
MediBot: An Ontology based Chatbot for Portuguese Speakers Drug’s
Users
Caio Viktor S. Avila
1
, Anderson B. Calixto
1
, Tulio Vidal Rolim
1
, Wellington Franco
2
,
Amanda D. P. Venceslau
2
, Vânia M. P. Vidal
1
, Valéria M. Pequeno
3
and Francildo Felix De Moura
1
1
Department of Computing, Federal University of Ceará, Campus do Pici, Fortaleza-CE, Brazil
2
Federal University of Ceará, Campus de Crateús, Crateús-CE, Brazil
3
TechLab, Departamento de Ciências e Tecnologias, Universidade Autónoma de Lisboa Luís de Camões, Portugal
amanda.pires@ufc.br, vpequeno@autonoma.pt
Keywords:
Chatbot, Data Integration, Semantic Web, Medical Informatics, Drugs.
Abstract:
Brazil is one of the countries with the highest level of drug consumption in the world. By 2012 about 66%
claimed to practice self-medication. Such activity can lead to a wide range of risks, including death from
drug intoxication. Studies indicate that a lack of knowledge about drugs and their dangers is one of the main
aggravating factors in this scenario. This work aims to universalize access to information about medications
and their risks for different user profiles, especially Brazilian and lay users. In this paper, we presented
the construction process of a Linked Data Mashup (LDM) integrating the datasets: consumer drug prices,
government drug prices and drug’s risks in pregnant from ANVISA and SIDER from BIO2RDF. In addition,
this work presents MediBot, an ontology-based chatbot capable of responding to requests in natural language
in Portuguese through the instant messenger Telegram, smoothing the process to query the data. MediBot acts
like a native language query interface on an LDM that works as an abstraction layer that provides an integrated
view of multiple heterogeneous data sources.
1 INTRODUCTION
Brazil is the fifth country with the highest consump-
tion of drugs in the world, being the first in Latin
America (Sousa et al., 2008). In the nation, drugs
occupy the first position among the agents causing in-
toxication, in front of toxic drugs such as pesticides,
illicit drugs, rodenticides, insecticides and foodstuffs
improper for consumption (Corrêa et al., 2013). In
2012, according to Pinto et al. (2012), 66% of the pop-
ulation said he had practiced self-medication in his
lifetime. It is understood as self-medication the use
of drugs without any intervention by a physician, or
other qualified professional, neither in diagnosis, nor
in prescription, nor the follow-up of treatment. Only
in 2016, there were approximately 56,937 registered
cases of human intoxication, of which, 226 evolved to
death, 20,527 being generated due to the use of drugs
(SINITOX, 2016).
The pharmaceutical professional is of great im-
portance in combating the risks resulting from self-
medication. However, in many cases access to a phar-
macist perhaps impracticable, either because of the
low number of professionals relative to the population
size or because of the distance in more remote areas.
Also, there is a need for public awareness campaigns
about the risks of adverse effects of certain drugs.
Authors claim that actions such as improving the
quality of prescriptions and preventing inappropriate
self-medication could reduce the occurrence of ad-
verse reactions (Queneau et al., 2007). Another prob-
lem is that the health terminologies used are rep-
resented using two perspectives: the end-user and
the health professional, making it difficult to under-
stand medical terminologies and their classifications.
Therefore, a common vocabulary is necessary, mak-
ing possible the understanding of medical terminolo-
gies and their classifications with a single meaning.
In the web, there is a wide variety of data about
drugs, such data originating from either government
organizations, such as regulatory agencies and public
data portals, makers, on-line drugstores and electronic
bulletin. However, the vast majority of these data are
in the proprietary format, such as spreadsheets, rela-
tional database backups or are available only through
web pages. Also, such data are isolated in data silos,
Avila, C., Calixto, A., Rolim, T., Franco, W., Venceslau, A., Vidal, V., Pequeno, V. and Felix De Moura, F.
MediBot: An Ontology based Chatbot for Portuguese Speakers Drug’s Users.
DOI: 10.5220/0007656400250036
In Proceedings of the 21st International Conference on Enterprise Information Systems (ICEIS 2019), pages 25-36
ISBN: 978-989-758-372-8
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
25

with no direct connection between resources. More-
over, each dataset perhaps represented in different vo-
cabularies, where the same concept in the real world
is represented with varying terms through different
datasets.
An example of information that can not be re-
trieved directly in the current state of data is given in
next: “What are the risks of the drug XX?”. This sim-
ple query requires information from a different set of
sources. The retrieval of such information has some
challenges, such as heterogeneity of the format of the
sources, where some that are founded in XLS, CSV,
RDF formats or web pages which requires different
query techniques. Besides, there’s the need to per-
form the information junction manually, since such
databases usually aren’t integrated. Moreover, it is
necessary for the user to have technical knowledge
about the area, as there may be a need to resolve
vocabulary inconsistencies. These problems can be
solved by using Semantic Web technologies, such as
ontologies that allow the data’s representation in the
same target vocabulary.
Semantic Web (Shadbolt et al., 2006) and Linked
Data (Bizer et al., 2011) technologies are used to han-
dle such limitations. This technology allows a seman-
tic integration between resources in different sources,
representing the data in a unified vocabulary through
the use of ontologies. The ontology provides a uni-
form vocabulary for data access, acting as an abstrac-
tion layer for data access. Besides, it allows its publi-
cation openly in a non-proprietary format that allows
retrieval of information by both computerized agents
and human users via SPARQL queries endpoints.
The main aim of this work is to facilitate access to
information about drugs and their risks easily and di-
rectly to users. For this purpose we performed the
integration of drug data with a focus on Brazilian
data, finally producing a Linked Data Mashup (Hoang
et al., 2014), and as an application an ontology-based
chatbot, called MediBot is presented to the Telegram
instant messenger to answer questions in natural lan-
guage about the integrated data, providing a more in-
tuitive user interface. The main contributions are:
• A data’s integration of different data sources about
drugs. We add the information on drug risks con-
tained in the SIDER dataset to the products sold
only in Brazil;
• A mashup model to integrate drug bases;
• A Natural Language Interface (NLI) for end-user
through the use of a ChatBot.
The rest of this paper is organized as follows: Sec-
tion 2 presents the related works. Section 3 describes
the data integration process. Section 4 introduces
Medibot, a chatbot application for queries about the
integrated data. In section 5 we present the evaluation
of MediBot. Finally, in section 6, are discussed the
conclusions and future work.
2 RELATED WORKS
In (Jovanovik, 2017), it showed to perform the inte-
gration and publication of data about drugs of twenty
and three countries. However, Brazil is not found be-
tween these. Also, the vocabulary defined for the inte-
gration does not fit the databases selected in this work.
(Natsiavas et al., 2017) present a model for use of
several databases integrated through Linked Data, in
particular, datasets of the project BIO2RDF for the
mining of signs of adverse reactions of drugs, show-
ing the potential of linked databases in the drug do-
main. However, the model was designed to be used as
part of a data mining platform, so it does not address
how users will access data. In addition to this work,
others have the same goal as (Nová
ˇ
cek et al., 2017)
and (Natsiavas et al., 2015).
(Vega-Gorgojo et al., 2016) presents PepeSearch,
a system that facilitates searching between different
sources Linked Data in the field of drugs and health,
such as Drug Bank(DB) and Sider. The system pro-
vides a faceted query interface that allows the user
to search on multiple data sources. However, the
system is best suited for use by specialists and re-
searchers, besides it has a powerful yet complex query
interface. Moreover, the system does not have data
on drug risks in pregnancy (DRP), despite having
the SIDER side-effects data. Lastly, “MedChatBot”
is a chatbot for medical students based on the open
source AIML UMLS to generate responses to queries
through knowledge extraction. Table 1 presents an
analysis of the aspects present in this work and the
others related (Kazi et al., 2012).
Table 1: Comparison of Related Work.
DB
Sider
DRP NLI
(Jovanovik, 2017) X X - -
(Natsiavas et al., 2017) X X - -
(Nová
ˇ
cek et al., 2017) X X - -
(Natsiavas et al., 2015) X X - -
(Vega-Gorgojo et al., 2016) X X - -
(Kazi et al., 2012) - - - X
This paper - X X X
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
26

3 LINKED DATA MASHUP
CONSTRUCTION
In this section, we describe the process of construc-
tion and publication of Linked Data Mashup (LDM).
The integration process was based on the Linked
Data Integration Framework (LDIF). LDIF suggests
the following execution flow: i) Extraction of data
sources; ii) Transformation of data (Triplification)
and construction of exported of views; iii) Resolu-
tion of the identity through links owl:sameAs; iv)
Data quality assessment and fusion and v) Data output
(Schultz et al., 2012).
3.1 Selected Sources
In this work, the following criteria that were used for
the selection of datasets: The data should have in-
formation about drugs description, commercial drugs,
drugs risks, drug’s indications and finally, the data
must have relevance for non-specialist users. Also,
preference is given to Brazilian or Portuguese data,
especially for drugs sold only in Brazil and its risks.
Based on the previously listed criteria, four differ-
ent datasets were selected, three of which are avail-
able from the Agência Brasileira de Vigilância San-
itária(ANVISA)
1
or Brazilian Sanitary Surveillance
Agency in English, and the last one belongs to the
BIO2RDF project (Belleau et al., 2008). From AN-
VISA, we selected:
• Consumer Drug Prices (CDP)
• Government Drug Prices (GDP)
• Drug’s risks in pregnant and breastfeeding (RPB)
CPD and GPD are found in the XLS and PDF file
formats in website
2
, wherein this work the XLS ver-
sion was used. Both datasets have information about
allopathic drugs, such as drug name, producer, bar-
code, therapeutic class, presentation, the active ingre-
dient, and prices. The only difference between them
is because the former has maximum selling prices for
the average consumer, while the latter has maximum
selling prices for government agencies.
The dataset RPB contains the risk categories of
substances during the period of pregnancy and breast-
feeding. This dataset can be found in webdocument
3
and is only available in unstructured PDF.
The last dataset selected was the SIDER made
available by the BIO2RDF project, and can be found
1
http://portal.anvisa.gov.br/
2
http://portal.anvisa.gov.br/listas-de-precos
3
http://portal.anvisa.gov.br/documents/33880/2561889
/116.pdf/2292b730-2bd5-4acc-b378-
10682b1fc344?version=1.0
in website
4
already in the RDF format. The dataset
SIDER contains data about drugs, their indications,
side effects, and different labels. However, the
database only has data in English, not containing in-
formation about Brazilian drugs, making it neces-
sary to translate it into Portuguese. This dataset has
been selected because it contains information about
the side effects of active principles, such information
is needed to inform the risks of a drug.
3.2 Vocabulary
Because datasets have different structures, physical
formats, and vocabularies, a mean was required to
standardize access to information. The Linked Data
approach uses ontologies to address such a problem.
In the Linked Data paradigm, ontologies OWL
5
are
used, which provide a representation of knowledge in
a taxonomic way by through of a hierarchy of classes
and properties, one of the objectives of OWL is to
structure data in a semantically understandable way
by the machine allowing the inference of implicit in-
formation based on defined axioms.
The OWL ontology provides a layer of semantic
abstraction, allowing access to the integrated data in a
transparent way to the user, in addition to using terms
closer to their daily life, abstracting coded fields and
giving definitions about terms, giving so the possibil-
ity of a greater understanding to the lay user.
In our work, a vocabulary was developed based on
the data dictionaries of the original datasets, as well as
other sources of knowledge about the domain such as
sites, books, and manuals, having, in particular, the
booklet “What we should know about drugs”
6
made
available by ANVISA. In developing the vocabulary,
it was always sought to conceptualize verbatim each
term used, in addition to providing different alter-
native nomenclatures following the non-ontological
sources cited before.The OWL implementation can be
found in
7
3.3 Exported Views
The exported view of a dataset consists of its repre-
sentation using the vocabulary of the target ontology.
The exported view represents an RDF view of the data
4
http://download.bio2rdf.org/files/release/3/sider/sider.htm
5
https://www.w3.org/OWL/
6
http://www.vigilanciasanitaria.sc.gov.br/index.php/
download/category/112-medicamentos?download=102:
cartilha-o-que-devemos-saber-sobre-medicamentos-anvisa
7
https://datahub.io/linkeddatamashupeducacional/data-
med/v/2
MediBot: An Ontology based Chatbot for Portuguese Speakers Drug’s Users
27

contained in the source, and this view can be material-
ized or virtual. In both views, there is a need for map-
ping rules to translate terms from the original dataset
into the target vocabulary. The difference between
materialized or virtual view lies in the fact that in the
first case the data is physically converted to the RDF
format and stored in a triplestore on which SPARQL
queries will be made. While in the second the data
is kept in its original format, where SPARQL queries
will be made through a mediator that translates the
SPARQL query into the native query language of the
data (e.g., SQL) through the mappings. In this work
we materialized the exported views
In this paper we will use the term triplification to
refer to the process of generating RDF
8
triples that
represent the original data using the defined target vo-
cabulary.
For the triplification of datasets CDP and GDP,
we imported the data into the relational database man-
agement system PostgreSQL
9
, due to the existence of
matured tools in the conversion of relational databases
for RDF. In this work, we used the tool D2RQ (Bizer
and Seaborne, 2004) that performs the transformation
of relational bases to RDF by mapping the original
schema to the desired target vocabulary. The mapping
language used was R2RML
10
.
For the triplification of the RPB dataset, manual
conversion of the PDF file to CSV was required on
account of the file’s internal structure. But finally, the
same process previously described was used for its
triplification.
Finally, since the SIDER source already exists in
the RDF format, it was only necessary to use SPARQL
CONSTRUCT to mapping the original dataset to the
desired vocabulary.
The result of this step was a set of four RDF
graphs with the target vocabulary representing each
original dataset. However, it is still necessary to find
out which resources represent the same object be-
tween the different RDF graphs and merge them into
one.
Figure 1 shows the exported views of the infor-
mation contained in the original sources to existing
concepts in the defined target ontology.
3.4 Identity Resolution
This step is responsible for discovering which differ-
ent resources represent the same object in the real
world to connect them via owl: sameAs links. These
resources can be found on a single source or between
8
https://www.w3.org/RDF/
9
https://www.postgresql.org/
10
http://www. w3. org/TR/r2rml/
the different sources, where the direct comparison be-
tween sources are only possible because there is al-
ready a guarantee that all sources have uniform struc-
ture and vocabulary because of the ontology.
In figure 2 is possible to see an example of iden-
tity resolution. In this example, two resources are
representing the drug “Reopro”, the CDP/Reopro (in
left) representing it in the dataset CDP and GDP/Re-
opro (on the right) in the dataset GDP. Each of the
resources has its properties, including its active prin-
ciples, being these CDP/abciximab and GDP/abcix-
imab. The active principle “abciximab” is also rep-
resented by different resources between the different
datasets, and there is no connection between the re-
sources. Without there being a connection between
the resources of different datasets, there is no possi-
bility, for example, to answer the risks of the drug
“Reopro”, since no dataset has such information indi-
vidually.
The identity resolution will be responsible for dis-
covering that the CDP/Reopro and GDP/Reopro re-
sources represent the same object, thus constructing
an owl:sameAs link between them. Furthermore, the
links between the CDP/abciximab, GDP/abciximab,
Sider/abciximab, and RPB/abciximab resources are
also discovered at this stage. After creating the
owl:sameAs links, it is already possible to merge in-
formation from several datasets about the drug “Reo-
pro” so that it is possible to discover its risks.
For this step, the tool Silk Volz et al. (2009) was
used. Silk uses user-specified rules to discover and
generate links between resources. For this work, sim-
ple rules, such as strings treatment, comparison of
values and averages, were used in general. For the
most part, the defined rules have used the dc:title at-
tribute that represents the title or nomenclature of the
resource.
Links were generated between resources of the
Drug, Laboratory, Therapeutic Class, Substance and
Presentation classes. The other classes were not taken
into account because their mappings themselves al-
ready guarantee the creation of resources with the
same URI. Table 2 illustrates the number of links gen-
erated between sources.
3.5 Quality Evaluation and Data Fusion
After the identity resolution step, it is already possi-
ble to know through the owl: sameAs links which dis-
tinct resources represent the same real-world object.
However, such resources are still represented by dis-
tinct URIs, so there is a need to merge such resources
into a single one that will encompass all the proper-
ties and relationships of the originals. However, this
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
28
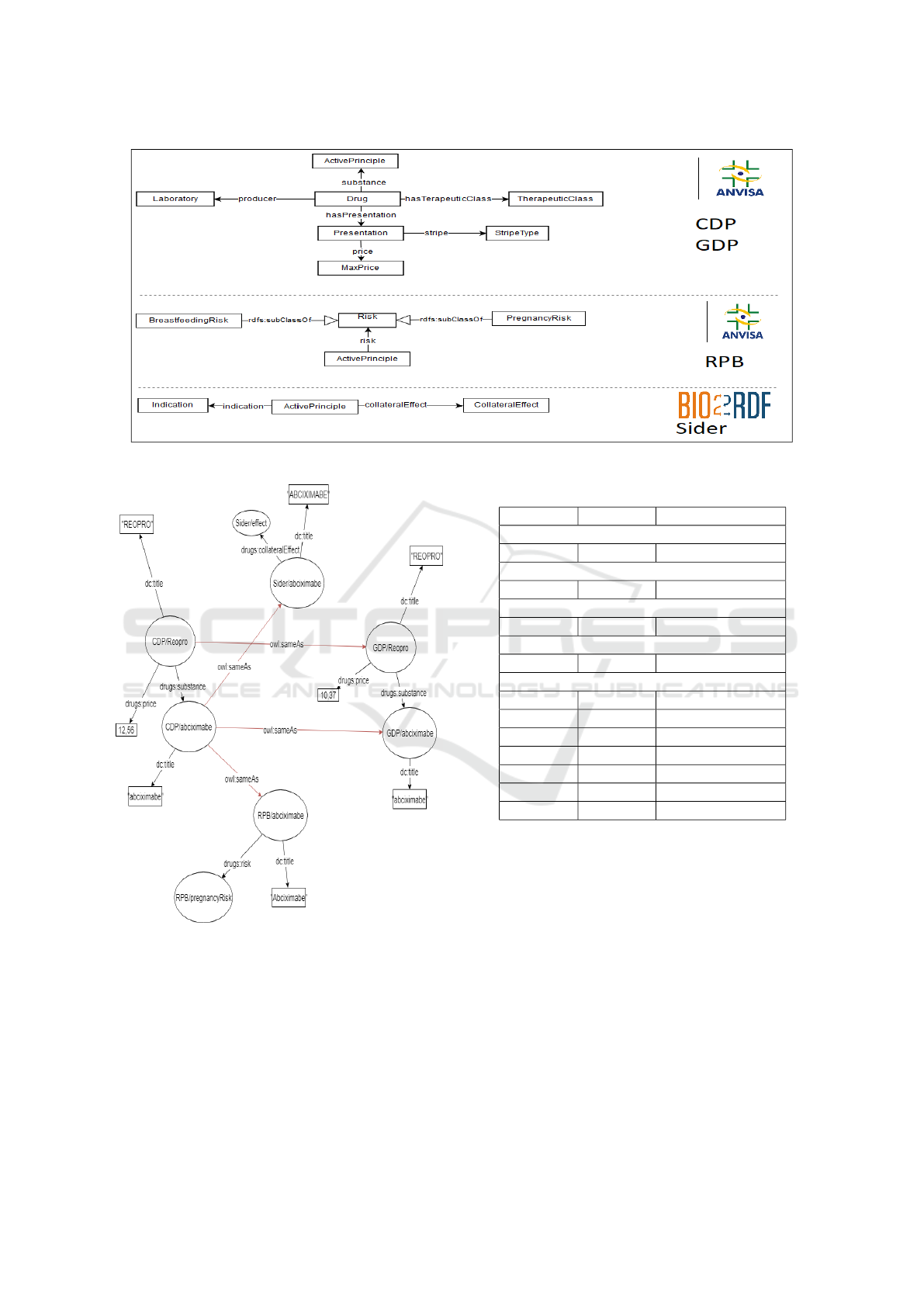
Figure 1: Datasets Exported Views.
Figure 2: Example of Identity Resolution.
merge may cause some problems, such as repeated
values for properties, conflicting between properties
values, or incorrect values. So it’s necessary to have
the quality evaluation of the sources, this process de-
fines the degree of priority of each source. After the
quality evaluation, the data fusion and data cleaning
process will be performed, choosing which informa-
tion should be kept or deleted, based on the priority
Table 2: Generated sameAs Links by Class.
Source A Source B Generated Links
Drug
CDP GDP 8839
Laboratory
CDP GDP 25623
Therapeutic Class
CDP GDP 485
Presentation
CDP GDP 25623
Substance (Active Principle )
CDP GDP 2164
CDP Sider 6401
CDP RPB 374
GDP Sider 6401
GDP RPB 323
RPB Sider 8956
Sider Sider 828175
of each source.
In figure 3, it is possible to observe the result
of the fusion process of the drug “Reopro”. After
the fusion, the drug “Reopro” will be represented
by a single resource, as well as its active principle.
The resulting merged entities will contain all the in-
formation, previously scattered among the different
datasets, where redundancy and inconsistency have
already been solved.
For this step, we used the tool Sieve (Mendes et al.,
2012). For the quality evaluation phase, the metric
ScoredPrefixList was used, where for each source a
weight was given in the following order: CDP, GDP,
RPB and finally Sider. Such order was selected us-
ing manual analysis taking into account the quality of
MediBot: An Ontology based Chatbot for Portuguese Speakers Drug’s Users
29

Figure 3: Example of resources fused.
how the data is structured, the scope of data, number
of conflicts in original data and the number of links
created between resources of the same source. This
last rule comes from the intuition that if a source has
many different resources that represent the same ob-
ject, then the source was constructed less rigorously,
so it will have a lower priority.
Fusion rules have been defined for cleaning only
the classes Drug, Laboratory, Therapeutic Class, Sub-
stance, and Presentation, due possible fusion of such
resources.
3.6 Publication of the Linked Data
Mashup
At the end of the semantic integration process, we
generated a dataset RDF containing the integrated
view of the four original datasets, now following the
same vocabulary and resources merged. This final
dataset is called Linked Data Mashup.
The resulting dataset was then hosted in the virtu-
oso triplestore
11
, which provides a SPARQL endpoint
capable of responding to SPARQL queries via HTTP.
This endpoint is accessed directly by the Medibot ap-
plication. Moreover, the dump of the final dataset rep-
resenting the LDM, in addition to the mapping files
and the OWL implementation of the ontology can be
accessed publicly via datahub
12
.
11
https://virtuoso.openlinksw.com/
12
https://datahub.io/linkeddatamashupeducacional/data-
med/v/2
4 MediBot
Although the ontology provides a layer of semantic
abstraction with terms closer to the user, there are
still problems in its access. To have access to the
data, before it is necessary to know about the ontol-
ogy’s schema and knowledge about Semantic Web
technologies, such as RDF, OWL and SPARQL. A
SPARQL query can be overly complicated, requir-
ing technical expertise on the part of the user which
would go against the purpose of this work which is to
universalize knowledge about drugs for users of dif-
ferent profiles. Therefore, in this work, a data access
interface was developed via natural language through
a chatbot, called MediBot.
MediBot is a chatbot for the instant messenger
Telegram, so that it can be used both via mobile appli-
cation and via the web interface on the PC. MediBot
was implemented in JavaScript using NodeJS. Cur-
rently, MediBot is able to answer questions in Por-
tuguese. MediBot can is contacted via Telegram by id
@websemantica_bot.
In subsection 4.1 the MediBot architecture is pre-
sented. While subsection 4.2 addresses the workflow
of the query process.
4.1 MediBot’s Architecture
Figure 4 shows MediBot’s architecture. Its architec-
ture is divided into three layers, user interface,server,
and linked data mashup.
The first layer, the user interface layer is the in-
teraction medium with the user, been represented by
Telegram instant messenger. The user interface can be
via mobile application in Android and IOS Telegram
app or via web browser in Telegram website. Besides
the mobile application and the website Telegram can
also be accessed via a desktop application on PC. The
reason for choosing the Telegram as a channel for the
chatbot is because the tool already has a large user
base and a robust infrastructure, besides having an
easy API for the creation and use of chatbots.
The second layer is the server layer. This layer
is responsible for the processing of requests and re-
sponses and is composed of two main components.
The first component of this layer is the application
server, which is responsible for receiving and pro-
cessing the user’s requests. The application server is
directly connected to the Telegram access API; this
component is the central module of the MediBot ar-
chitecture. The application server receives the user
request and builds the SPARQL query responsible
for retrieving the desired information. Also, the ap-
plication server is also responsible for sending the
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
30
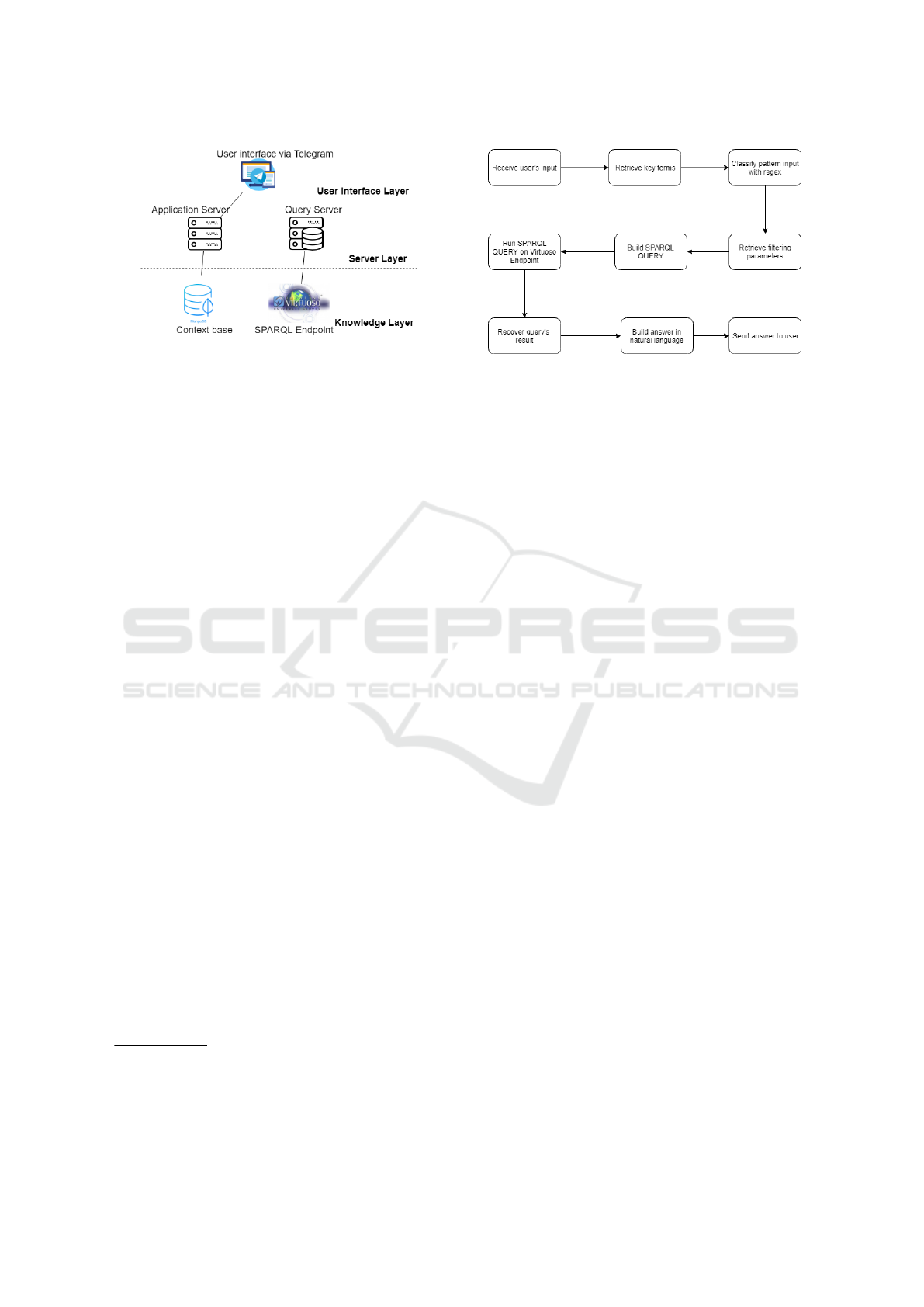
Figure 4: MediBot’s architecture.
SPARQL query to the query server and sending the
response message to the user when it is ready.
The query server is responsible for receiving the
SPARQL query and executing it on the SPARQL end-
point made available by the virtuoso. Besides, this
component is also responsible for obtaining the re-
sponse from the SPARQL query and constructing the
response message to the user. The reason for split-
ting the server layer into two is because of the poten-
tially long time to run the SPARQL query and gen-
erate the response message to the user. By dividing
the application server into two components, it will not
be locked during the processing of time-consuming
queries, which would prevent it from receiving new
requests and interactions that do not require queries.
The third and last layer is the Knowledge layer.
This layer is responsible for storing the knowledge
necessary for the chatbot to respond and interact with
the user. This layer is composed of two components:
• Context Base: It is responsible for storing rel-
evant context information during the interactive
mode, such as personal information about the user
such as name, surname and language, as well as
information about the conversation flow as cur-
rent interaction state, list of options presented, last
message, term and object selected for consulta-
tion. Context information is stored in an instance
of the non-relational object-oriented mongoDB
13
database, which allows fast storage and quick re-
trieval of data stored in JSON, allowing direct pro-
cessing by javascript code without the need for
pre-processing the data.
• Linked Data Mashup: its description is detailed
in chapter 3, loaded in the virtuoso triplestore that
provides a SPARQL endpoint for the realization
of queries.
13
https://www.mongodb.com/
Figure 5: MediBot’s workflow in quick response mode.
4.2 Query Workflow
MediBot has two modes of operation, the first being
the quick response mode and the second the interac-
tive one. In quick response mode, MediBot has a set
of SPARQL queries predefined and uses a simple reg-
ular expression evaluation approach to mapping the
entry in one those predefined queries. During the in-
put evaluation process, key terms and filtering param-
eters are retrieved. Key terms help classify into which
type of query the entry should be mapped to while
filtering parameters are used in FILTER clauses to re-
strict the query result to the specific intent of the user.
Finally, the SPARQL query is built and performed via
HTTP on the Virtuoso endpoint. Moreover, the build-
ing answer process also uses answers patters prede-
fined. Figure 5 shows the workflow performed by
MediBot.
Seven types of queries have been defined, which
are shown in Table 3. While Figure 6 presents an im-
age of MediBot responding to query 1 performed in
natural language in Portuguese through the Telegram.
In addition to the quick response mode, MediBot
has an interactive mode. While the former provides
quick and easy access to information, the latter pro-
vides a versatile form of access to the knowledge con-
tained in the sources. The main difference between
the two modes lies in the interactive and conversa-
tional character of the interactive mode, while the in-
teraction of the quick responses mode is summed up
to individual questions and answers, the interactive
mode performs information retrieval tasks in a con-
versational way where the context of the conversation
and previous interactions influences the results of fu-
ture interactions.
The interactive mode is oriented to finite tasks,
where the user starts one task at a time through the
sending of messages containing pre-established key-
words; moreover, a task remains active as long as
the user does not explicitly intend to finish it. How-
MediBot: An Ontology based Chatbot for Portuguese Speakers Drug’s Users
31
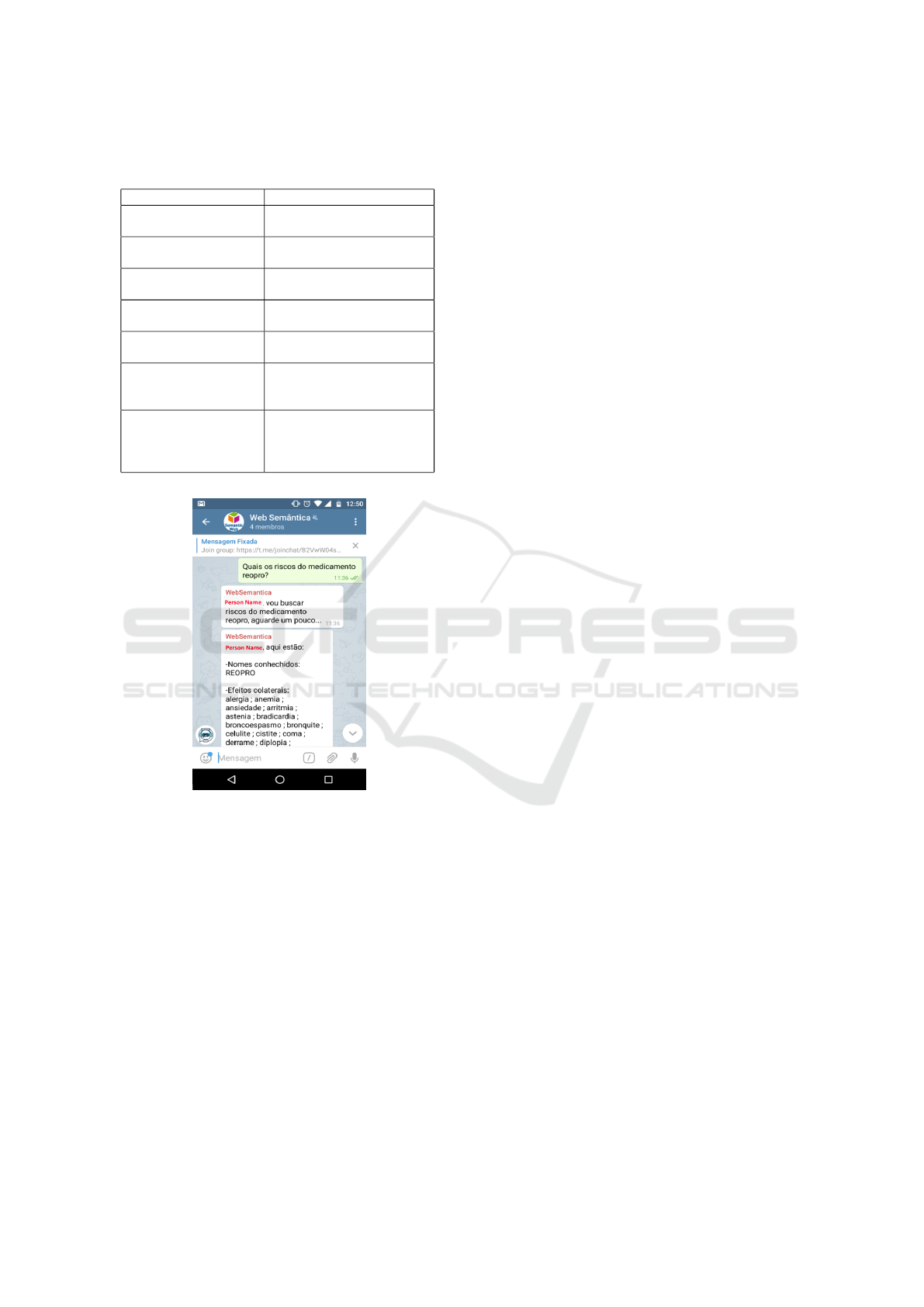
Table 3: Types of queries answered by MediBot in quick
response mode.
Type of query Example
Drugs with
a principle active
What are the drugs with
the substance dipyrone?
Definition
of terms in domain
Define therapeutic class
Informations about
certain drug
Talk about the drug aspirin
Indicate the risks
of a drug
What are the risks of the
drug reopro?
List of drugs’s
presentation
What are the presentations
of the drug reopro?
Information about
barcode presentation
Give information about bar
code presentation
7896382701801
Price of a presentation
with ICMS tax in
one State
What are the price with
ICMS tax of presentation
7896382701801
in the state of Ceará?
Figure 6: Example of Query in Portuguese on Telegram
about the Drug’s risks.
ever, during any point of an interactive mode task, the
user can perform a quick response mode query, mean-
while, the already started task remains in stand by to
be resumed any time the user wishes.
During interactive mode, two types of tasks can
be performed, browser and query, which will be de-
scribed later. However, the flow followed during the
conversation is not free, following a well-defined pat-
tern flow, where chatbot and user alternate their ques-
tions and answers. For decision of which next steps
to be followed during a specific task’s conversation,
the MediBot uses information about past interactions
during the same task (in this case called context), the
current point of the task (in this case called state) and,
in cases where the chatbot expects a response from
the user, the message received message (in this case
called input).
To enable continuous interaction between chatbot
and user the interactive mode was implemented fol-
lowing a variation of the pushdown automaton ap-
proach, where the current state of the conversation or
simply state is represented as a state of the automa-
ton, the context of the conversation is represented as
the auxiliary memory of the automaton, and the input
of the user is represented by the input signal of the
automaton. A remarkable aspect of the implementa-
tion of MediBot is the fact of the possibility of state
change without an explicit input sequence, this is due
to the fact that previous user responses may already
have the information necessary for the state change
without that there is a need for the user to reissue his
intention. Due to simplicity and space constraints in
this paper, the formal definition of the MediBot au-
tomaton will not be presented, however, in the figures
8 and 7 flowcharts are presented representing the tasks
performed in the interactive mode, being possible to
derive the automaton to from these flowcharts.
There are two types of possible tasks in interactive
mode, which are:
• Browser Task: This task allows the user to
navigate recursively on the existing terms in the
knowledge base, including the ontology schema
and its instances. When the user starts the task of
browsing over a term, MediBot presents the differ-
ent names, types and definitions of the term, be-
sides presenting its properties and allow the user
to select one of this to be the new pivot of the
task, being able to navigate on the concepts of the
sources. In figure 8 is presented the browser task’s
flow. In figure 10 is presented as an example of a
browser task’s interaction on Telegram.
• Query Task: This task allows the user to view
data about instances contained in the knowledge
base. Likewise the browser task, the query task is
also recursively interactive. During this task, the
user can ask to query the properties of a given in-
stance. Upon receiving a query from a user, Med-
iBot returns a list of instance’s properties, giv-
ing the user the option to select one of these to
view its values. If the selected property is a re-
lation (owl:ObjectProperty) MediBot displays the
list of instances as values for the property se-
lected, which can be selected as the new pivot
for the query task. If the selected property is a
simple attribute (owl:DatatypeProperty), it sim-
ply displays a list with the constant values for the
property. In figure 7 is presented the query task’s
flow. In figure 9 is presented as an example of a
query task’s interaction on Telegram.
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
32
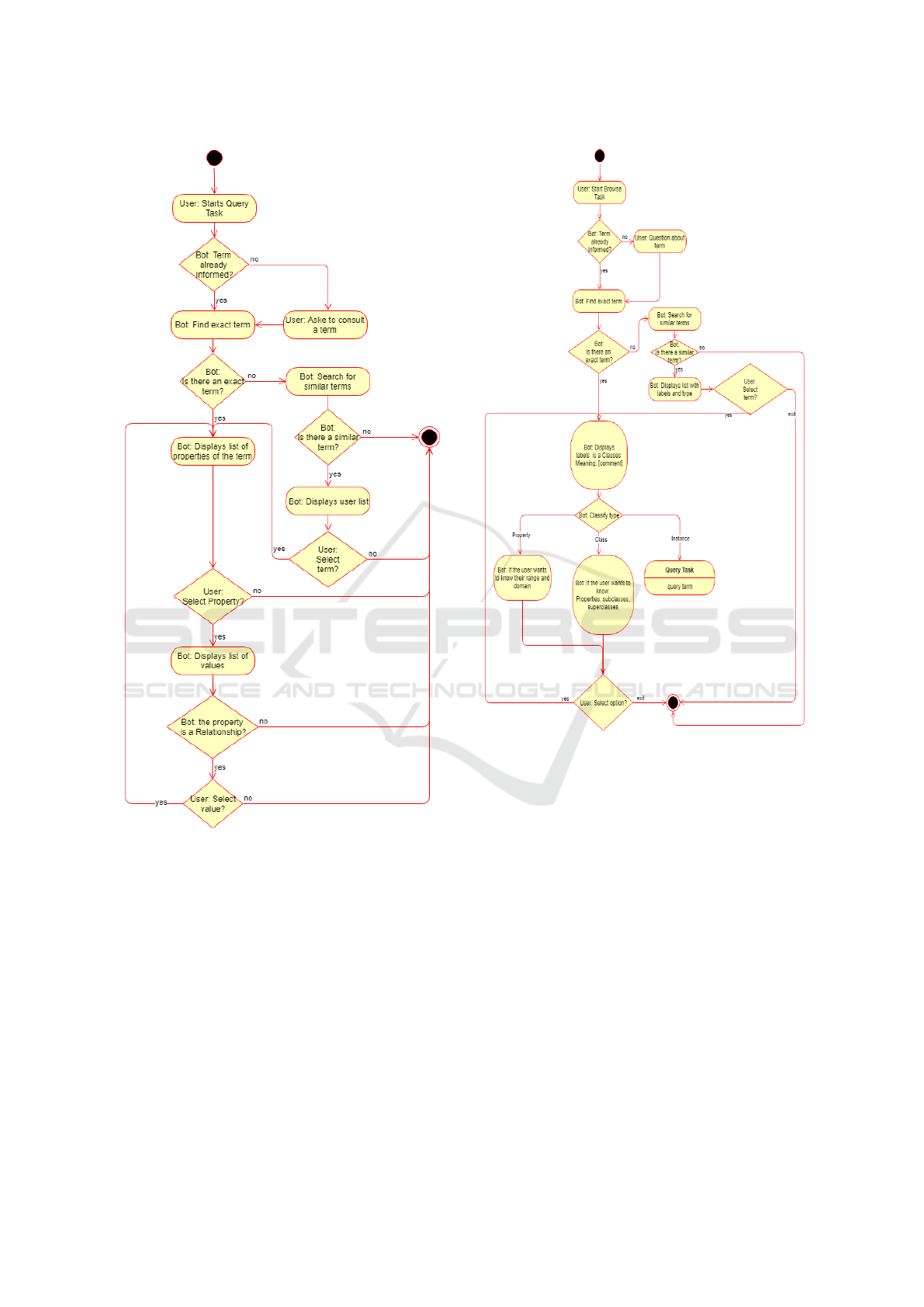
Figure 7: Query task’s flowchart.
It is interesting to note that some steps of the query
and browsers tasks are similar, such as terms list’s dis-
play, these steps were implemented as a single state in
the automaton, with the context being the only point
of differentiation them. Another important implemen-
tation element lies in context’s persistence since for
being an online application and multi-user isn’t feasi-
ble to keep the context in memory, so each time the
context is changed or necessary it has resorted to the
persistence mean MongoDB as described in subsec-
tion 4.1.
Figure 8: Browser task’s flowchart.
5 EVALUATION
In this work, we select the task-based evaluation
method described in (Konstantinova and Orasan,
2013) to measure the MediBot’s usability degree. In
this method are defined sets of tasks that are then re-
quested to be performed by volunteers.
To evaluate the practical usefulness of MediBot,
a set of 6 questions about information contained in
the sources was elaborated. The questions were asked
for not involved with the project volunteer users who
had to use the tool to respond to them. The ques-
tions can be divided into three levels of difficulty: (1)
Easy, can be answered with a single interaction with
the chatbot; (2) Medium, and can be answered with
at least two interactions; and (3) Difficult, requiring
more than two interactions with the chatbot.
The questions were asked through face-to-face in-
MediBot: An Ontology based Chatbot for Portuguese Speakers Drug’s Users
33

Figure 9: Example query task in telegram.
terviews with volunteers. Ten volunteers, four men
and six women participated in the study. The average
age of the participants was 31 years, having a min-
imum age of 22 years and a maximum of 63 years.
While none of them has technical knowledge about
the field of medicines. Also, two of the participants
had technical knowledge in information technology,
among them one knowing about ontologies.
As evaluation criteria, It was using three aspects.
The first criteria were the time needed to solve the
questions, the second was the rate of correctness, and
finally, the personal opinion of each evaluated that en-
tered as a subjective criterion. In table 4 the result of
the evaluation for each question and the final average
Figure 10: Example browser task in telegram.
is presented.
In the criterion of success rate dropouts were
counted as an error, however, the time of such cases
did not enter the meantime because it would cause
distortions.
It is noteworthy that only queries Q001 and Q002
were able to be performed without prior knowledge
about the ontology and the types of queries and their
flows, while the others needed queries on such infor-
mation. This fact was already expected, since Medi-
Bot has a limited set of pre-defined questions (which
includes queries Q001 and Q002), whereas query and
browser tasks require a correct starting point to be
useful.
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
34
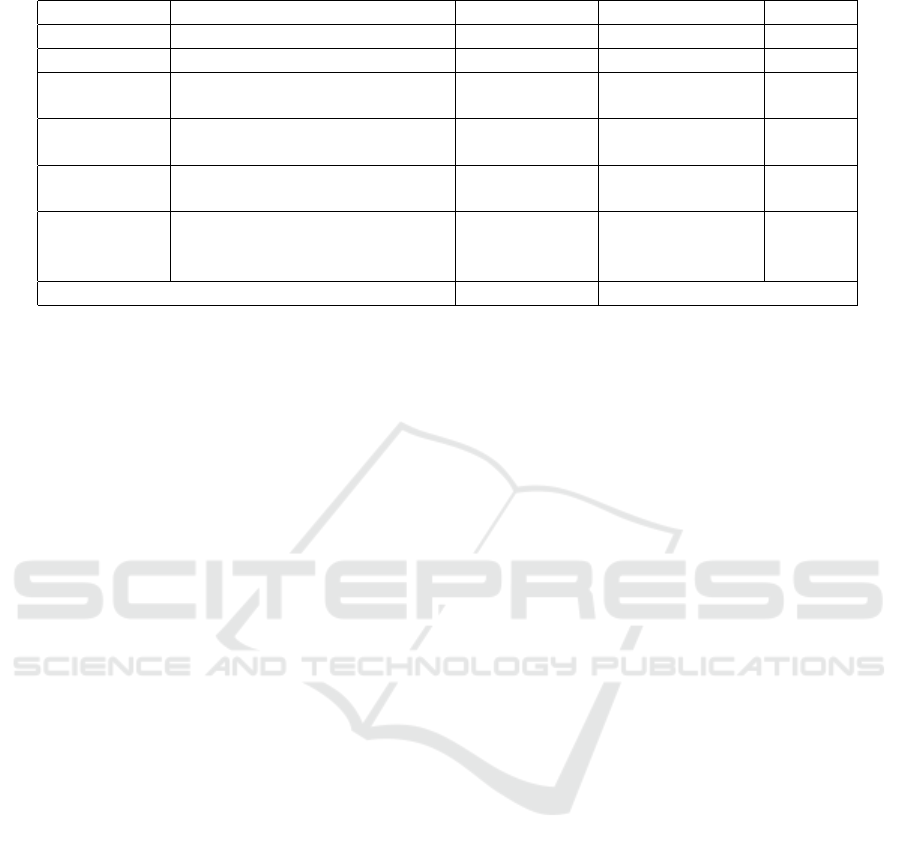
Table 4: Evaluation result.
ID_Question Question Mean Time(s) Success Rate(%) Difficult
Q001 What is a black box remedy? 30,53 100 1
Q002 What are the risks of Tylenol? 51,2 100 1
Q003
Which drugs have the same
active ingredient as Buscopan?
142,10 80 2
Q004
What is Buscopan’s maximum
price?
358,96 20 2
Q005
What is the relationship between
a substance and a presentation?
152,05 70 3
Q006
Which state has the lowest
maximum price for the
prescription drug orencia?
320,34 10 3
Mean 175,86 63,33
Another interesting point is that queries Q004 and
Q006 took considerably longer, in addition to having
a lower success rate. This fact can be explained be-
cause their answers are not represented in a factual
way at the base. In the case of Q004, there was a
need for a comparison operation, which in general
users tried to compare all existing prices, which re-
quired many interactions, leading to a high dropout
rate. There was the possibility to remove the number
of possibilities taking in characteristics of the presen-
tation, such as quantity, route of administration and
others. Already in the case of query Q006, there was
a need to have an understanding of how the state was
related to price, where once again users attempted
to make all comparisons. However, such a question
could be resolved only by looking at the lowest value
for taxes and which states adopted it.
During the reporting of opinions about the use of
the tool the main points presented were that ontol-
ogy’s image and examples of queries were very use-
ful. In addition to the preference for quick questions
about query and browser tasks.
6 CONCLUSION AND FUTURE
WORK
The main objective of this work is to universalize
access to information about drugs and their risks to
users of different profiles, principally Brazilian and
lay users in the domain.
To fulfill this goal, in this paper we presented
MediBot, an ontology-based chatbot capable of re-
sponding to requests in natural language in Por-
tuguese via the instant messenger Telegram. Medibot
provides a data access interface without the need for
the user to have prior knowledge about the structure
of the ontology or technical knowledge about Seman-
tic Web technologies.
Besides, in this work, we carried out the process
of semantic data integration in the domain of drug’s
data, where the selection of data related to Brazil was
preferred. Four separate datasets for integration were
selected: “Consumer Drug Prices” (CDP), “Govern-
ment Drug Prices” (GDP) and “Drug’s risks in preg-
nant and breastfeeding” (RPB) made available by the
National Agency Sanitary Surveillance (ANVISA in
Portuguese), and Sider available from the BIO2RDF
project.
Each of the datasets was originally in a different
physical format, such as spreadsheets, RDF or PDF.
In addition, each original dataset had a different vo-
cabulary. These facts show the justification of the use
of technologies of Linked Data and Semantic Web to
offer a layer of abstraction that allows transparent ac-
cess to the data in an integrated way and with a vo-
cabulary closer to that used by the user in their daily
lives.
During the usability evaluating the process of
MediBot, we realized that tasks that require compar-
ison operations, such as Q004 and Q006, present a
challenge for users because they require a great repet-
itive manual effort, so as future work we intend to im-
plement automated ways to perform such operations,
besides increasing the number of quick questions.
Still, as future work, we intend to develop an auto-
matic and incremental data update mechanism to en-
sure access to updated information efficiently. More-
over, we aim to integrate with a larger number of
datasets, such as global dataset such as drug bank
and registries from other countries, and use e-SUS
datasets with microdata on the use of drugs.
MediBot: An Ontology based Chatbot for Portuguese Speakers Drug’s Users
35

REFERENCES
Belleau, F., Nolin, M.-A., Tourigny, N., Rigault, P., and
Morissette, J. (2008). Bio2rdf: towards a mashup to
build bioinformatics knowledge systems. Journal of
biomedical informatics, 41(5):706–716.
Bizer, C., Heath, T., and Berners-Lee, T. (2011). Linked
data: The story so far. In Semantic services, inter-
operability and web applications: emerging concepts,
pages 205–227. IGI Global.
Bizer, C. and Seaborne, A. (2004). D2rq-treating non-
rdf databases as virtual rdf graphs. In Proceed-
ings of the 3rd international semantic web con-
ference (ISWC2004), volume 2004. Proceedings of
ISWC2004.
Corrêa, A. D., Caminha, J. d. R., Souza, C. A. M. d., and
Alves, L. A. (2013). Uma abordagem sobre o uso de
medicamentos nos livros didáticos de biologia como
estratégia de promoção de saúde. Ciência & Saúde
Coletiva, 18:3071–3081.
Hoang, H. H., Cung, T. N.-P., Truong, D. K., Hwang, D.,
and Jung, J. J. (2014). Retracted: Semantic informa-
tion integration with linked data mashups approaches.
International Journal of Distributed Sensor Networks,
10(4):813875.
Jovanovik (2017). Consolidating drug data on a global scale
using linked data. Journal of biomedical semantics,
8(1):3.
Kazi, H., Chowdhry, B., and Memon, Z. (2012). Med-
chatbot: An umls based chatbot for medical stu-
dents. International Journal of Computer Applica-
tions, 55(17).
Konstantinova, N. and Orasan, C. (2013). Interactive ques-
tion answering. In Emerging Applications of Natural
Language Processing: Concepts and New Research,
pages 149–169. IGI Global.
Mendes, P. N., Mühleisen, H., and Bizer, C. (2012). Sieve:
linked data quality assessment and fusion. In Proceed-
ings of the 2012 Joint EDBT/ICDT Workshops, pages
116–123. ACM.
Natsiavas, P., Koutkias, V., and Maglaveras, N. (2015). Ex-
ploring the capacity of open, linked data sources to as-
sess adverse drug reaction signals. In SWAT4LS, pages
224–226.
Natsiavas, P., Maglaveras, N., and Koutkias, V. (2017).
Evaluation of linked, open data sources for mining ad-
verse drug reaction signals. In International Confer-
ence on Internet Science, pages 310–328. Springer.
Nová
ˇ
cek, V., Vandenbussche, P.-Y., and Muñoz, E. (2017).
Using drug similarities for discovery of possible ad-
verse reactions. In AMIA Annual Symposium Proceed-
ings. AMIA.
Pinto, M. C. X., Ferré, F., and Pinheiro, M. L. P. (2012).
Potentially inappropriate medication use in a city of
southeast brazil. Brazilian Journal of Pharmaceutical
Sciences, 48(1):79–86.
Queneau, P., Bannwarth, B., Carpentier, F., Guliana, J.-M.,
Bouget, J., Trombert, B., Leverve, X., Lapostolle, F.,
Borron, S. W., and Adnet, F. (2007). Emergency de-
partment visits caused by adverse drug events. Drug
Safety, 30(1):81–88.
Schultz, A., Matteini, A., Isele, R., Mendes, P. N., Bizer,
C., and Becker, C. (2012). LDIF - A Framework for
Large-Scale Linked Data Integration. In 21st WWW,
Developers Track, page to appear.
Shadbolt, N., Berners-Lee, T., and Hall, W. (2006). The
semantic web revisited. IEEE intelligent systems,
21(3):96–101.
SINITOX (2016). Sistema Nacional de Informações Toxi-
cológicas registro de intoxicações no brasil. Accessed:
2018-09-17.
Sousa, H. W., Silva, J. L., and Neto, M. S. (2008). A im-
portância do profissional farmacêutico no combate à
automedicação no brasil. Revista eletrônica de far-
mácia, 5(1).
Vega-Gorgojo, G., Giese, M., Heggestøyl, S., Soylu, A.,
and Waaler, A. (2016). Pepesearch: Semantic data for
the masses. PloS one, 11(3):e0151573.
Volz, J., Bizer, C., Gaedke, M., and Kobilarov, G. (2009).
Silk-a link discovery framework for the web of data.
LDOW, 538.
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
36
