fajarda et al., 2018).
Concerning the Data Custodian stage of the
pipeline, two main approaches can be followed. The
first one using a common data model, and the second
using the SW principles.
3.2 Common Data Model Approach
The Command Data Model approach requires that
Data Custodians’ databases use a shared schema to
all. Therefore, a model must be delineated, which is
currently not a problem, since there are already some
common data models defined for observational stud-
ies, e.g. the OMOP CDM. This common data model
is, already used in several countries (Hripcsak et al.,
2015). However, to be used the Data Custodians need
to convert their database into the OMOP CDM, using
Extract, Transform and Load (ETL) methodologies.
The OHDSI provides documentation of best practices
to perform the transformation, including several tools
to support the data migrations. In the early stages,
the OMOP CDM migration process was very com-
plex, however, currently, this procedure is optimised,
and OHDSI created several tools to guide the different
specialised entities involved. These specialised enti-
ties are:
• Local data experts and CDM experts, which to-
gether design the ETL transformation, without
creating the migration script;
• People with medical knowledge, which define the
code mappings;
• A technical person, which creates and implements
the ETL scripts following the specifications de-
fined previously.
In the final stage of the migration, all the entities in-
volved need to ensure the quality control of the imple-
mentation, this validates the process and ensures that
the data is consistent. However, despite all of these
tools and protocols design to help these entities, it is
still impossible to fully automate this process. An-
other disadvantage of this process is the need for peo-
ple with medical knowledge, which can be an expen-
sive resource.
Assuming that this procedure was done in all the
available databases, the Study Manager and the Data
Custodians can use some tools to extract and analyse
the data, e.g. ATLAS. With a local installation of AT-
LAS, the Study Manager can define a cohort and send
the resulting extraction script to all the Data Custodi-
ans involved in the study. The Data Custodians can,
then, execute the script received, in their local AT-
LAS installation, which provides a result, that can be
analysed and filtered, before being sent to the Study
Manager. This procedure ensures that Data Custodi-
ans have full control over their data and keeps non-
authorised users away from patients data, preserving
data privacy.
3.3 Semantic Web Approach
An alternative to using a common data model is the
use of Semantic Web technologies. This approach re-
Strategies to Access Patient Clinical Data from Distributed Databases
469
quires that Data Custodians use a common ontology
to specify the knowledge of the domain. The Web
Ontology Language (OWL) (Hitzler et al., 2012) is
the W3C semantic language to describe the entities
of domains, providing classes, properties, individuals,
and data values. Ontologies have been used by several
communities to structure knowledge domains. Just to
give an example, the Gene Ontology (GO)
9
defines
concepts to describe gene function along three differ-
ent aspects: molecular function, cellular component,
and biological process. Many more biomedical on-
tologies and terminologies can be found on the NCBO
BioPortal
10
(Whetzel et al., 2011).
The Semantic Web approach relies on the con-
version of the original data into RDF format. This
conversion must be performed for each distributed
database by using a convenient solution, such as the
SCALEUS tool. After the creation of the semantic
model for the domain of interest and the data con-
version into RDF, the SPARQL query language is
the convenient tool for extracting knowledge from the
created semantic database.
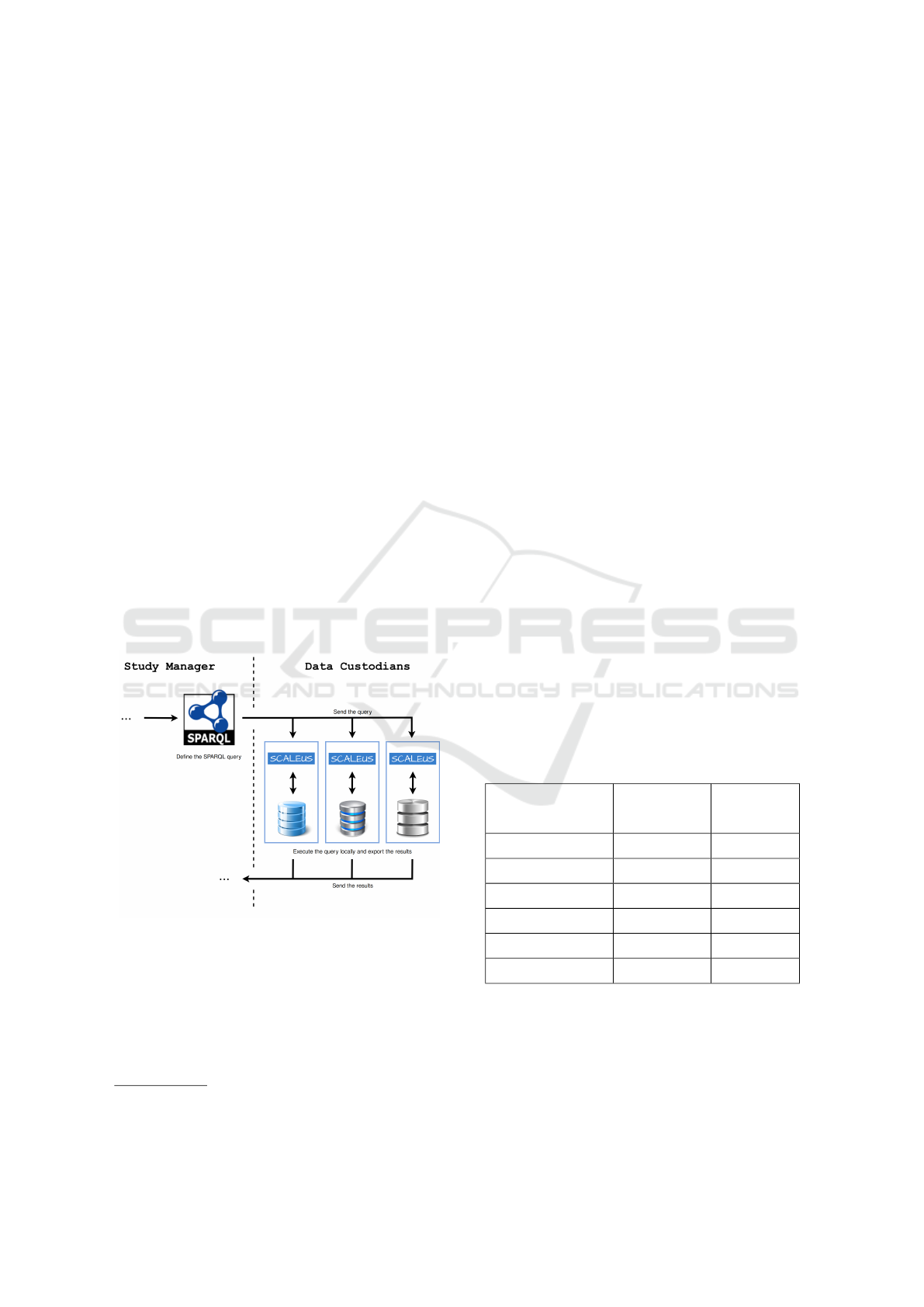
Figure 2 presents a snippet of the query pipeline.
In this scenario, the Study Manager uses SPARQL to
create the desired query and send it to the Data Cus-
todians. Then, the Data Custodians sends back the
patients’ data to the Study Manager, for interpretation
and compilation of results.
Figure 2: Semantic Web approach.
The SCALEUS solution provides a set of data
connectors and interfaces that help in the translation
process to a user pre-defined model (ontology) to be
negotiated between the Data Custodians. Although
migration tasks are simplified, the need for consensus
among Data Custodians may make it difficult to use
this option.
9
http://www.geneontology.org/
10
https://bioportal.bioontology.org/
4 DISCUSSION
Choosing the best strategy for using data from het-
erogeneous and distributed repositories is a task that
impacts across the course of an entire project. As no
universal formulas are covering all kinds of possibil-
ities, it is desirable for decision-makers to be aware
of the strengths and weaknesses of the most widely
used and documented available options. Some selec-
tion criteria can be pointed out from the authors’ ex-
perience based on their collaboration in the EMIF and
RD-Connect projects:
• Cost - Sum of costs of implementation and train-
ing of users;
• Data Quality - Ability to serve the purpose of
users;
• Interoperability - Interoperability relates with
machine-readability and machine-actionability,
describing the extent to which solutions can au-
tomatically exchange and interpret data;
• Extendibility - Possibility to extend and add fur-
ther information a-posteriori;
• Consistency - Refers to the data structure co-
herence between the different Data Custodians’
databases;
• Efficiency - Efficiency in producing an answer to
a research question.
Table 1 presents a summary of the evaluation of
both methodologies according to the considered se-
lection criteria, indicating the most favourable ap-
proach for each criterion.
Table 1: Assessment of the methodologies.
Common
Data Model
Semantic
Web
Cost + -
Data quality +/- +/-
Interoperability - +
Extendibility - +
Consistency + -
Efficiency + -
Legend. + : better; +/- : tied; - : worst
Data migration requires significant investments
in infrastructures, software solutions and human re-
sources. Considering that for both approaches the
infrastructure is already in place and that solutions
are open and free (e.g. OHDSI, SCALEUS), hu-
man effort become more relevant in the cost equation.
HEALTHINF 2019 - 12th International Conference on Health Informatics
470

Both approaches require medical knowledge. How-
ever, since the OMOP CDM migration is more pop-
ular, this pipeline is already optimised, reducing the
costs. Furthermore, the Semantic Web approach has
less adhesion in this scenario, causing more costs to
support this transition.
This data transition demands a great understand-
ing of the institutional data and its structure, which is
a requirement to produce a solid migration. During
this process, the data owners need to specify how to
deal with poor quality data. This is done in an ini-
tial stage of the data migration, where Data Custodi-
ans have the responsibility to ensure the data quality
of their databases, thus optimising the analyst’s work,
which avoids errors during the migration.
The ability of systems and applications to collab-
orate at a machine-machine level is a requirement for
automating the extraction of knowledge from hetero-
geneous and distributed data repositories. For this
collaboration to be possible between the different sys-
tems, they must communicate using a set of standards
that enable the intelligible communication of infor-
mation using, preferably, the Internet. For the first
approach, machine-machine interoperability is more
difficult. The data model used in this approach is not
as appropriate as that defined for SW solutions. On
the other hand, the existence of a series of standards
ensures that the second approach meets those require-
ments in an easier way to implement.
After putting a system in production, its data
model may need changes to reflect the changes in
the reality of interest. This need can, in the limit,
lead to the whole system having to be changed in
depth. The first approach is based on the use of entity-
relationship data models that scale poorly comparing
to SW solutions. In fact, changes to semantic data
models do not significantly change the systems in pro-
duction, ensuring a good extendibility.
Regardless of the approach chosen, Data Custodi-
ans will have a shared data structure. The Common
Data Model approach has already a well-defined data
structure. Additionally, the different data representa-
tion formats are normalised in the OMOP CDM ap-
proach, keeping the same conventions consistent in
the data model. Another aspect is the vocabulary def-
inition and mapping due to the existence of several
clinical terms. Those have been mapped onto OMOP
Vocabularies, improving the ability to analyse and
search the databases. Furthermore, the vocabulary
definition helps researchers find relevant drug codes.
For instance, if a researcher wants to find a drug by
its National Drug Code (NDC), s/he can do it easily
searching for it in ATLAS or ATHENA, which is also
a standardised process, due to the consistency in the
cohort definition, analysis design and results report-
ing. Succinctly, using OHDSI tools in OMOP CDM
databases, allows the observational research to be per-
formed by institutional groups, generating systematic
scientific practices, where research guidelines can just
be followed. In contrast, in the SW approach, vocab-
ulary and relationships are not standardised, needing
to be negotiated in advance.
The efficiency in observational studies is mainly
based on the Data Custodians’ response delay. This
lack of response’s speed can be turned in months or
even years of waiting to get a final answer which may
be one of the biggest challenges that researchers need
to deal, due to data accessing permissions restriction.
The pipeline presented, intends to reduce this delay,
mainly due to all the technology involved, which fa-
cilitates the querying process. Furthermore, the role
division reduces some boundaries that existed due to
the lack of agreements and rules. A Data Custodian
can quickly and easily query his/her database, anal-
yse the result, make the necessary adjustments, by fil-
tering some sensitive data, and sent it to the Study
Manager. This is possible mainly due to data classifi-
cation, and tools prepared to work with it. We could
also analyse the efficiency of both approaches individ-
ually. However, the most significant delay is the coor-
dination of people, which is enriched in the pipeline.
5 CONCLUSIONS
Observational data research offers the opportunity to
chart empirically-demonstrated scientific work and si-
multaneously produces an empirical evaluation of the
quality of the evidence generated, useful for mean-
ingfully informing decision-making processes. In or-
der to support such studies, while ensuring data pri-
vacy and security, a strategy for querying different
databases in a mediated way is needed. In this paper,
we analysed two different strategies to perform dis-
tributed queries in health databases. Both approaches
use open-source solutions and can offer alternative
pipelines to help researchers answer their questions
without the need for direct access to data. The first
approach is based on the use of a common data model
and the second on the application of Semantic Web
principles.
The approaches were evaluated based on a set
of selection criteria created from the authors’ expe-
rience in the application of each of the approaches.
Both approaches are similar when we consider the
data quality criteria. The Common Data Model so-
lution is more performant for the cost, consistency,
and efficiency. When considering interoperability and
Strategies to Access Patient Clinical Data from Distributed Databases
471

extendibility, the Semantic Web approach is more
favourable.
ACKNOWLEDGEMENTS
This work was partially funded by the NETDI-
AMOND project, grant number POCI-01-0145-
FEDER-016385. AP is supported by FCT, grant
PD/BD/142877/2018.
REFERENCES
Alamri, A. (2018). Semantic health mediation and access
control manager for interoperability among health-
care systems. Journal of Information Technology Re-
search, 11:87–98.
Almeida, J., Ribeiro, R., and Oliveira, J. L. (2018). A mod-
ular workflow management framework. In Proceed-
ings of the 11th International Conference on Health
Informatics (HealthInf 2018).
Berners-Lee, T., Hendler, J., and Lassila, O. (2012). The
semantic web. Scientific American, 284:34–43.
Brown, J., Balaconis, E., Mazza, M., Syat, B., Rosen, R.,
Kelly, S., Swan, B., and Platt, R. (2012). Ps1-46:
Hmornnet: shared infrastructure for distributed query-
ing by hmorn collaboratives. Clinical medicine & re-
search, 10(3):163–164.
Chen, H., Yu, T., and Chen, J. Y. (2013). Semantic web
meets integrative biology: a survey. Briefings in
Bioinformatics, 14:109–125.
Cheng, H. G. and Phillips, M. R. (2014). Secondary analy-
sis of existing data: opportunities and implementation.
Shanghai archives of psychiatry, 26(6):371.
Doolan, D. M., Winters, J., and Nouredini, S. (2017). An-
swering research questions using an existing data set.
Medical Research Archives, 5(9).
Fajarda, O., Silva, L. A. B., Rijnbeek, P. R., Van Spey-
broeck, M., and Oliveira, J. L. (2018). A methodology
to perform semi-automatic distributed ehr database
queries. In HEALTHINF, pages 127–134.
Ganz, M. L., Wintfeld, N., Li, Q., Alas, V., Langer, J., and
Hammer, M. (2014). The association of body mass
index with the risk of type 2 diabetes: a case–control
study nested in an electronic health records system in
the united states. Diabetology & metabolic syndrome,
6(1):50.
Geissbuhler, A., Safran, C., Buchan, I., Bellazzi, R.,
Labkoff, S., Eilenberg, K., Leese, A., Richardson, C.,
Mantas, J., Murray, P., et al. (2013). Trustworthy reuse
of health data: a transnational perspective. Interna-
tional journal of medical informatics, 82(1):1–9.
Hitzler, P., Kr
¨
otzsch, M., Parsia, B., Patel-Schneider, P. F.,
and Rudolph, S. (2012). Owl 2 web ontology language
primer (second edition). w3c recommendation.
Hripcsak, G., Duke, J. D., Shah, N. H., Reich, C. G., Huser,
V., Schuemie, M. J., Suchard, M. A., Park, R. W.,
Wong, I. C. K., Rijnbeek, P. R., et al. (2015). Obser-
vational health data sciences and informatics (ohdsi):
opportunities for observational researchers. Studies in
health technology and informatics, 216:574.
Kahn, M. G., Batson, D., and Schilling, L. M. (2012).
Data model considerations for clinical effectiveness
researchers. Medical care, 50.
Lasierra, N., Schweitzer, M., Gorfer, T., Toma, I., and
Hoerbst, A. (2017). Building a semantic model
to enhance the user’s perceived functionality of the
ehr. Studies in Health Technology and Informatics,
228:137–141.
Lopes, P., Silva, L. B., and Oliveira, J. L. (2015). Chal-
lenges and opportunities for exploring patient-level
data. BioMed research international, 2015.
Nass, S. J., Levit, L. A., Gostin, L. O., et al. (2009). The
value, importance, and oversight of health research.
Ogunyemi, O. I., Meeker, D., Kim, H.-E., Ashish, N.,
Farzaneh, S., and Boxwala, A. (2013). Identifying
appropriate reference data models for comparative ef-
fectiveness research (cer) studies based on data from
clinical information systems. Medical care, 51:S45–
S52.
Ohmann, C. and Kuchinke, W. (2007). Meeting the chal-
lenges of patient recruitment. International Journal
of Pharmaceutical Medicine, 21(4):263–270.
Overhage, J. M., Ryan, P. B., Reich, C. G., Hartzema, A. G.,
and Stang, P. E. (2011). Validation of a common data
model for active safety surveillance research. Jour-
nal of the American Medical Informatics Association,
19(1):54–60.
Pakhomov, S., Weston, S. A., Jacobsen, S. J., Chute, C. G.,
Meverden, R., Roger, V. L., et al. (2007). Electronic
medical records for clinical research: application to
the identification of heart failure. Am J Manag Care,
13(6 Part 1):281–288.
Pisani, E. and AbouZahr, C. (2010). Sharing health data:
good intentions are not enough. Bulletin of the World
Health Organization, 88(6):462–466.
Piwowar, H. A. and Chapman, W. W. (2010). Public shar-
ing of research datasets: a pilot study of associations.
Journal of informetrics, 4(2):148–156.
Reisner, S. L., Vetters, R., Leclerc, M., Zaslow, S., Wol-
frum, S., Shumer, D., and Mimiaga, M. J. (2015).
Mental health of transgender youth in care at an ado-
lescent urban community health center: a matched ret-
rospective cohort study. Journal of Adolescent Health,
56(3):274–279.
Ross, T. R., Ng, D., Brown, J. S., Pardee, R., Hornbrook,
M. C., Hart, G., and Steiner, J. F. (2014). The hmo
research network virtual data warehouse: a public data
model to support collaboration. EGEMS, 2(1).
Schneeweiss, S. and Avorn, J. (2005). A review of uses
of health care utilization databases for epidemiologic
research on therapeutics. Journal of clinical epidemi-
ology, 58(4):323–337.
Schreiber, G. and Raimond, Y. (2015). Rdf 1.1 primer. w3c
working group note.
Sernadela, P., Gonz
´
alez-Castro, L., and Oliveira, J. (2017).
Scaleus: Semantic web services integration for
HEALTHINF 2019 - 12th International Conference on Health Informatics
472

biomedical applications. Journal of Medical Systems,
41:1–11.
Silva, L. B., Trifan, A., and Oliveira, J. L. (2018). Montra:
An agile architecture for data publishing and discov-
ery. Computer methods and programs in biomedicine,
160:33–42.
Speicher, S., Arwe, J., and Malhotra, A. (2015). Linked
data platform 1.0. w3c recommendation.
Trifan, A., D
´
ıaz, C., Oliveira, J., et al. (2018). A method-
ology for fine-grained access control in exposing
biomedical data. Studies in health technology and in-
formatics, 247:561–565.
Trifir
`
o, G., Coloma, P., Rijnbeek, P., Romio, S., Mosseveld,
B., Weibel, D., Bonhoeffer, J., Schuemie, M., Lei, J.,
and Sturkenboom, M. (2014). Combining multiple
healthcare databases for postmarketing drug and vac-
cine safety surveillance: why and how? Journal of
internal medicine, 275(6):551–561.
Whetzel, P. L., Noy, N. F., Shah, N. H., Alexander, P. R.,
Nyulas, C., Tudorache, T., and Musen (2011). Biopor-
tal: enhanced functionality via new web services from
the national center for biomedical ontology to access
and use ontologies in software applications. Nucleic
Acids Research, 39:W541–W545.
Wilkinson, M., Dumontier, M., Aalbersberg, I., Appleton,
G., Axton, M., Baak, A., Blomberg, N., Boiten, J., da
Silva Santos, L., Bourne, P., Bouwman, J., Brookes,
A., Clark, T., Crosas, M., Dillo, I., Dumon, O., Ed-
munds, S., Evelo, C., Finkers, R., Gonzalez-Beltran,
A., Gray, A., Groth, P., Goble, C., Grethe, J., Heringa,
J., ’t Hoen, P., Hooft, R., Kuhn, T., Kok, R., Kok,
J., Lusher, S., Martone, M., Mons, A., Packer, A.,
Persson, B., Rocca-Serra, P., Roos, M., van Schaik,
R., Sansone, S., Schultes, E., Sengstag, T., Slater, T.,
Strawn, G., Swertz, M., Thompson, M., Van Der Lei,
J., Van Mulligen, E., Velterop, J., Waagmeester, A.,
Wittenburg, P., Wolstencroft, K., Zhao, J., and Mons,
B. (2016). The fair guiding principles for scientific
data management and stewardship. Scientific Data, 3.
Zaveri, A. and Ertaylan, G. (2017). Linked data for life
sciences. Algorithms, 10:126.
Strategies to Access Patient Clinical Data from Distributed Databases
473
