
GIANA Polyp Segmentation with Fully Convolutional Dilation
Neural Networks
Yun Bo Guo and Bogdan J. Matuszewski
a
Computer Vision and Machine Learning (CVML), Research Group, School of Engineering,
University of Central Lancashire, Preston, U.K.
Keywords: Fully Convolutional Neural Networks, Dilation Convolution, Polyp Segmentation, Video Colonoscopy,
Segmentation Quality.
Abstract: Polyp detection and segmentation in colonoscopy images plays an important role in early detection of
colorectal cancer. The paper describes methodology adopted for the EndoVisSub2017/2018 Gastrointestinal
Image ANAlysis – (GIANA) polyp segmentation sub-challenges. The developed segmentation algorithms are
based on the fully convolutional neural network (FCNN) model. Two novel variants of the FCNN have been
investigated, implemented and evaluated. The first one, combines the deep residual network and the dilation
kernel layers within the fully convolutional network framework. The second proposed architecture is based
on the U-net network augmented by the dilation kernels and “squeeze and extraction” units. The proposed
architectures have been evaluated against the well-known FCN8 model. The paper describes the adopted
evaluation metrics and presents the results on the GIANA dataset. The proposed methods produced
competitive results, securing the first place for the SD and HD image segmentation tasks at the 2017 GIANA
challenge and the second place for the SD images at the 2018 GIANA challenge.
1 INTRODUCTION
Colorectal cancer is one of the leading causes of
cancer deaths worldwide. Often, it arises from benign
polyps which with time become malignant. To
decrease mortality, an early detection and assessment
of polyps is essential. For an initial evaluation, an
image of a segmented polyp could provide important
evidence to describe polyp characteristics. In the
current routine clinical practice, polyps are detected
and delineated in colonoscopy images manually by
highly trained clinicians. To automate these
processes, machine learning and computer vision
techniques have been considered to improve polyps’
detectability and segmentation objectivity (Bernal et
al., 2015).
An automatic polyp segmentation is a very
challenging task. This is because polyps’ appearance,
shape and size are highly variable (see Figure 1). In
the early stages, a colorectal polyp is small and could
have no obvious differentiating texture appearance,
and therefore could be easily confused with other
intestinal tissue. In the later stages polyps
a
https://orcid.org/0000-0001-7195-2509
progressively change, often significantly increasing
in size and could develop more distinctive texture and
colour patterns. Some of the polyps grow so large,
that they will take most of the camera field of view,
possibly not fitting entirely into the image frame.
Additionally, illumination used in the colon screening
can cause image artefacts, with pattern of shadows,
highlights and occlusions, making the segmentation
task even harder. A single polyp could look
significantly different depending on the camera
position. Furthermore, for some polyp types there is
no apparent boundary between the polyp and the
surrounding tissue. As in most cases of manual
delineation, polyp segmentation is affected by the
lab’s guidelines and experience of the clinician. It is
therefore hard to determine the gold standard for the
automatic segmentation procedures dealing with all
possible types of polyps.
This paper proposes novel fully convolutional
neural networks to accomplish this challenging
segmentation task. The FCNN methods that were
developed produce the polyp occurrence confidence
map (POCM). The polyp position in the image frame
632
Guo, Y. and Matuszewski, B.
GIANA Polyp Segmentation with Fully Convolutional Dilation Neural Networks.
DOI: 10.5220/0007698806320641
In Proceedings of the 14th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2019), pages 632-641
ISBN: 978-989-758-354-4
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
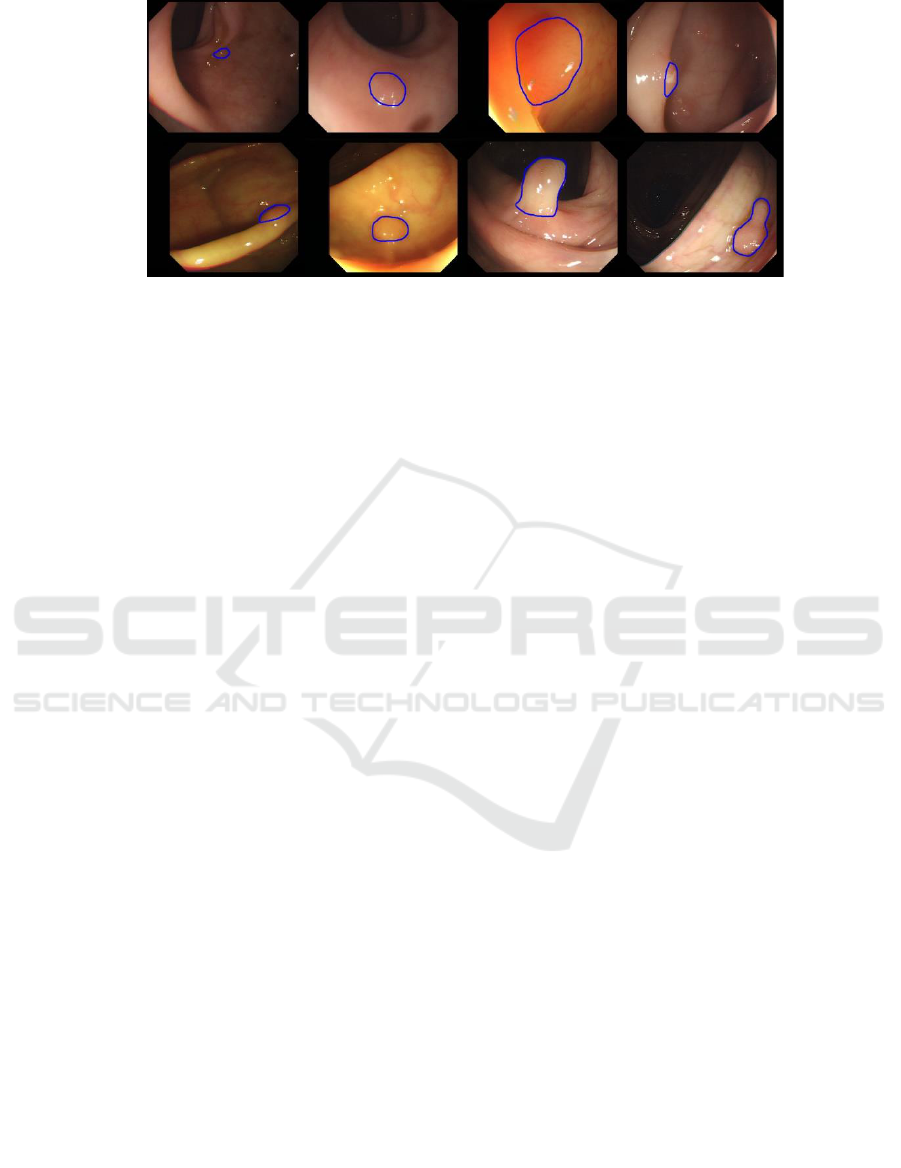
Figure 1: Examples, from the GIANA SD training dataset, showing polyps with different size, position, shape and colour.
The blue contour is the ground truth marked by clinicians.
is indicated by higher values of the POCM. In the
post-processing, the final polyp delineation is either
obtained by simple thresholding or the hybrid-level
set (Zhang et al. 2008, 2009) is used on the POCM to
smooth the polyp contour and eliminate small noisy
network responses.
2 RELATED WORK
Most of the existing polyp segmentation methods can
be divided into two main approaches based either on
polyp apparent edge or texture. Due to the fact that in
many cases, polyps have well-defined shapes, some
of the early approaches attempted to fit predefined
polyp shape models. Hwang et al. (2007) used ellipse
fitting techniques based on image curvature, edge
distance and intensity values. Gross et al. (2009) used
the Canny edge detector to process prior-filtered
images, identifying the relevant edges using a
template matching technique. Breier et al. (2011a,
2011b) investigated applications of active contours
for the polyp segmentation. Although these methods
perform well for typical polyps, they require manual
contour initialisation.
The above mentioned techniques rely heavily on a
presence of complete polyp contours. To improve the
robustness, further research was focused on the
development of robust edge detectors. Bernal et al.
(2012) presented a “depth of valley” concept to detect
more general polyp shapes, then segment the polyp
through evaluating the relationship between the
pixels and detected contour. Further improvements of
this technique are described in (Bernal et al., 2013)
and (Bernal et al., 2015). In the subsequent work,
Tajbakhsh et al. (2013) put forward a series of polyp
segmentation method based on edge classification,
utilising the random forest classifier and Haar
descriptor features. In the follow-up work (Tajbakhsh
et al. 2014a, 2014b) segmentation was refined via use
of several sub-classifiers.
Another class of polyp segmentation methods is
based on texture descriptors, typically operating on a
sliding window. Karkanis et al. (2003) combined
Grey-Level Co-occurrence Matrix (GLCM) and
wavelet. Using the same database and classifier,
(Iakovidis et al., 2005) proposed a method which
provided the best results in terms of area under the
curve (AUC) metric. Local Binary Pattern and the
original GLCMs methods are also tested in
(Alexandre et al. 2008), however, because of a
different dataset, and values of the design parameters,
the results cannot be directly compared. More
recently, with advances in deep learning, hand-
crafted feature descriptors are gradually being
replaced by convolutional neural networks (CNN)
(LeCun et al. 1998) and (Krizhevsky et al. 2012).
Park et al. (2015) formulated a pyramid CNN to
learn the scale-invariant polyps’ features. The
features are extracted from the same patch at three
different scales through three CNN paths. Ribeiro et
al. (2016) evaluated CNN comparing it with other
state-of-art hand-crafted features used for polyp
classification, and found that CNN has superior
performance. CNN is not only used for recognition
but also for feature extraction. R. Zhang et al. (2017)
designed a transfer learning scheme. They used a pre-
trained CNN to extract low-level polyp features and
SVM for classification. It illustrates that CNN can
learn informative and robust low-level features.
However, the general problem with the sliding
window approach is that it is harder to use image
contextual information and it is inefficient in the
prediction mode (i.e. segmentation of the test
images). This problem has been addressed by the so
called fully convolutional networks (FCN), with the
GIANA Polyp Segmentation with Fully Convolutional Dilation Neural Networks
633

Figure 2: The proposed Dilated ResFCN polyp segmentation network. Frome left to right, Blue: Feature extraction part;
Yellow: Dilation convolution; Green: Skip connection.
two key architectures (Long et al. 2015) and
(Ronneberger et al., 2015). These methods can be
trained end-to-end and output complete segmentation
results, without a need for any post-processing.
Vázquez et al. (2017) and Akbari et al. (2018) directly
segmented the polyp image by standard FCN.
L. Zhang et al. (2017) use the same FCN, but they add
a random forest to decrease the false positive. The U-
net (Ronneberger et al., 2015) is one of the most
popular architectures for biomedical image
segmentation. It has been also used for polyp
segmentation. Li et al. (2017) designed a U-net
architecture for polyp segmentation with smooth
contours.
In recent years, it has been noticed that there is a
close relationship between receptive fields and
segmentation results of convolutional networks. As
for generic image segmentation, a new layer called
dilation convolution has been proposed (Yu et al.
2015) to control the CNN receptive field in a more
flexible way. Chen et al. (2018) also utilised dilation
convolution and developed further network changes
called atrous spatial pyramid pooling (ASPP) to learn
the multi-scale features. The ASPP module consists
of four parallel convolutional layers with different
dilations.
In summary, polyp segmentation is becoming more
and more automated and integrated. Deep feature
learning and end-to-end architectures are gradually
replacing the hand-crafted features operating on a
sliding window. Polyp segmentation can be seen as a
semantic instance segmentation problem and
therefore, a large number of techniques developed in
computer vision for generic semantic segmentation
could possibly be adopted, providing effective and
more accurate methods for polyp segmentation.
3 METHOD
3.1 Pre-processing
The first step in the proposed processing pipeline is
the removal of black borders in the images. The
border pixels have small random intensity variations,
and therefore CNN could be “distracted” and learn
unnecessary image border patterns. It has been found
that the border pixels obtained from the same video
sequence have always the same value. After video
sequence detection, images from the same video are
stacked and the border can be easily located via
analysis of local variance. To save the memory and
training computational load, all the input images are
re-scaled to 250x287x3 in size.
GIANA 2019 - Special Session on GastroIntestinal Image Analysis
634

3.2 Dilated ResFCN
The first proposed network architecture, Dilated
ResFCN, is shown in Figure 2. It is derived from the
architecture proposed by (Peng et al., 2017). The
proposed network consists of three sub-structures
preforming different tasks, these are: feature
extraction layers, multi-resolution classification
layers and the deconvolution layers. The feature
extraction part of the network is based on the
previously proposed ResNet50 model (He et al. 2016).
It can be divided into five sub components. Res1 –
Res5. The Res1 represents the first convolutional and
pooling layers. Res2 – Res5 represents the sub-
networks having respectively 9, 12, 18, 9
convolutional layers with 256, 512, 1024, 2048
feature maps. Each of these sub-networks operates on
the gradually spatially reduced feature maps, down-
sampled with a stride of 2 when moving from sub-
network Resi to the sub-network Resi+1, the size of
corresponding feature map is 62*72, 31*36, 16*18,
8*9. Excluding the regular connection, the outputs
from the Res2 to Res5 is being directed to parallel
classification paths consisting of a dilation
convolutional layer, 1x1 convolutional layer, dropout
layer and final 1x1 convolutional layer with two
outputs corresponding to the polyp and background
confidence maps. There are four such parallel paths
fed from the outputs of Res2- Res5, with each path
using different dilations. The outputs of these four
paths are subsequently combined by skip connection
which includes the deconvolution layers and fusion
layer.
In the proposed Dilated ResFCN network, the
deconvolution layers perform bilinear interpolation
without training. The initial weights of the proposed
architecture have two sources: The feature extraction
part is initialized by a publicly available ResNet-50
model, which was trained on the ImageNet. The
convolutional layers in the four parallel paths are
initialized by the Xavier method (Glorot and Bengio,
2010). The network is trained with softmax cross-
entropy loss using Adam optimizer.
3.3 SE-Unet
The second proposed network, SE-Unet, is shown in
Figure 3. It is design to segment polyps which have
been missed by the ResFCN as it more “sensitive” in
some cases than ResFCN, however overall tends to
produce more false positive pixels. This method is
inspired by the U-net and SE-net (Hu et al., 2017).
The whole network can be divided into four parts,
consisting of feature learning, up-sampling, Atrous
spatial pyramid pooling (ASPP) and SE-modules.
The VGG16 network is used as an encoder with the
decoder being a mirrored VGG16. The resolution of
the last encoder layer is 16×18. The ASPP is used to
learn the multi-scale high-level features, it consists of
Figure 3: The structure of the proposed SE-Unet architecture.
GIANA Polyp Segmentation with Fully Convolutional Dilation Neural Networks
635
1×1 kernel, 3×3 kernel, and two dilation kernels with
dilation rates 2 and 4. Each component of ASPP
outputs 256 feature maps, so the total number of
feature maps is 1024.
Pixels at the same position are fused by a 1×1×256
kernel. The SE-module is added behind each
concatenation layer in the up-sampling module. For
each feature map in the concatenation layer it assigns
a coefficient between zero and one. Large coefficients
indicate that the corresponding features have more
significance.
The up-sampling layers implement bilinear
interpolation and the initial weights are selected using
the Xavier method. The network is trained with the
sigmoid cross-entropy loss using Adam optimizer.
4 IMPLEMENTATION
4.1 Dataset
The proposed polyp segmentation methods are
developed and evaluated on the database from the
EndoVisSub2017 GIANA Polyp Segmentation
Challenge. The training database, with the ground
truth segmented polyps, has two subsets: (i) SD
(CVC-ColonDB), consisting of 300 low resolution
500-by-574 pixels polyp images, and (ii) HD, with 56
high resolution 1080-by-1920 pixels images. The test
database has 612 SD (CVC-ClinicDB) images with
reduced 384-by-288 resolution and 108 full
resolution HD images. For selection of the methods’
design parameters, 4-fold tests have been performed
on the training data. The SD subset consist of the
images extracted from a few video sequences, with
images from the same sequence being highly
correlated (i.e. showing the same polyp). Therefore,
when constructing the validation data folds, care was
taken not to include any images from the same video
simultaneously in the training and test subsets for any
of the folds. This paper only reports the results
obtained for the SD images.
4.2 Data Augmentation
Data augmentation is a standard technique, used to
enlarge training data sets. It is frequently used,
particularly in cases when the available dataset is
relatively small. More recently, it has been reported
that data augmentation can play an important role in
controlling the generalisation properties of deep
networks, e.g. Hernández and König (2018) has
experimentally demonstrated that the augmentation
alone could provide better results on test data, than in
combination with the weight decay and dropout.
Whereas the training data augmentation is now
commonly accepted methodology, data augmentation
during the test time is not yet extensively used.
However it is gradually growing in popularity. It is
anticipated that it can further improve generalisation
properties of the deep architectures.
4.2.1 Training Data Augmentation
From a perspective of a typical training set used in a
context of the deep learning, the training data
available for the polyp segmentation (see section 4.1)
is rather small. Therefore, available data were heavily
augmented with random rotation, translation, scale
changes as well as colour and contrast jitter. In total,
after augmentation, the training data include more
than 90,000 images. Based on ablation tests with the
FCN8 and Dilated ResFCN networks, it has been
concluded that rotation and colour jitter have the most
significant effect on improvement of the
segmentation performance. Although intuitively not
necessary obvious, the colour jitter plays an important
role. This can be explained by the fact that the
network is trained on the data from a small number of
subject and the original images don’t reflect all
possible variations of tissue pigmentation, vascularity
or indeed instrument setup, including illumination
and camera parameters. Results of comprehensive
ablation tests are to be reported in a separate
publication. A sample of the augmented images using
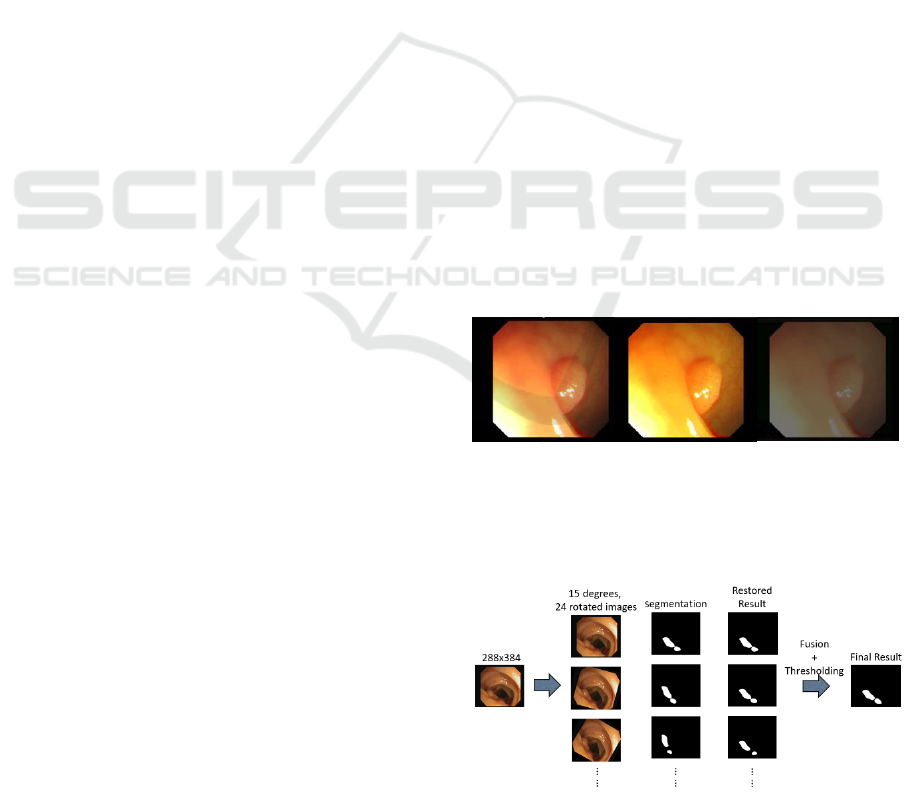
colour and contrast jitter is shown in figure 4.
Figure 4: A sample of the augmented images using the
colour and contrast jitter. From left: original, colour jitter
and contrast jitter images.
4.2.2 Test-time Data Augmentation
Figure 5: Test-time rotation based data augmentation.
GIANA 2019 - Special Session on GastroIntestinal Image Analysis
636
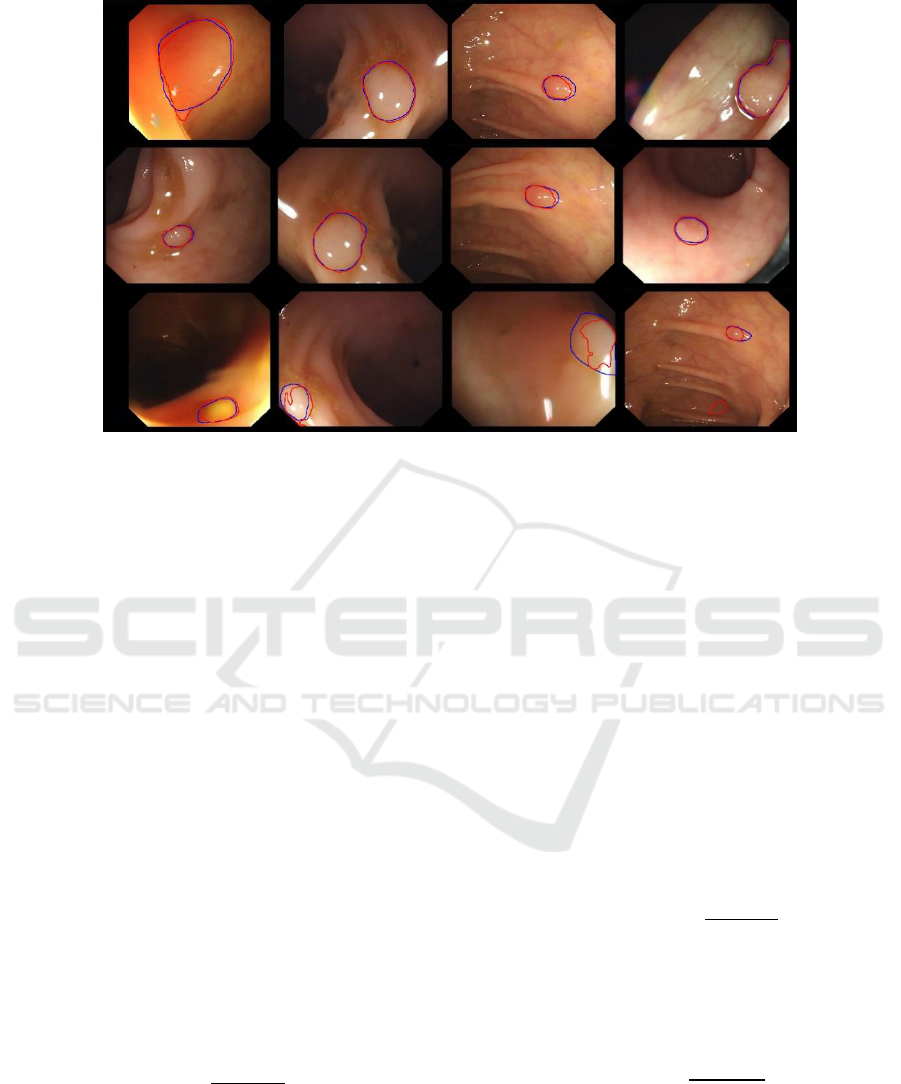
Figure 6: Typical results, with red and blue contours representing respectively segmentation results and the ground truth. Top:
results obtained using the Dilated ResFCN network. Middle: results obtained using SE-Unet. Bottom: The segmented polyps
using SE-Unet, which were not detected using the Dilated ResFCN.
Since the implemented CNNs don’t have built in
rotation invariance, one possible way to further
improve the accuracy of the segmentation is to
perform the rotation data augmentation during the test
time. For this purpose rotated versions of the original
test image are presented to the network and the
corresponding outputs are averaged to better utilise
generalisation capabilities of the network. The whole
process is explained in figure 5. The test-time data
augmentation implemented for the Dilated ResFCN
uses 24 rotated images.
4.3 Evaluation Measures
4.3.1 Dice Index
For a single segmented polyp the Dice coefficient
(also known as F1 score) is used as the base
evaluation metric. It was also adopted as a metric by
the GIANA challenge. This metric is used to compare
the similarity between the binary segmentation results
and the ground truth. It is calculated as follows:
where: S represents the result of the segmentation, G
represents the corresponding segmentation ground
truth and |A| represent number of pixels in object A.
As for the overall results obtained on the all test
images, the mean and the standard deviation of the
Dice coefficients calculated for each image are used.
Jaccard similarity index, also known as Intersection
over Union, is another popular similarity metric often
used in literature. However, as the Dice coefficient
and Jaccard index have monotonic relation, only Dice
coefficient results are reported in this paper.
4.3.2 Precision and Recall
Precision and recall are standard measures used in a
context of binary classification. For image
segmentation, precision is calculated as the ratio
between the number of correctly segmented pixels
and the number of all segmented pixels:
Recall is calculated as a ratio between the number of
the correctly segmented pixels and the number of
pixels in the ground truth:
In the context of image segmentation precision and
recall could be used as indicators of over- and under-
segmentation.
GIANA Polyp Segmentation with Fully Convolutional Dilation Neural Networks
637

Figure 7: The typical segmentation results of a hybrid method for images form the test set.
4.3.3 Hausdorff Distance
In this work the Hausdorff distance is used to evaluate
how closely the contour of the segmented polyp
matches the shape of the corresponding ground truth.
The Hausdorff distance is a common metric used to
measure the similarity between contours of two
objects. It is defined as:
where: d(x, y) denotes the distance between points
x ∈ G and y ∈ S. The smaller the value of the
Hausdorff distance the better the two contours match,
with the 0 indicating the perfect overlap between
contours. It should be noted that the Hausdorff
distance is complementary to the Dice coefficient as
these metrics measure different properties of the
segmented objects. It is quite possible to have
segmentation results with the Dice coefficient close
to 1 (with 1 indicating the perfect match) and the
Hausdorff distance having a large value, indicating a
poor contour match.
5 RESULTS
5.1 Validation Results
As part of selection of the design parameters the
developed methods were tested on the 4-fold
validation data (see section 4.1). A number of
parameters have been tested, including parameters of
the backpropagation training algorithm (e.g. learning
rate, momentum, number of epochs, etc.) or post
processing such as polyp occurrence confidence map
(POCM) threshold. This section shows only the
results used to select which output of the Dilated
ResFCN network should be used. The results from the
proposed networks are also compared against the
well-known FCN8 network (Long et al. 2015) and the
hybrid method. The hybrid method uses the Dilated
ResFCN as the base segmentation method and
switches to the SE-Unet when the base network does
not detect any polyp.
Table 1: Mean values obtained for different metrics on
4-fold validation data using FCN8s, Dilated ResFCN
(DPFCN), SE-Unet and hybrid segmentation.
Dice
Precision
Recall
Hausdorff
FCN8s
Foreground
0.6321
0.6922
0.6497
271
FCN8s
Background
0.6682
0.6767
0.6524
193
DRFCN
Foreground
0.7789
0.8038
0.8099
56
DRFCN
Background
0.7860
0.8136
0.8060
54
SE-Unet
0.6969
0.7477
0.7138
109
Hybrid
0.8014
0.8349
0.8210
62
GIANA 2019 - Special Session on GastroIntestinal Image Analysis
638
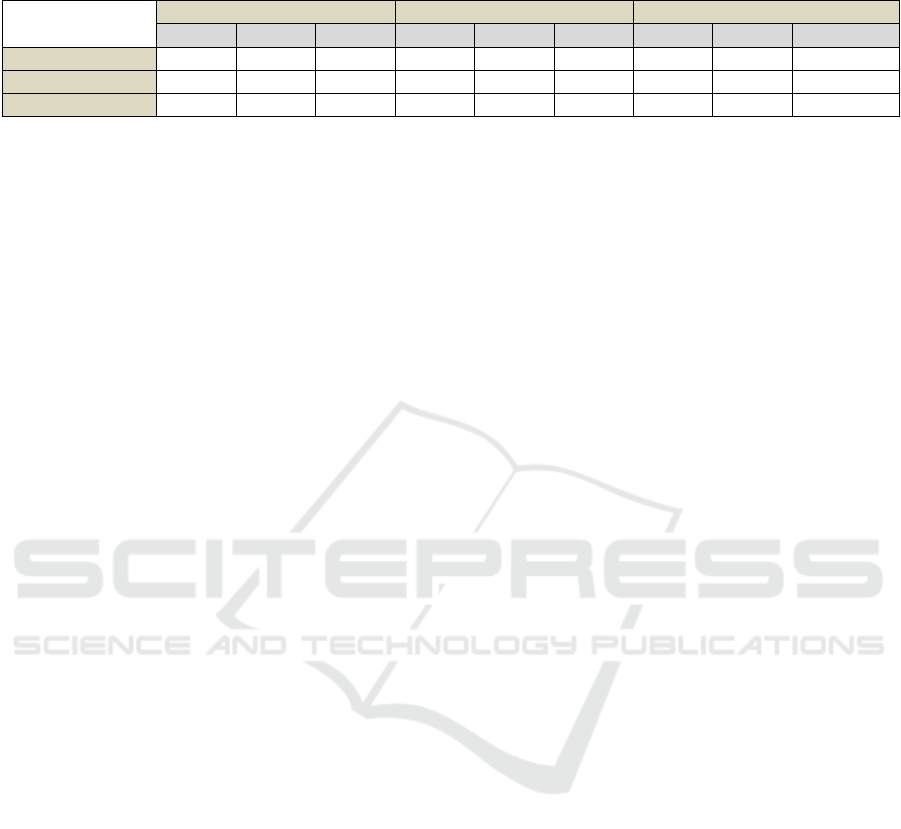
Table 2: Statistics of the Dice coefficient obtained on the test data.
Method
Foreground
Background
Rotation test data augmentation
Mean
Std
Missing
Mean
Std
Missing
Mean
Std
Missing
Dilated ResFCN
0.7717
0.2394
17
0.8126
0.2043
9
0.8293
0.1956
9
SE-Usnet
0.8019
0.2240
14
N/A
N/A
N/A
0.8102
0.2207
13
Hybrid
0.7825
0.2204
6
0.8169
0.1904
6
0.8343
0.1837
3
As it can be seen from the Table 1 the overall best
results are provided by the hybrid method followed by
the Dilated ResFCN. Both proposed methods
outperform the FCN8 segmentation network. A
selection of typical results obtained on the validation
data with different types of polyps is shown in Figure 6.
This figure also demonstrates typical differences in the
segmentation results generated by the two proposed
methods.
5.2 Test Data Results
Table 2 shows the Dice coefficient’s mean and
standard deviation as well as the number of missed
polyps obtained on the test dataset. The results for the
both proposed methods, and the hybrid method (see
section 4.4) are shown. With the Dilated ResFCN used,
the DICE segmentation statistics are reported when the
foreground or the background network outputs are
used. The results obtained with the test-time data
augmentation are also reported. It can be seen that the
best results, with biggest mean Dice coefficient,
smallest Dice standard deviation and the smallest
number of missed polyps are achieved by the hybrid
method with implemented test-time data augmentation.
A sample of the typical segmentation results obtained
by the hybrid method with the test-time data
augmentation is shown in Figure 7. It should be noted
that the method is able to successfully segment polyps
of various size, shape and appearance. The Dilated
ResFCN was used to generate results submitted to the
GIANA 2017 challenge. The Dilated ResFCN clearly
outperformed other submissions with the highest mean
and the smallest standard deviation of the Dice
coefficient. The results generated by the hybrid method
with some added post processing (not reported in this
paper) were submitted to the GIANA 2018 challenge.
The submission secured second place, with small
standard deviation and only slightly smaller Dice
coefficient compared to the wining submission.
6 CONCLUSIONS
The paper describes two novel fully convolutional
neural network architectures specifically designed for
segmentation of polyps in video colonoscopy images.
The networks have been developed and tested on the
Gastrointestinal Image ANAlysis – (GIANA) polyp
segmentation database. The available training dataset
with 300 low resolution and 56 high resolution
images, is very limited from a perspective of a typical
training set used in a context of the deep learning.
Therefore, available data were heavily augmented
with random rotation, translation, scale changes as
well as colour and contrast jitter, with the rotation and
colour jitter havening the most significant effect on
the quality of the segmentation. In total, after
augmentation the training data include more than
90,000 images. The output from the network was
optionally processed using the hybrid level set
method. However, it should be noted that the DICE
similarity scores obtained using a simple thresholding
of the network outputs are very similar to the values
of this measure obtained after applying the level set
method. Nevertheless the level set could be used as it
provides a simple mechanism to control smoothness
of the segmented polyp boundaries. The proposed
architectures provide competitive results, as is
evident from the fact that they achieved the best
results for the polyp segmentation task at the GIANA
2017 challenge and second place for polyp
segmentation in the SD (low resolution) images at the
GIANA 2018 challenge.
To the best knowledge of the authors, temporal
dependencies in the colonoscopy video have not yet
been used for polyp detection or segmentation within
context of the deep architectures. The authors are
aiming to examine various scenarios to test if such
information could improve the overall performance
of the polyp segmentation FCNNs. Two possible
processing pipelines are to be investigated, with the
explicit image warping obtained with a help of image
registration (Shen et al., 2005) and implicit temporal
fusion as part of the deep architecture.
GIANA Polyp Segmentation with Fully Convolutional Dilation Neural Networks
639

ACKNOWLEDGEMENTS
The authors would like to acknowledge the organisers
of the Gastrointestinal Image ANAlysis – (GIANA)
challenges for providing video colonoscopy polyp
images.
REFERENCES
Akbari, M., Mohrekesh, M., Nasr-Esfahani, E.,
Soroushmehr, S. M., Karimi, N., Samavi, S., &
Najarian, K. (2018). Polyp Segmentation in
Colonoscopy Images Using Fully Convolutional
Network. arXiv preprint arXiv:1802.00368.
Alexandre, L. A., Nobre, N., and Casteleiro, J. (2008).
Color and position versus texture features for
endoscopic polyp detection. In BioMedical
Engineering and Informatics, 2008. BMEI 2008.
International Conference on (Vol. 2, pp. 38-42). IEEE.
Bernal, J., Sánchez, F. J., Fernández-Esparrach, G., Gil, D.,
Rodríguez, C., & Vilariño, F. (2015). WM-DOVA
maps for accurate polyp highlighting in colonoscopy:
Validation vs. saliency maps from
physicians. Computerized Medical Imaging and
Graphics, 43, 99-111.
Bernal, J., Sánchez, J., & Vilarino, F. (2012). Towards
automatic polyp detection with a polyp appearance
model. Pattern Recognition, 45(9), 3166-3182.
Bernal, J., Sánchez, J., & Vilarino, F. (2013). Impact of
image preprocessing methods on polyp localization in
colonoscopy frames. In Engineering in Medicine and
Biology Society (EMBC), 2013 35th Annual
International Conference of the IEEE (pp. 7350-7354).
IEEE.
Bernal, J., Tajkbaksh, N., Sánchez, F. J., Matuszewski, B.
J., Chen, H., Yu, L., ... & Pogorelov, K. (2017).
Comparative validation of polyp detection methods in
video colonoscopy: results from the MICCAI 2015
Endoscopic Vision Challenge. IEEE transactions on
medical imaging, 36(6), 1231-1249.
Breier, M., Gross, S., and Behrens, A. (2011a). Chan-Vese-
segmentation of polyps in colonoscopic image data.
In Proceedings of the 15th International Student
Conference on Electrical Engineering POSTER (Vol.
2011).
Breier, M., Gross, S., Behrens, A., Stehle, T., and Aach, T.
(2011b). Active contours for localizing polyps in
colonoscopic nbi image data. In Medical Imaging
2011: Computer-Aided Diagnosis (Vol. 7963, p.
79632M). International Society for Optics and
Photonics.
Chen, L. C., Papandreou, G., Kokkinos, I., Murphy, K., and
Yuille, A. L. (2018). Deeplab: Semantic image
segmentation with deep convolutional nets, atrous
convolution, and fully connected crfs. IEEE
transactions on pattern analysis and machine
intelligence, 40(4), 834-848.
Glorot, X., and Bengio, Y. (2010). Understanding the
difficulty of training deep feedforward neural networks.
In Proceedings of the thirteenth international
conference on artificial intelligence and statistics (pp.
249-256).
Gross, S., Kennel, M., Stehle, T., Wulff, J., Tischendorf, J.,
Trautwein, C., and Aach, T. (2009). Polyp
segmentation in NBI colonoscopy. In Bildverarbeitung
für die Medizin 2009 (pp. 252-256). Springer, Berlin,
Heidelberg.
Hernández-García, A., and König, P. (2018). Do deep nets
really need weight decay and dropout?.
arXiv:1802.07042v3
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep
residual learning for image recognition. In Proceedings
of the IEEE conference on computer vision and pattern
recognition (pp. 770-778).
Hu, J., Shen, L., and Sun, G. (2017). Squeeze-and-
excitation networks. In Proceedings of the IEEE
Conference on Computer Vision and Pattern
Recognition (pp. 7132-7141).
Hwang, S., Oh, J., Tavanapong, W., Wong, J., and De
Groen, P. C. (2007). Polyp detection in colonoscopy
video using elliptical shape feature. In Image
Processing, 2007. ICIP 2007. IEEE International
Conference on (Vol. 2, pp. II-465). IEEE.
Iakovidis, D. K., Maroulis, D. E., Karkanis, S. A., and
Brokos, A. (2005). A comparative study of texture
features for the discrimination of gastric polyps in
endoscopic video. In Computer-Based Medical
Systems, 2005. Proceedings. 18th IEEE Symposium
on (pp. 575-580). IEEE.
Karkanis, S. A., Iakovidis, D. K., Maroulis, D. E., Karras,
D. A., and Tzivras, M. (2003). Computer-aided tumor
detection in endoscopic video using color wavelet
features. IEEE transactions on information technology
in biomedicine, 7(3), 141-152.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012).
Imagenet classification with deep convolutional neural
networks. In Advances in neural information
processing systems (pp. 1097-1105).
LeCun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998).
Gradient-based learning applied to document
recognition. Proceedings of the IEEE, 86(11), 2278-
2324.
Li, Q., Yang, G., Chen, Z., Huang, B., Chen, L., Xu, D., ...
and Wang, T. (2017). Colorectal polyp segmentation
using a fully convolutional neural network. In Image
and Signal Processing, BioMedical Engineering and
Informatics (CISP-BMEI), 2017 10th International
Congress on (pp. 1-5). IEEE.
Long, J., Shelhamer, E., and Darrell, T. (2015). Fully
convolutional networks for semantic segmentation.
In Proceedings of the IEEE conference on computer
vision and pattern recognition (pp. 3431-3440).
Park, S., Lee, M., and Kwak, N. (2015). Polyp detection in
colonoscopy videos using deeply-learned hierarchical
features. Seoul National University.
GIANA 2019 - Special Session on GastroIntestinal Image Analysis
640

Peng, C., Zhang, X., Yu, G., Luo, G., and Sun, J. (2017).
Large kernel matters—improve semantic segmentation
by global convolutional network. In Computer Vision
and Pattern Recognition (CVPR), 2017 IEEE
Conference on (pp. 1743-1751). IEEE.
Ribeiro, E., Uhl, A., and Häfner, M. (2016). Colonic polyp
classification with convolutional neural networks.
In Computer-Based Medical Systems (CBMS), 2016
IEEE 29th International Symposium on (pp. 253-258).
IEEE.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image
segmentation. In International Conference on Medical
image computing and computer-assisted
intervention (pp. 234-241). Springer, Cham.
Shen, J. K., Matuszewski, B. J., and Shark, L. K. (2005).
Deformable image registration. In Image Processing,
2005. ICIP 2005. IEEE International Conference
on (Vol. 3, pp. III-1112). IEEE.
Tajbakhsh, N., Chi, C., Gurudu, S. R., and Liang, J.
(2014a). Automatic polyp detection from learned
boundaries. In Biomedical Imaging (ISBI), 2014 IEEE
11th International Symposium on (pp. 97-100). IEEE.
Tajbakhsh, N., Gurudu, S. R., and Liang, J. (2013). A
classification-enhanced vote accumulation scheme for
detecting colonic polyps. In International MICCAI
Workshop on Computational and Clinical Challenges
in Abdominal Imaging (pp. 53-62). Springer, Berlin,
Heidelberg.
Tajbakhsh, N., Gurudu, S. R., and Liang, J. (2014b).
Automatic polyp detection using global geometric
constraints and local intensity variation patterns.
In International Conference on Medical Image
Computing and Computer-Assisted Intervention (pp.
179-187). Springer, Cham.
Vázquez, D., Bernal, J., Sánchez, F. J., Fernández-
Esparrach, G., López, A. M., Romero, A., ... and
Courville, A. (2017). A benchmark for endoluminal
scene segmentation of colonoscopy images. Journal of
healthcare engineering, 2017.
Yu, F., and Koltun, V. (2015). Multi-scale context
aggregation by dilated convolutions. arXiv preprint
arXiv:1511.07122.
Zhang, L., Dolwani, S., and Ye, X. (2017). Automated
polyp segmentation in colonoscopy frames using fully
convolutional neural network and textons. In Annual
Conference on Medical Image Understanding and
Analysis (pp. 707-717). Springer, Cham.
Zhang, R., Zheng, Y., Mak, T. W. C., Yu, R., Wong, S. H.,
Lau, J. Y., and Poon, C. C. (2017). Automatic Detection
and Classification of Colorectal Polyps by Transferring
Low-Level CNN Features From Nonmedical
Domain. IEEE J. Biomedical and Health
Informatics, 21(1), 41-47.
Zhang, Y., and Matuszewski, B. J. (2009). Multiphase
active contour segmentation constrained by evolving
medial axes. In Image Processing (ICIP), 2009 16th
IEEE International Conference on (pp. 2993-2996).
IEEE.
Zhang, Y., Matuszewski, B. J., Shark, L. K., and Moore, C.
J. (2008). Medical image segmentation using new
hybrid level-set method. In Fifth International
Conference BioMedical Visualization: Information
Visualization in Medical and Biomedical
Informatics (pp. 71-76). IEEE.
GIANA Polyp Segmentation with Fully Convolutional Dilation Neural Networks
641
