
Identification of Diseases in Corn Leaves using Convolutional Neural
Networks and Boosting
Prakruti Bhatt, Sanat Sarangi, Anshul Shivhare, Dineshkumar Singh and Srinivasu Pappula
TCS Research and Innovation, Mumbai, India
Keywords:
Disease Classification, Adaptive Boosting, Ensemble Classifier, CNN Features.
Abstract:
Precision farming technologies are essential for a steady supply of healthy food for the increasing population
around the globe. Pests and diseases remain a major threat and a large fraction of crops are lost each year
due to them. Automated detection of crop health from images helps in taking timely actions to increase yield
while helping reduce input cost. With an aim to detect crop diseases and pests with high confidence, we use
convolutional neural networks (CNN) and boosting techniques on Corn leaf images in different health states.
The queen of cereals, Corn, is a versatile crop that has adapted to various climatic conditions. It is one of the
major food crops in India along with wheat and rice. Considering that different diseases might have different
treatments, incorrect detection can lead to incorrect remedial measures. Although CNN based models have
been used for classification tasks, we aim to classify similar looking disease manifestations with a higher
accuracy compared to the one obtained by existing deep learning methods. We have evaluated ensembles
of CNN based image features, with a classifier and boosting in order to achieve plant disease classification.
Using an ensemble of Adaptive Boosting cascaded with a decision tree based classifier trained on features from
CNN, we have achieved an accuracy of 98% in classifying the Corn leaf images into four different categories
viz. Healthy, Common Rust, Late Blight and Leaf Spot. This is about 8% improvement in classification
performance when compared to CNN only.
1 INTRODUCTION
Convolutional Neural networks (CNN) based deep
learning methods are proving quite useful for im-
age classification tasks as they can learn the high
level features effectively. CNN’s have made tremen-
dous advances in computer vision tasks especially
in object classification (He et al., 2016; Chollet,
2016; Szegedy et al., 2016; Simonyan and Zisser-
man, 2014). Considering Large Scale Visual Recog-
nition Challenge (Russakovsky et al., 2015) based
on ImageNet dataset (Deng et al., 2009), the bench-
mark for error rates, CNN models have achieved the
lowest error rate of 3.57% (He et al., 2016) which
is comparable to human error rate. It is also ob-
served (Sharif Razavian et al., 2014) that extracting
features of a new dataset from a deep network pre-
trained on ImageNet database (Deng et al., 2009) and
training Support Vector Machine (SVM) (Cortes and
Vapnik, 1995) using these features performs better
classification than other complex supervised classi-
fication approaches. This motivates us to leverage
the high level features extracted from the trained con-
volutional neural networks which have been recently
indicated to be very robust (Yosinski et al., 2014).
Sharada Mohanty et. al in (Mohanty et al., 2016)
have performed supervised leaf disease classification
with 99.35% accuracy by fine tuning the top layer of
CNN models with a dataset taken in near ideal condi-
tions. Erika Fujita et. al in (Fujita et al., 2016) have
proposed a CNN based classifier trained on cucum-
ber viral diseases and achieved 82.3% average classi-
fication accuracy. We have explored the possibility of
using deep CNN model pre-trained on the ImageNet
database with 1000 classes of over 14 million im-
ages to extract the features of corn leaf images. Var-
ious methods are evaluated to develop a solution that
gives the most accurate recognition results especially
in similar looking disease manifestations.
We propose a system where the images are clas-
sified using features from a convolutional neural net-
work pretrained on ImageNet data and then boosting
is applied to accurately differentiate between similar
looking classes in accordance with the confusion ma-
trix. Classification performance of features from dif-
ferent CNN architectures viz. VGG-16, Inception-v2,
894
Bhatt, P., Sarangi, S., Shivhare, A., Singh, D. and Pappula, S.
Identification of Diseases in Corn Leaves using Convolutional Neural Networks and Boosting.
DOI: 10.5220/0007687608940899
In Proceedings of the 8th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2019), pages 894-899
ISBN: 978-989-758-351-3
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

ResNet-50 and MobileNet-v1 used with 3 different
classifiers viz. Softmax, Random Forest and SVM
have been presented in this paper. We achieved a rea-
sonably good performance by using Adaptive boost-
ing (AdaBoost) after getting class probabilities from a
decision tree based classifier. Features extracted from
a pre-trained Inception-v2 network used along with
this ensemble gave the highest accuracy. The ten-
sorflow (Abadi et al., 2015) implementation of CNN
models has been used to extract the feature vector
of the images. Scikit learn library (Pedregosa et al.,
2011) has been used for application of Random forest
and AdaBoost methods.
2 DATASET AND
PREPROCESSING
We have utilized corn leaf images from PlantVillage
dataset (Hughes and Salath
´
e, 2015) for 4 health con-
ditions, viz. Healthy, Common Rust, Late Blight and
Leaf Spot. 500 images have been randomly taken
from each class. Data augmentation with rotation,
flipping and addition of salt and pepper noise have
been done to avoid overfitting of the model and get
better accuracy on the images taken in different con-
ditions. This resulted in 2000 images of each class.
Augmentation helped increase the data quality as well
as quantity of images for training the classification
models. Figure 1 shows the four classes of the im-
ages we have considered from the database. Images
were resized according the the input size of the neu-
ral networks. We train and evaluate the models after
performing normalization on the image data. Nor-
malization of every image is performed for scaling
the data to an acceptable range for the network. Im-
age normalization results in contrast stretching, so it
also enhances the poor contrast images in the dataset.
Mean subtraction centers the data around zero mean
for each channel and normalization binds the range
of the image data values, thus helping the network
to learn faster since gradients act uniformly for each
channel as well as for all image data values. The im-
ages are resized according to the input size require-
ments of the CNN models. For VGG-16, MobileNet-
v1 and ResNet-50, images are resized to 224x224x3,
and for Inception-v2 they are resized to 299x299x3.
(a) (b) (c) (d)
Figure 1: Corn leaf Images from the PlantVillage database
(a) Common Rust (b) Healthy (c) Late Blight (d) Leaf Spot.
3 CNN FEATURES OF IMAGE
DATA
A CNN is made up of an arrangement of convolu-
tional layers that can be seen as a linear transforma-
tion over the image, followed by activation layer to
add non linearity in the network and then the pooling
layer to reduce the propagation of the redundancy in
the image in consecutive layers. A convolution layer
in CNN extracts features of an input image while
preserving spatial relation between pixels by using a
small matrix that strides over the input image. This
resulting image is called an Activation map or a Fea-
ture map. Rectified Linear Unit (ReLU), an element
wise activation function max(0,x) replaces all negative
pixel values in the feature map by zero. Activation
functions introduce non-linearity in the CNN as most
of real-world data that CNN would be used to learn
is non-linear. Spatial Pooling, i.e. downsampling is
applied on the feature map after ReLU to reduce the
dimensionality. This reduces the number of parame-
ters and computations in the network thereby reduc-
ing overfitting (Krizhevsky et al., 2012). It makes the
feature invariant to scaling and small distortions in the
input image. The last layer of a CNN is a Fully Con-
nected (FC) neural network layer. Adding FC helps
the network to learn the non-linear combination of
features computed from convolutional layers. The FC
layer is followed by an average or a max pooling layer
for a classification task.
We evaluate performance of features from VGG-
16 (Simonyan and Zisserman, 2014), Inception-
v2 (Szegedy et al., 2016), ResNet-50 (He et al., 2016)
and MobileNet-v1 (Howard et al., 2017) trained on
ImageNet database for classification of corn leaf
health state, as it has been seen that the models trained
on this vast database generalize well on other datasets
too after transfer learning (Zeiler and Fergus, 2014).
VGG-16 is a sequential CNN with 8 convolutional
layers having different number of filters with 3 × 3
receptive fields. Inception-v2 has blocks of multiple
filters that are applied on the same tensor and then
concatenated at the output of each block. It can be
termed as a CNN made up of small convolutional
Identification of Diseases in Corn Leaves using Convolutional Neural Networks and Boosting
895

Figure 2: Proposed ensemble method of classification.
modules. ResNet-50 is 50 layered deep CNN with
residual blocks which can be termed as shortcut con-
nections between the layers. These residual connec-
tions help in combating the problem of vanishing gra-
dient in case of networks with large number of lay-
ers, thus helping in better training of the network and
increasing the accuracy. In MobileNet, the normal
convolution is replaced by depthwise convolution fol-
lowed by pointwise convolution. This is called depth-
wise separable convolution and significantly reduces
the number of parameters compared to the normal
convolutions for a network with the same depth.
4 METHOD
We perform feature extraction by forward passing
an image through a trained convolutional neural net-
work. These features are then fed to the classification
module in order to accurately classify it into one of the
four classes of corn health condition. In case the con-
fidence level of the classification result i.e. the prob-
ability of predicted class is not satisfactorily high, the
boosting method is utilized in the cascade to confi-
dently predict the correct class. Figure 2 illustrates
the classification approach that we have used in or-
der to get maximum accuracy in predicting the cor-
rect health condition from the corn leaf image. If the
class label is denoted as {c
i
}
C
i=1
where C is the total
number of classes, the classification output would be
the C length array P of probabilities with which the
image belongs to each class. It can be denoted as P
= {p
i
}
C
i=1
for {p
i
= p(c
i
/ f
x
,W )}
C
i=1
where f
x
are the
features of the image, p is probability of each class
and W denotes the classifier parameters. As f
x
are
obtained using a neural network, considering all the
network layers as a non-liner transformation of im-
age pixels x, we can denote f
x
= W
n
x + b
n
where W
n
and b
n
represent CNN model parameters. Hence the
probability or the confidence of classification depends
on the CNN models for feature extraction as well as
classification.
4.1 Feature Extraction
Robust feature extraction is one of the most impor-
tant steps in order to achieve high classification ac-
curacy in crop images because there can be a lot of
variations within the images of single class. These
variations can be due to different severity levels of
diseases or pests, changes in light conditions, varia-
tions in size of the leaves and different growth stages
of the crops. Hence, we evaluate different types of
convolutional neural networks as feature extractors
and different classifiers to classifiy health condition of
corn leaves. Augmentation of image dataset is done
in order to incorporate the variations in the images
that would be captured in uncontrolled conditions.
The top layers of deep CNNs - VGG-16, Inception-
v2 and MobileNet-v1 pre-trained on ImageNet data
have been re-trained with images from each class cor-
responding to Healthy, Common Rust, Late Blight,
Leaf Spot conditions of corn leaves. As the con-
sidered CNN models have been trained on a large
and varied database, they are seen to generalize well
on other datasets too for classification using trans-
fer learning (Zeiler and Fergus, 2014)). This helps
us to utilize the optimal weights of deep architec-
tures learned through large visual data. While train-
ing these CNN models, the softmax layer in each of
these is replaced by a 4-neuron softmax layer. Then
the weights of lower layers are fixed and the top lay-
ers of the network are fine-tuned by the corn leaf im-
ages. In case of VGG-16, for example, output of the
pooling layer on top of the other networks is taken
as the feature vector because higher levels of network
learn generalized features. The topmost convolutional
block before the max pooling and the three FC lay-
ers in VGG-16 were retrained, and output of topmost
FC layer with 1000 neurons is taken as feature vec-
tor. For Inception-v2, ResNet-50 and MobileNet-v1,
the last convolutional block and the FC layer are re-
trained and output of average pooling layer before the
last FC layer is taken as the feature vector.
4.2 Classification
The standard classification method used with a CNN
is using a dense layer of Fully Connected (FC) neu-
rons and a softmax layer in order to get the probabil-
ity that the given image belongs to a particular class.
Adding FC layer helps the network to learn the non
linear combination of features computed from convo-
lutional layers followed by pooling for classification.
The softmax layer at output of FC layer ensures that
sum of output probabilities is 1. The softmax function
takes arbitrary-sized real-valued vector and outputs
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
896

(a) (b) (c) (d)
Figure 3: Confusion matrices for CNN based classifiers without boosting: (a) VGG-16 (b) Inception-v2 (c) ResNet-50 (d)
MobileNet-v1.
a probability vector of size [1x number of classes].
Number of neurons of softmax layer is equal to num-
ber of classes C. However, features from the trained
CNN can also be fed into other classification algo-
rithms like Support Vector Machine (SVM) (Hearst,
1998) or Random Forest (RF) (Breiman, 2001) in
place of using a softmax classifier. We evaluated three
methods for classification on the features extracted
from the neural network: Softmax, Random Forest,
and Support Vector Machine.
Random Forest classifier builds multiple decision
trees and merges them to get a more accurate and
stable prediction. While training, it sets a stopping
criteria for node splits resulting in utilization of the
entire feature space with a control in correlation be-
tween the trees. This helps in managing the trade-off
between bias and variance. While RF is based on de-
cision trees, SVM is a linear classifier, based on the
idea of getting a best hyperplane to divide the data in
two classes. The hyperplane with the greatest possible
margin between itself and any point within the train-
ing set is considered to be best as it has a higher proba-
bility of new data being classified correctly. Different
kernels like Radial Basis Function (RBF) can be used
to map a higher dimensional data in a space where
a linear separation is possible. The best performing
classifer is then followed by boosting for increasing
the overall accuracy when used as an ensemble.
4.3 Boosting
Boosting helps the base classifier to form a strong
rule for separation between the classes. We have used
Adaptive booosing (AdaBoost) (Freund and Schapire,
1997) on top of the base classification algorithm i.e.
softmax, RF or SVM in this case to increase the accu-
racy between the two weakly classified classes. Ad-
aboost is best used to boost the performance of de-
cision trees as it is a sequential ensemble that aims
to convert a set of weak classifiers or learners into a
strong one. Each learner is added sequentially while
training and trained using adaptively weighted train-
ing data. Every learner is assigned a weight and
a more accurate one is given a higher weight. It-
eratively, the learner(s) are added till the limit is
reached or the accuracy stops increasing. Initially,
equal weight is given to each image feature and if the
prediction is incorrect in the first stage then a higher
weight is given to such an image in the next iterations.
So the idea is that the weights of classifers as well as
the data points are set in such a way that the weightage
of the classifiers is more on the points that are difficult
to classifiy. If none of the output probability using the
base classifier (CNN model with RF) exceeds the con-
fidence level of 50%, i.e. if max(p) < 0.5, we use the
next level of adaptive boosting to increase the confi-
dence of classification and assign a probable health
condition to the input image.
5 RESULTS AND EVALUATION
For our experiments, we retrain the models using
transfer learning as mentioned in Sec. 3. The dataset
has been split into 3 sets viz. training, cross vali-
dation and test. Using test images which the neural
network has never seen, we get a more generalized
measure of classification accuracy whereas cross val-
idation data is used to tune the network parameters to
prevent over-fitting or bias while training. The CNN
models have been fine-tuned with the corn leaf im-
ages in a batch of 32 for every iteration using SGD
optimizer with the learning rate of 0.001 with Ne-
strov momentum. We evaluated these re-trained CNN
models for classification accuracy obtained on same
test image set for all 4 classes with corn leaf health
conditions. A total of 2000 images were taken from
PlantVillage dataset where 1600 were used for train-
ing and the rest for validation. The 244 test images
were taken randomly and not used for training and
validation of the classification models. The test data
Identification of Diseases in Corn Leaves using Convolutional Neural Networks and Boosting
897
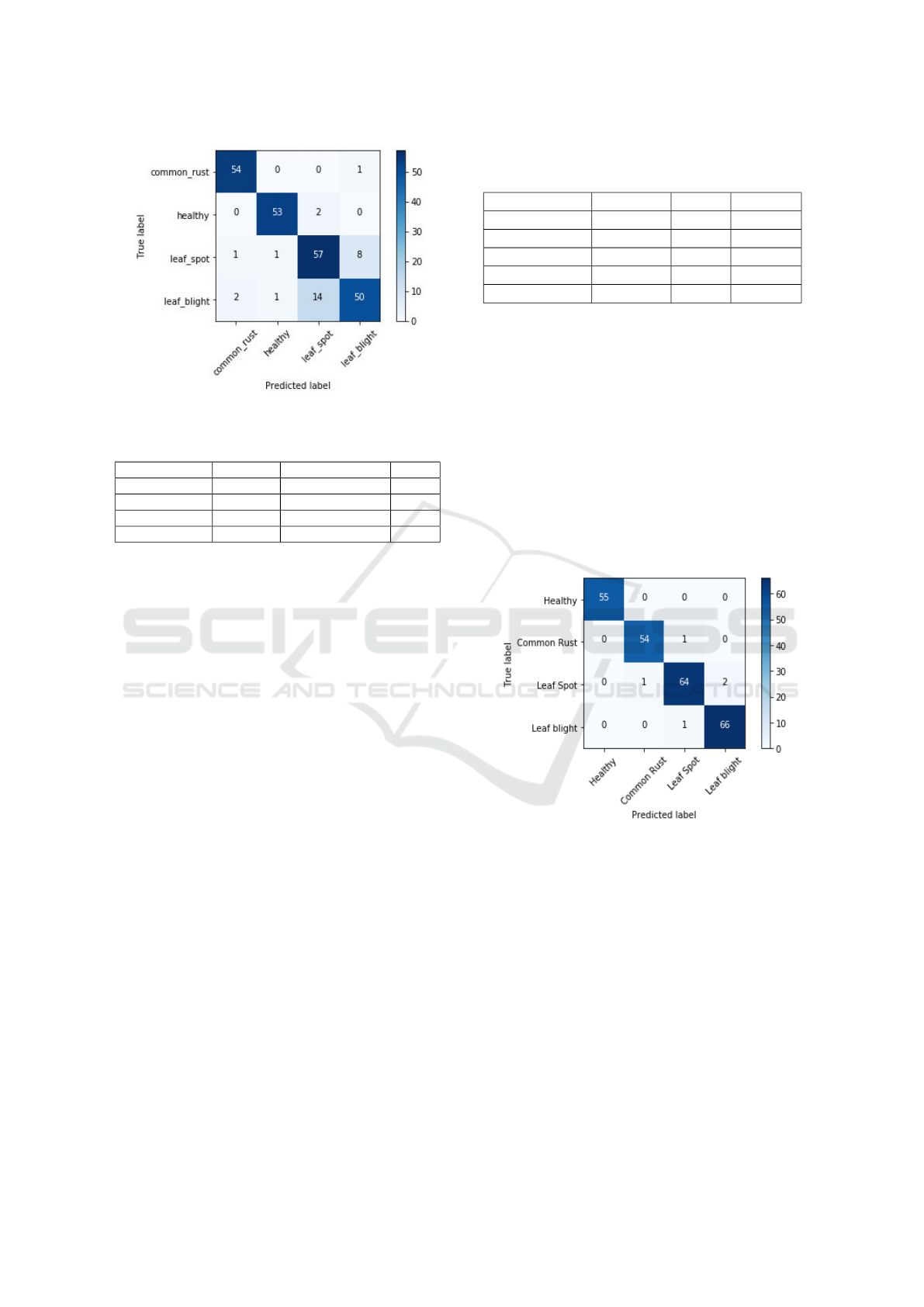
Figure 4: Confusion matrix: SVM on Inception-v2 features.
Table 1: Classification accuracy (%) for different methods.
Architecture Softmax Random Forest SVM
VGG-16 85 87 85
Inception-v2 86 90 87
ResNet-50 73 72 80
MobileNet-v1 80 82 82
had 55 images of Healthy and Common Rust condi-
tion each, 67 images affected by Late Blight and Leaf
Spot each. Equivalent scores for such test data also
show that the models do not suffer from bias or over-
fitting.
Apart from Softmax, we evaluated RF and SVM
that take features extracted from CNNs as input and
classify them into 4 classes. Table 1 shows the
average accuracy obtained on same test data when
we used VGG-16, Inception-v2, MobileNet-v1 and
ResNet-50 for classification using Softmax, RF and
SVM classifiers. It is seen that for images taken in
different conditions like size, resolution, angle and
brightness the solution that uses Inception-v2 for fea-
ture extraction and Random Forest with 100 decision
trees for classification gives the average classfication
accuracy of 90% which is maximum of all. Once
we observed that RF performs better than Softmax,
we also experimented to classifiy CNN features with
SVM classifier. We used RBF kernel SVM with pa-
rameters ‘C = 1.0’ and ‘gamma = 0.1’ values and
obtained about 87% accuracy with Inception-v2 fea-
tures, which is lower than that of RF. The confusion
matrix for SVM classifer over Inception-v2 features
for comparison is shown in Figure 4 while that for RF
over same features is shown in Figure 3(b). Hence
we selected a model based on features from Inception
and RF to develop an ensemble for classification.
It was observed from confusion matrix for every
classifier as seen in Figure 3 as well as Figure 4 that
for all of the classification methods, there is most
Table 2: Classification scores for corn crop health using Ad-
aboost.
Leaf state Precision Recall f1-score
Healthy 1 1 1
Common Rust 0.98 0.98 0.98
Leaf Spot 0.96 0.95 0.94
Late Blight 0.97 0.98 0.96
Total 0.97 0.98 0.97
confusion in differentiating between Late Blight and
Leaf Spot. We have evaluated accuracy over differ-
ent CNN architectures as well as classification meth-
ods and then selected the one with highest accuracy
to ensemble with AdaBoost. After adding Adaboost
on the next level after Inception-v2 features classified
by Random Forest, the accuracy increased to 98% be-
cause the classification accuracy between Leaf Spot
and Late Blight increased through boosting as seen in
Figure 5. We used adaptive boosting with 100 deci-
sion tree based estimators with learning rate of 1.0.
Table 2 shows the precision, recall and F1-score of
the proposed ensemble method.
Figure 5: Confusion matrix: Classification with the pro-
posed ensemble with CNN, RF and AdaBoost.
6 CONCLUSION AND FUTURE
WORK
Through the proposed system utilizing transferability
of CNN features along with boosting, we achieved
a test accuracy of 98% with classification score of
{precision, recall, f1-score} = {0.97, 0.98, 0.97} in
automated crop state diagnosis of corn leaves. Data
augmentation to increase the variety in the training
image set also helped in extraction of robust features,
thus resulting in better classification accuracy on dif-
ferent images. Hence, along with the basic classifica-
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
898

tion techniques, using the features from CNN trained
on augmented data, and ensembling with AdaBoost
on similar looking classes seems to be a promising
solution to automate the crop health diagnosis. This
would help farmers and agriculture experts to take
faster actions. Appropriate models can be selected
based on the accuracy and computational efficiency.
REFERENCES
Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z.,
Citro, C., Corrado, G. S., Davis, A., Dean, J., Devin,
M., Ghemawat, S., Goodfellow, I., Harp, A., Irving,
G., Isard, M., Jia, Y., Jozefowicz, R., Kaiser, L., Kud-
lur, M., Levenberg, J., Man
´
e, D., Monga, R., Moore,
S., Murray, D., Olah, C., Schuster, M., Shlens, J.,
Steiner, B., Sutskever, I., Talwar, K., Tucker, P., Van-
houcke, V., Vasudevan, V., Vi
´
egas, F., Vinyals, O.,
Warden, P., Wattenberg, M., Wicke, M., Yu, Y., and
Zheng, X. (2015). TensorFlow: Large-scale machine
learning on heterogeneous systems. Software avail-
able from tensorflow.org.
Breiman, L. (2001). Random forests. Mach. Learn.,
45(1):5–32.
Chollet, F. (2016). Xception: Deep Learning with
Depthwise Separable Convolutions. arXiv preprint
arXiv:1610.02357.
Cortes, C. and Vapnik, V. (1995). Support-vector networks.
Machine learning, 20(3):273–297.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei,
L. (2009). Imagenet: A large-scale hierarchical image
database. In IEEE Conference on Computer Vision
and Pattern Recognition (CVPR), pages 248–255.
Freund, Y. and Schapire, R. E. (1997). A decision-theoretic
generalization of on-line learning and an application
to boosting. Journal of Computer and System Sci-
ences, 55(1):119 – 139.
Fujita, E., Kawasaki, Y., Uga, H., Kagiwada, S., and Iy-
atomi, H. (2016). Basic investigation on a robust and
practical plant diagnostic system. In 15th IEEE Inter-
national Conference on Machine Learning and Appli-
cations (ICMLA), pages 989–992.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE Conference on Computer Vision and Pattern
Recognition, pages 770–778.
Hearst, M. A. (1998). Support vector machines. IEEE In-
telligent Systems, 13(4):18–28.
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D.,
Wang, W., Weyand, T., Andreetto, M., and Adam,
H. (2017). Mobilenets: Efficient convolutional neu-
ral networks for mobile vision applications. CoRR,
abs/1704.04861.
Hughes, D. P. and Salath
´
e, M. (2015). An open ac-
cess repository of images on plant health to en-
able the development of mobile disease diagnostics
through machine learning and crowdsourcing. CoRR,
abs/1511.08060.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). Im-
ageNet Classification with Deep Convolutional Neu-
ral Networks. In Advances in Neural Information Pro-
cessing Systems 25, pages 1097–1105.
Mohanty, S. P., Hughes, D. P., and Salath
´
e, M. (2016). Us-
ing Deep Learning for Image-Based Plant Disease De-
tection. Frontiers in Plant Science, 7:1419.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer,
P., Weiss, R., Dubourg, V., Vanderplas, J., Passos,
A., Cournapeau, D., Brucher, M., Perrot, M., and
Duchesnay, E. (2011). Scikit-learn: Machine learning
in Python. Journal of Machine Learning Research,
12:2825–2830.
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S.,
Ma, S., Huang, Z., Karpathy, A., Khosla, A., Bern-
stein, M., et al. (2015). Imagenet large scale visual
recognition challenge. International Journal of Com-
puter Vision, 115(3):211–252.
Sharif Razavian, A., Azizpour, H., Sullivan, J., and Carls-
son, S. (2014). Cnn features off-the-shelf: an as-
tounding baseline for recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition workshops, pages 806–813.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
arXiv preprint arXiv:1409.1556.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wo-
jna, Z. (2016). Rethinking the inception architecture
for computer vision. In Proceedings of the IEEE Con-
ference on Computer Vision and Pattern Recognition,
pages 2818–2826.
Yosinski, J., Clune, J., Bengio, Y., and Lipson, H. (2014).
How transferable are features in deep neural net-
works? In Advances in neural information processing
systems, pages 3320–3328.
Zeiler, M. D. and Fergus, R. (2014). Visualizing and under-
standing convolutional networks. In European confer-
ence on computer vision, pages 818–833. Springer.
Identification of Diseases in Corn Leaves using Convolutional Neural Networks and Boosting
899
