
MobileECG: An Ubiquitous Heart Health Guardian
Jos
´
e Maria Monteiro
1
, Jo
˜
ao Paulo do Vale Madeiro
2
, Angelo Brayner
1
and Narciso Arruda
1
1
Department of Computing Science, Federal University of Cear
´
a, Fortaleza, Cear
´
a, 60440-900, Brazil
2
Institute for Engineering and Sustainable Development,
University for the International Integration of the Afro-Brazilian Lusophony, Redenc¸
˜
ao, Cear
´
a, 62790-970, Brazil
Keywords: ECG Signal Processing, Pervasive and Mobile Platform, ECG Feature Extraction, Prediagnosis, Data
Integration.
Abstract:
Electrocardiogram (ECG) is a widespread and efficient medical procedure for monitoring heart health. ECG is
a fast, low-cost and non-invasive examination. Its output allows anomaly analysis by health experts. Despite
its application in clinical environments, ECG acquisition and analysis as a daily routine is far from being a
reality for a large part of the world’s population. In this context, we present here a mobile and pervasive
platform, named MobileECG, which provides ECG signal acquisition, automatic feature extraction and real-
time prediagnosis. Furthermore, MobileECG implements the ubiquitous computing features. Hence, it runs
on mobile devices (smartphone or tablet), assuring this way anytime and anywhere access for anyone to its
functionalities. MobileECG is in fact an ubiquitous Heart Health Guardian. Besides, MobileECG supports
ECG data integration and publication using Linked Data technology, providing a public knowledge base,
which may be used to support complex queries, run mining algorithms and yield collaboration among experts.
1 INTRODUCTION
Electrocardiography is a technique used to record
electrical potential oscillations produced by cardiac
activity. Temporal evolution of such oscillations is
called the electrocardiogram (ECG) signal, which is
the most widespread test in cardiology for the diag-
nosis of cardiac diseases and anomalies. The human
body by itself acts as a conductor of electrical cur-
rent, and any two points on the surface can be con-
nected by electrodes to record an ECG or monitor
the heart’s rhythm. The time-series obtained by the
electrocardiographic record contains a series of wave-
forms and complexes, which are called the P-wave,
the QRS complex, and the T-wave, which are sepa-
rated by regular intervals. The analysis of ECG signal
behavior (waveform pattern, duration and interval) al-
lows the extraction of a variety of information, which
can subsidize the identification of a great variety of
heart diseases.
Chronic non-communicable diseases (NCD), in-
cluding heart disease and cancer, are responsible for
approximately 70% of all deaths worldwide, accord-
ing to the World Health Organization (Daar et al.,
2007). Allied to this fact, the world population is ag-
ing. Such a context requires a significant need to mon-
itor patient’s health status, while she/he is in her/his
personal environment. Consequently, a plethora of
wearable systems for health monitoring has been de-
veloped and applied for providing real-time feedback
information on individual’s health condition, either to
the user himself, to a medical center, or directly to a
health professional.
Over the last decades, ubiquitous computing has
gained critical importance for implementing health
care applications. Ubiquity provides the means to
make health care services available for anyone at any-
time and anywhere. Thus, patient data may be ac-
quired by biosensors containing embedded micropro-
cessors, coupled up with bluetooth modules. Further-
more, a mobile application may execute signal pro-
cessing or data analytic techniques on collected data,
or even simply forward collected data to a physician’s
or a clinic’s computers.
In this paper, an innovative pervasive and mobile
platform for ECG signal acquisition, processing and
prediagnostic extraction is presented. The proposed
platform, denoted MobileECG, is able to collect and
preprocess ECG raw data in a mobile device (smart-
phone or tablet). Thereafter, prediagnosis algorithms
are executed in order to trigger alerting messages to
the patient, physician, or emergency station, which
should be sent by means of a WiFi or cellular net-
work.
Monteiro, J., Madeiro, J., Brayner, A. and Arruda, N.
MobileECG: An Ubiquitous Heart Health Guardian.
DOI: 10.5220/0007703705790586
In Proceedings of the 21st International Conference on Enterprise Information Systems (ICEIS 2019), pages 579-586
ISBN: 978-989-758-372-8
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
579

Besides, MobileECG maintains an integrated
ECG data repository, which stores an integrated view
of ECG data coming from heterogeneous sources
(with different ECG data standards) and data of pa-
tient, treatment and drug data sources. Such a repo-
sitory can be used by machine learning algorithms to
automatize prediagnosis.
Empirical experiments on real ECG datasets have
been conducted to evaluate MobileECG efficiency
and effectiveness. The results reveal that MobileECG
can be an important tool for helping health experts to
identify heart diseases more quickly in a more reliable
way.
The paper is structured as follows. Section 2 an-
alyzes approaches similar to the one presented in this
paper. Section 3 details the different steps of the pro-
posed platform. In Section 4, results of empirical ex-
periments are presented to highlight the benefits and
feasibility of the proposed approach. Finally, section
5 concludes the paper and points out future work.
2 RELATED WORK
A literature review on pervasive health care systems
is presented in (Orwat et al., 2008). According to
the authors, 60% of investigated systems provide ana-
lytical and diagnostic functionality. Only 46% im-
plement automatic alerting, and from this, only 14%
support an alerting function generated by a mobile
device. Since pervasive health care systems encom-
pass a wide variety of components, one can classify
them by the following criteria: ECG data acquisi-
tion systems (Venkatachalam et al., 2011), real-time
ECG digital signal processing (Madeiro et al., 2012;
do Vale Madeiro et al., 2017), feature extraction (El-
haj et al., 2016; Martis et al., 2014), ECG remote
monitoring systems (Worringham et al., 2011), col-
laborative databases (Gonc¸alves et al., 2011) and ma-
chine learning techniques (Luz et al., 2016; Elhaj
et al., 2016).
In (da Silva et al., 2015), the authors describe
a biosensor for sensing ECG signals, with minimal
electrical contact points with users, a so-called off-
person ECG. The key goal is to support ECG signal
acquisition on palms or fingers. Thus, the proposed
biosensor can be embedded in any object with which
the user interacts.
In (Wen et al., 2008), the authors propose an ECG
telemonitoring system. According to their approach,
the Holter Monitor will record the ECG signal of the
patient continuously up to 48 h. The monitored data
is transmitted to the server through the Internet when
a wired network is available. The Holter also con-
tains a software program performing real-time ECG
classification. When specific abnormal heartbeats are
detected, the Holter transmits them with the GPS
(global positioning system) information to the server
via MMS (multimedia messaging service) in real-
time. The physician at the server side can communi-
cate with the patient also by using MMS. In the server,
a GIS (geographic information system) is used for lo-
cating the patient in an emergency case by using the
GPS information packaged in the MMS message.
In (Worringham et al., 2011), the authors propose
a system to enable walking-based cardiac rehabili-
tation in which the patient’s single-lead ECG, heart
rate, and GPS-based speed and location are transmit-
ted by a programmed smartphone to a secure server
for real-time monitoring by physicians. According to
the authors, the feasibility of this approach was evalu-
ated in 134 remotely-monitored exercise assessment
and exercise sessions in cardiac patients unable to
undertake hospital-based rehabilitation. Completion
rates, rates of technical problems, detection of ECG
changes, pre- and post-intervention six minute walk
test (6 MWT), cardiac depression and Quality of Life
(QOL) could be measured. Several exercise and post-
exercise ECG changes were detected.
In (Ngo and Veeravalli, 2014), DuyHoa Ngo et
al. have proposed a platform based on Web Semantic
technologies which provides the storage of features
extracted from the ECG signal within a database fol-
lowing Linked Data patterns. The authors emphasize
that the proposed platform is part of a healthcare sys-
tem based on cloud data storage, which also captures
ECG signals and other vital signs through biosensors
located on the surface of the patient’s body.
Thus, one can enumerate two important strengths
of the proposed platform: the reproducibility of the
methodology is notorious due to the fact that the pro-
posed hardware and software are open-source; Mo-
bileECG provides basis for integrating multiple ECG
data sources, including data from biosensors and data
from public or institutional databases which may po-
tentialize the accuraccy of pattern recognition pro-
cesses.
3 THE MobileECG PLATFORM
The MobileECG architecture presents the following
components: acquisition module, mobile application
module, ECG signal processing and feature extraction
module, ECG data extraction module, and finally, the
data integration and publishing module. Fig. 1 de-
picts an abstract model of the proposed architecture,
emphasizing the physiological data flow, from the pa-
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
580
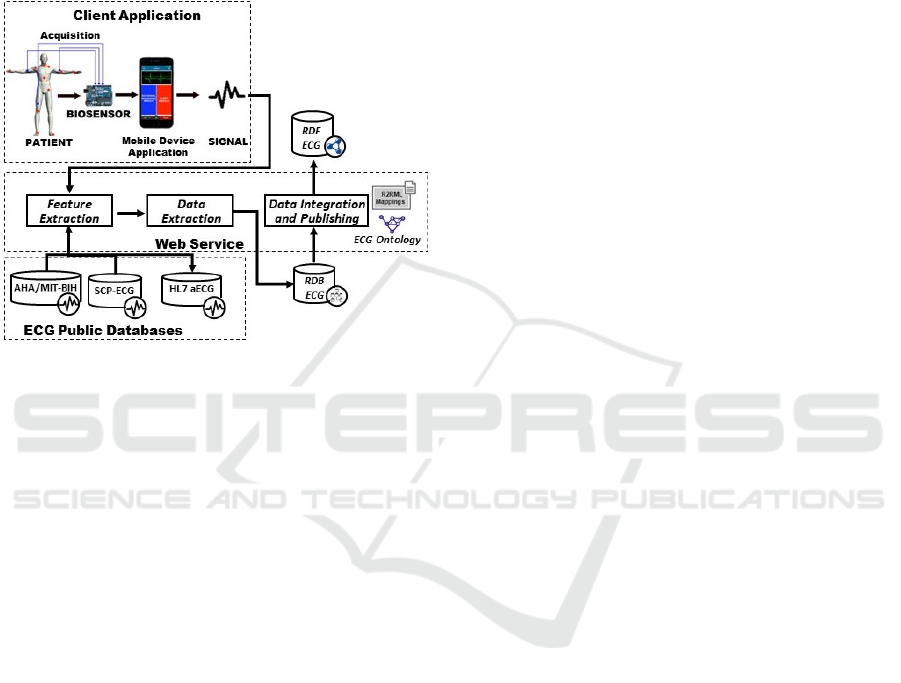
tient to the ECG data repository. For the sake of clar-
ity, the MobileECG components are grouped by their
behavior in a client-server architecture. Thus, the ac-
quisition and mobile application modules are located
in the client area, whereas the other modules are in the
server area, as web servers. Each one of the aforemen-
tioned components is described in detail and analyzed
next.
Figure 1: Overview of the MobileECG platform.
3.1 Acquisition Module
The acquisition module is composed of a biosensor
coupled with an Arduino microcontroller and it is lo-
cated in the client area. The capture of the ECG
biopotentials is performed by the biosensor, which
measures and amplifies electrical potentials derived
from the electrical activity of the heart, and then
transmits the analog signal to Arduino. An open-
source hardware that allows direct communication
(SHIELD) with an Arduino microcontroller has been
deployed as biosensor. The electrodes used for sig-
nal acquisition consist of three leads which should be
placed on the wrists and right ankle of the patient. The
shield converts the analog differential signal (ECG
biopotentials) into a single stream of data as output.
A third order “Besselworth” analog filter’s cutoff fre-
quency is set to f
c
= 40Hz, and the output analog sig-
nal is discretized via a dedicated ADC embedded in
the Arduino board. Default values for discretizing pa-
rameters are: 10-bit ADC with 256 Hz sampling rate.
The Arduino board is responsible for receiving the
analog signal and for converting it into a digital sig-
nal. Each discretized sample from each lead or chan-
nel is split into two bytes, named high byte and low
byte, and each byte is stored in packets. Thereafter,
each individual byte from the packet is serially trans-
mitted to the bluetooth module, which in turn sends
ECG raw data to a mobile device.
3.2 Mobile Device Application Module
The mobile application is in charge of establishing
connections with a paired device (bluetooth module
associated to a given biosensor and Arduino board)
and for searching for unpaired devices. Whenever a
device is selected, the mobile application collects the
bluetooth identification of the connected device and
its Mac address.
As soon as the mobile application receives ECG
raw data from a given device, it is able to plot ECG
signal buffers on screen as long as a 5-second buffer
of ECG signal is completely filled. A 5-second
buffer of ECG signal contains 21,760 bytes. The mo-
bile application triggers a thread, named Connection-
Thread, which receives the Mac address of the paired
device. Within the Connection-Thread, we define a
number of bytes (or packet length) transferred from
the Arduino bluetooth module to the mobile device
on each transfer. Thus, an Android device continu-
ously receives packets of 289 bytes from the Arduino
bluetooth module. Each received packet is stored into
a byte array (length of 21, 760 bytes). Thereafter, the
mobile application triggers the ECG data buffer de-
coding process. This process searches for each pair
of synchronizing bytes (A5 and 5A) and for the pair
of bytes corresponding to the ECG sample of a given
channel. Since the second channel has been defined as
default, high and low bytes are located, respectively,
five bytes and six bytes after each pair of synchroniz-
ing bytes. Thus, a given ECG sample is computed as
the sum of the low byte and high byte shifted by 8
bits left. Each computed ECG sample is stored in a
buffer array which will contain a 5-second ECG win-
dow for subsequent display on screen and subsequent
sending to the Web Service. After filling a 5-second
ECG signal buffer, the mobile application forwards
the pre-processed ECG buffer to the Web Service. Af-
ter sending each ECG buffer to the Web Service, the
class Plot-Real-Time finally performs the plot of the
buffer.
3.3 ECG Signal Processing and ECG
Feature Extraction Module
As soon as the mobile app finishes the ECG buffer
transmission, the Web Service (Apache Maven) joins
all received signal buffers and stores them in a sin-
gle file. A set of algorithms for digital signal pro-
cessing is performed, including signal filtering for
denoising, wavelet transform (time-frequency analy-
sis) for selective enhancement of the QRS complex,
P and T waves, wave peak detection and waveform
delineation. The main goal is to extract the following
MobileECG: An Ubiquitous Heart Health Guardian
581
parameters: P-wave amplitude, P-wave duration, PR
interval, QRS amplitude, QRS duration, T-wave am-
plitude, T-wave duration, QT and ST segments, and
intervals between beats. Those algorithms have been
implemented using Matlab, which is an IDE devel-
opment environment widely applied for digital signal
processing (Islam et al., 2012).
The first stage for ECG feature extraction is re-
sponsible for detecting the QRS complex, which is
the most expressive waveform of the ECG signal re-
garding amplitude and period of oscillation. The cor-
rect detection of the QRS complex and its precise de-
lineation are fundamental conditions for efficient de-
tection and segmentation of the other waves. Addi-
tionally, it provides the necessary support for algo-
rithms able to recognize patterns of cardiac arrhyth-
mias. After proceeding with the denoising process, a
sequence of filtering routines are established for en-
hancing the QRS complex and attenuating artifacts
and other physiologic waves. The Wavelet Trans-
form, the First-Derivative function, and the Hilbert
Transform are sequentially computed, and the mod-
ule of the resultant analytic signal is used at a decision
stage. An adaptive threshold algorithm is applied over
the resultant filtered signal for individual detection of
each QRS (Madeiro et al., 2012). Then, the computa-
tion of an indicator related to the area covered by the
QRS complex provides the detection of QRS onset
and offset (Madeiro et al., 2012).
Concerning P-wave and T-wave detection and seg-
mentation, firstly, we apply Wavelet Transform over
signal windows established between segmented QRS
complexes for estimating T-wave and P-wave peak lo-
cations. Then, we fit the parameters of a synthetic
function, computed as a composition of two Gaussian
functions, in order to model each waveform (T-wave
and P-wave) by evaluating the normalized root mean
square (RMS) error. This approach provides the de-
lineation of each waveform (by detecting onsets and
ends) and the computing of parameters related to the
wave width and to a distortion factor of the Gaussian
functions which also characterize their morphologies
(do Vale Madeiro et al., 2017). All the above ex-
plained algorithms, built as Matlab language scripts,
are compiled and converted in a single executable file.
After the Web Service receives all the ECG signal
buffers and converts them into a single file, the Web
Service itself calls a Java method that runs the re-
ferred executable file. Then, the set of all ECG ex-
tracted parameters (waveform peaks and widths, seg-
ments and intervals) are converted into text files. Af-
ter obtaining these text files, the Web Service calls a
Java method responsible for storing the extracted pa-
rameters within a relational database.
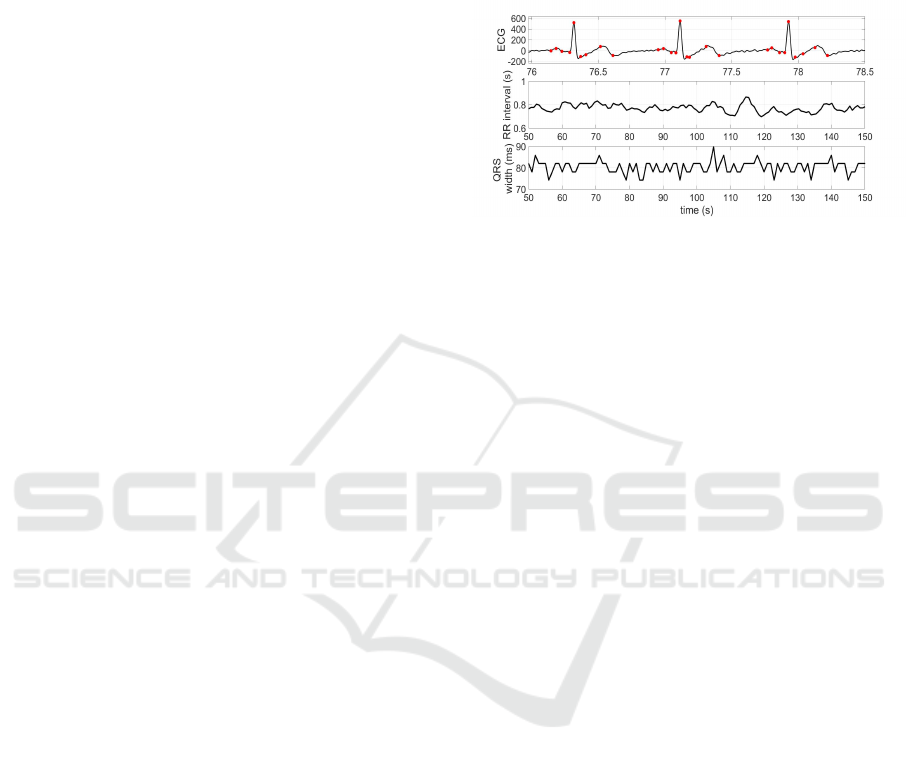
As an illustrative example, Fig. 2 presents a set of
results showing ECG beat cycles with all the char-
acteristic waves properly segmented, and two time
series of the extracted parameters intervals between
beats and QRS widths.
Figure 2: Practical example of ECG feature extraction by
applying algorithms over the signal sent by Android app.
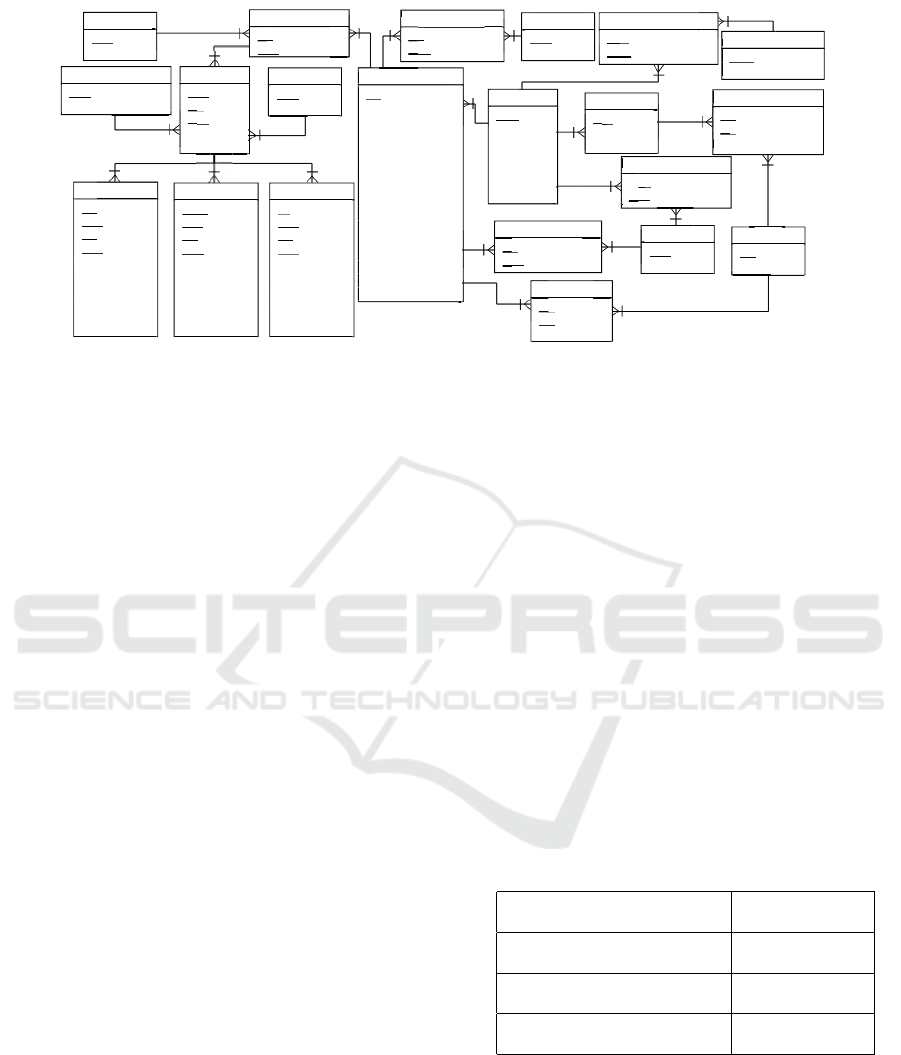
3.4 Data Extraction Module
This module consists of storing features extracted
from an ECG signal in a relational database. By do-
ing this, it is possible to formulate SQL queries in
order to relate ECG signals to properties of a given
diagnosis. Figure 3 brings the schema of the pro-
posed database. The ECG table contains an identi-
fication of a given ECG record (exam) and also some
clinic/diagnosis information from the corresponding
patient, such as his/her systolic and diastolic pres-
sure at the time of ECG acquisition. The table Pa-
tient contains some identification data of a given pa-
tient, such as document number, birth date and birth
place. The Prescription table stores information on
current medicines taken by a given patient. Drug is
fed with medicines related to treatments for the most
common cardiac diseases. The table Cycle relates the
PQRST complex to a unique cardiac beat. Tables re-
lated to P-wave, QRS complex and T-wave contain
the identifier of the cycle to which they belong. More-
over, they contain information derived directly from
the process for waveform detection and segmentation:
onsets, offsets and the corresponding amplitudes.
Tables Cycle
D
iagnosis, ECG
D
iagnosis and
ECG
C
ardiopathy provides to associate ECG wave-
form patterns to known cardiac conditions and/or
cardiac diseases, such as: conduction disorders,
arrhythmias, cardiomyopathies, and other events.
The content of such tables may result from Machine
Learning algorithms, or may derive from ECG public
databases, in the case of ECG signals originated from
those databases, or even may derive from manual
specialists’ annotations.
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
582

3.5 Data Integration and Publishing
Advances in heart disease research have been ham-
pered in part by the fragmented gathering and stor-
ing of data. Among the major ECG standards, we
can highlight: (i) AHA/MIT-BIH (Physionet) (Gold-
berger et al., 2000a), an ECG data format extensively
used in cardiac physiology research; (ii) SCP-ECG
(Mandellos et al., 2010), which is an ECG European
standard that specifies a data format and a transmis-
sion protocol for ECG records; and (iii) HL7 aECG
(Bond et al., 2011), which is an ECG data American
standard adopted by the Food and Drug Administra-
tion (FDA), from the USA, for clinical research.
This knowledge needs to be integrated. To make
matters worse, typically heart disease knowledge is
expressed in ambiguous terminology. Without the aid
of a well-defined knowledge representation, the ad
hoc data integration is hard work. In addition, another
fundamental challenge to heart research is querying
complex heart data.
In this context, we propose the designing of an
integrated repository of ECG data whose purpose
is: (i) to integrate data in heterogeneous ECG stan-
dards, that is, to store ECG data coming from vari-
ous sources with different data standards (AHA/MIT-
BIH, SCP-ECG and HL7 aECG); (ii) to integrate data
about cardiac diseases, cardiomyopathies, treatments
and drugs with LOD
1
(Linked Open Data); (iii) to
store ECG data coming from Patient’s smartphones;
(iv) To make possible the querying of complex heart
data; and (v) to enable running data mining algo-
rithms over integrated data.
Therefore, to support the design of the integrated
ECG repository, we propose the use of Semantic
Web technologies, such as: RDF, OWL, SKOS and
SPARQL. These technologies enable the explicit re-
presentation of knowledge and its processing to de-
duce new information. Besides, they provide an envi-
ronment where heterogeneous data can be combined
based on a common knowledge representation, the
schemata can be extended in an easily and dynamic
way, and applications can query that integrated data
and draw inferences using vocabularies.
In (Gonc¸alves et al., 2011), the authors proposed
an ECG reference ontology, that is, an ontology re-
sulting from an application-independent representa-
tion of the ECG domain. This reference ontology can
be used to support the design of interoperable versions
of ECG data formats like AHA/MIT-BIH, SCP-ECG,
and HL7 aECG.
This ECG reference ontology can be used to sup-
port the redesign and the possible unification of ex-
1
https://lod-cloud.net/
isting ECG data standards. In this sense, it can be
used to support the design of an interoperable and
collaborative ECG data repository, which can store
ECG signals from various data sources with differ-
ent ECG data standards. However, in order to build a
wide ECG repository that makes it possible to answer
complex queries, it is necessary to extend the refer-
ence ontology proposed in (Gonc¸alves et al., 2011),
adding concepts related to drugs, treatments, and dis-
ease causes, among others, which is one of the goals
of the MobileECG platform. In this way, we use the
vocabularies ecg
2
, available in the work (Gonc¸alves
et al., 2007), and heal th
3
(additional terms).
The ecgo vocabulary, contains terms created by
the proposed approach. Fig. 4 presents the ontology
applied in this step. As we can observe, it contains
proposed classes for providing inference and classi-
fying of cardiac rhythms (e.g. ecgo : SlowRhythm,
ecgo : NormalRhythm, ecgo : FastRhythm), types of
cardiac beats/cycles (e.g. ecgo : NormalBeat and
ecgo : JunctionalEscapeBeat) and possible ECG di-
agnosis (e.g. ecgo : PersistentAtrialFibrillation and
ecgo : LongQT Syndrome).
The mappings between the relational database
schema and the extended ontology have been exe-
cuted by means of the R2RML language and the
D2RQ tool (Priyatna et al., 2014). It is important to
emphasize that it is not necessary to use an ontology
to perform mappings with the D2RQ tool; however its
use greatly facilitates the mapping process.
The MobileECG platform allows you to import
ECG signals from any repository using one of the
following standards: AHA/MIT-BIH, SCP-ECG and
HL7 aECG. To support this import process, we have
implemented a wrapper for each one of these three
formats. Thus, to import a particular ECG signal,
from a public database, it is necessary to follow three
steps: (i) initially, download the files containing the
signal, from a public database; (ii) then, apply the set
of algorithms described in the Section 3.3 for extract-
ing the features of the signal; (iii) finally, the data of
the signal must be extracted, similar to the process
described in the Section 3.4.
The data integration and publishing module con-
sists of a Web Service which uses Java methods for
accessing the relational database containing the infor-
mation extracted from the ECG signals and exports
the relational data to the RDF format. We split the
process into two steps. In the first step, we create a
dump of the relational data in RDF format. To per-
form this step, we use the D2RQ tool together with the
mappings in the R2ML3 language, which associates
2
http://nemo.inf.ufes.br/biomedicine/ecg.html
3
Available at https://health-lifesci.schema.org
MobileECG: An Ubiquitous Heart Health Guardian
583
Pwave
- pid
pid
-
pid
cyid
cyid
cyid
-
cyid
eid
- chid
- onsetTime
- peakTime
peakT
- offsetTime
- onsetAmp
onsetAmp
onsetAmp
- peakAmp
peakAmp
- offsetAmp
Patient
- paid
paid
-
paid
name
- CPF
- gender
-
gender
birthDate
- deathDate
- birthPlace
ECG
- eid
- paid
paid
paid
paid
paid
-
paid
sinal
- datetime
- BMI
- pacemaker
pacemaker
-
pacemaker
pacemaker
systolicPressure
systolicPressure
- diastolicPressure
- cancer
- tobaccoism
- alcoholism
- syncope
-
syncope
sedentarism
- flutter
- channelsNumber
- CVA
-
Pwave
Prescription
- pri
pri
d
-
pri
pa
pri
id
pa
- date
-
p
pr
p
-
p
p
pa
p
-
da
Drug
- d
d
id
- description
Cycle
- cyid
cyid
cyid
-
cyid
eid
- chid
- peid
peid
- cdid
y
Cycle
did
QRSWave
- qrsid
qrsid
qrsid
-
qrsid
cyid
cyid
cyid
-
cyid
eid
- chid
- onsetTime
- peakTime
peakT
- offsetTime
- onsetAmp
onsetAmp
onsetAmp
- peakAmp
peakAmp
- offsetAmp
TWave
- tid
- cyid
cyid
cyid
-
cyid
eid
- chid
- onset
onset
Time
- peakTime
peakT
-
peakT
offsetTime
- onsetAmp
onsetAmp
onsetAmp
- peakAmp
peakAmp
- offsetAmp
PrescriptionDrug
- pi
pi
d
-
pi
did
- dosage
n
-
pi
pi
i
p
-
p
di
d
ECG_Drug
- ei
ei
d
- did
- dosage
E
-
i
ei
i
-
d
i
g
ug
Diagnosis
- daid
- description
ECG_Diagnosis
- eid
- daid
EC
-
i
ei
i
d
-
da
-
sis
Cardiopathy
- caid
- description
ECG_Cardiopathy
- ei
ei
d
- caid
EC
-
i
ei
i
-
ca
C
-
y
y
thy
ECG_Cardiopathy
- pai
pai
d
-
pai
caid
EC
-
p
pa
p
-
p
ca
ECG_Channel
- eid
- chid
chid
chid
chid
Channel
- chid
- description
E
-
e
i
-
c
h
el
Period
- peid
peid
-
peid
startTime
Cycle_Diagnosis
- cd
cd
cd
id
- description
-
ch
-
pe
p
cd
Institution
- ins
ins
id
- description
Patient_Institution
- pai
i
i
d
pa
-
pa
insid
I
n
s
t
i
t
u
on
-
p
description
-
-
ins
s
d
d
d
id
Figure 3: The relational database schema.
the relational database schema with the ontology vo-
cabulary adopted to represent the ECG signals. In the
second step, the RDF data present in the dump gen-
erated previously are materialized in an RDF triple-
store, more specifically in Virtuoso, in a semiauto-
matic way. Virtuoso provides a SPARQL endpoint
that is provided to perform semantic queries.
The RDF Repository is a collaborative database
for heart research. It makes possible the identification
of data cohorts (including patients, medicines, treat-
ments, and ECGs) for study, and the export of data
for statistical analysis. Besides, the RDF Repository
will make it possible to infer new knowledge through
the use of machine learning algorithms.
4 EMPIRICAL EVALUATION
In order to assess the potentials of the proposed ap-
proach, empirical evaluation have been carried out.
Firstly, we evaluate the performance of the Arduino
code for ECG signal acquisition, conditioning and for
sending ECG data to the bluetooth module.
In terms of memory consumption, using Olimex
SHIELD-EKG-EMG for analog signal acquisition,
Arduino Uno and IDE Arduino 1.8.5, we obtain that
sketch (code) uses 2272 bytes (7%) of program stor-
age space and global variables use 223 bytes (10%) of
dynamic memory, leaving 1825 bytes for local vari-
ables. Regarding the mobile application, we con-
duct the experiments using smartphone model LG-
M250ds and Android version 7.0. We have measured
the smartphone memory comsumption, immediately
before and immediately after the smartphone appli-
cation starts to run. We can observe a significant
increasing for memory consumption while the class
Plot-Real-Time performs the plot of the 5s-ECG sig-
nal buffer.
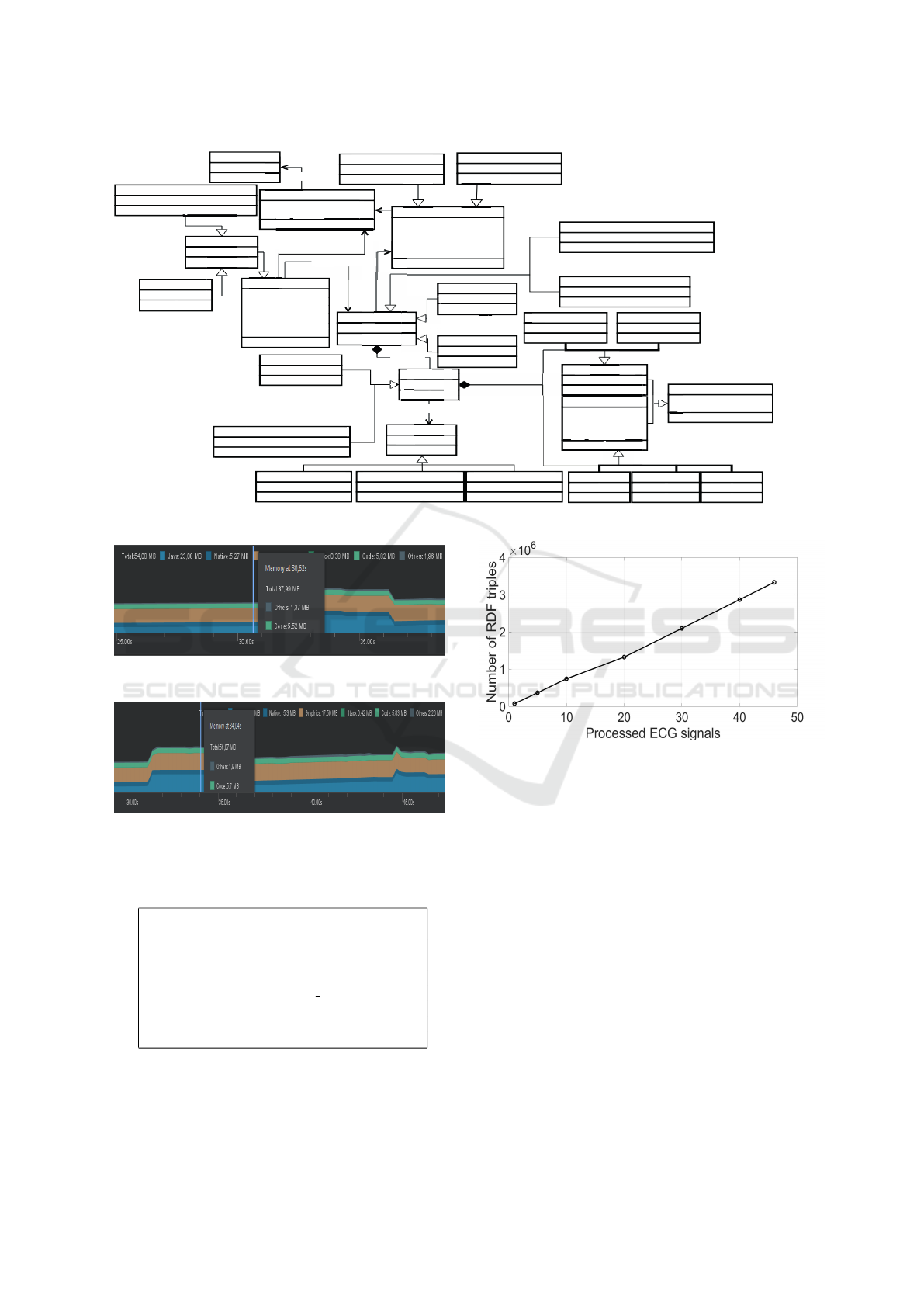
According to Fig. 5, the total use of memory is
37.99 MB immediately before the first ECG signal
buffer is exhibited on the mobile screen, increasing to
56.07 MB (see Fig. 6) while the ECG signal buffer is
being plotted, during 5 seconds, and again decreasing
to 37.99 MB.
As already mentioned, after the mobile app con-
cludes the ECG buffer transmission, the Web Service
joins all the received signal buffers and converts them
in a single file. We have measured the average time re-
quired for specific ECG signal processing tasks which
runs in the Web Service, in terms of the total duration
of a given record. Thus, for those experiments, we
have used a 2.70 GHz Intel(R) Core(TM) i7-7500U
CPU, 8 GB of RAM and Windows 10 Home, version
1803, for running Web Service. We have obtained the
results illustrated in Table 1.
Table 1: Performance evaluation (processing time) for ex-
tracting ECG signal features within Web Service (percent-
age of the signal duration).
ECG Signal processing
task
Processing time
ECG signal filtering
for denoising
0.129%
QRS complex detection
and delineation
15.43%
T-wave and P-wave detection
and delineation
26.638%
Querying over ECG signal data is a rather com-
plex task, since the data is stored in unstructured bi-
nary files. On the other hand, the MobileECG plat-
form enables the execution of semantic queries. To
demonstrate this fact, we present a query which was
executed on an RDF database generated through the
MobileECG approach.
Sparql Query 01: Which male patients over 60
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
584
ecg:Cycle
health:Drug
+ rdfs:label
ecg:Waveform
ecg:Patient
+ foaf:name
+ foaf:gender
gender
+ ecgo:CPF
ecgo
+ schema:birthDate
+ schema:birthPlace
+ schema:deathData
part_of
g
ecg
:
Patient
me
er
er
nder
F
CPF
:
e
birthDate
birthPla
ecgo:hasECG
ecgo:NormalBeat
ecg:Pwave
ecgo:ProlongQTSyndrome
ecg:Twave
ecg:Wave
+ ecg:peakTime
+ ecg:onsetAmp
+ ecg:offsetAmp
+ ecg:peakAmp
ecg:Segment
ecg:ElementaryForm
+ ecg:onsetTime
ecg
+ ecg:offsetTime
ecg:PRsegment ecg:STsegment
ecg:QRSwave
ec
+
ec
+
ec
g
ecg
:
Wave
+
ecg
:
peakTime
g
pe
+
ecg
:
onsetAmp
g
mp
+
ecg
:
offsetAmp
p
g
p
+
g
ecg
:
pe p
peakAmp
t
men
t
e
c
l
e
ec
g
g
:
Seg
m
e
cle
g
g
g
g
g
g
Cardiopathy
HypertrophicCardiomyopathy
ARVC
g
e
ecg
:
Prescription
+ ecgo:prescriptionDate
prescriptionDate
ecgo
prescriptionDate
+ ecgo:dosage
e
g
ug
health:drug
ecgo:PatientECG
+ ecgo:BMI
ecgo
+ ecgo:pacemaker
ecgo
pacemaker
+ ecgo:systolicPressure
ecgo
systolicPressure
+ ecgo:diastolicPressure
ecgo
+ ecgo:channelsNumber
p
Cardiopat
ecgo:Tobaccoism
ecgo:Alcoholism
g
ecgo
:
ntECG
g
ecgo
:
hasECG
g
g
+
ec
+
ec
G
ecgo:Canal1
ecgo:Canal12
ecgo:LongQTSyndrome
ecgo:SlowRhythm
ecgo:Beat
+ ecgo:beatRate
g
go
:
Bea
t
ecgo:hasBeat
ecgo:NormalRhythm ecgo:FastRhythm
.
.
.
.
.
.
.
.
.
.
ec
g
ecg
+
ecgo
g
ecg
avefor
e
g
ecgo
:
Canal1
rm
.
.
.
.
.
.
.
.
ecgo:PersistentAtrialFibrillation
.
.
.
.
.
.
Figure 4: Scheme of the ontology applied in the data integration and publishing module.
Figure 5: Memory consumption immediately before the
first ECG signal buffer is exhibited on mobile screen.
Figure 6: Memory consumption immediately after the first
ECG signal buffer starts to be exhibited on mobile screen.
years old have had accelerated beats (over 100 beats
per minute) within the ECG?
SELECT ?patient
WHERE {?patient a health:Patient ;
health:age ?age ;
health:gerner ?gender ;
ecga:hasECG ?ecg .
?ciclo ecgo:part of ?ecg .
?ciclo ecgo:hasBeat ?bat .
?bat a ecgo:FastBeat.
FILTER(?age >60 && ?gender = "male")}
Figure 7: Sparql Query 01.
Finally, we have computed the number of gener-
ated RDF triples as we increase the number of full-
Figure 8: Amount of generated RDF triples per number of
full-processed ECG records.
processed ECG records, e.g. ECG signals for which
we have published the corresponding data in the RDF
repository. For this experiment, we have considered
applying 30-min ECG records from MIT-BIH Ar-
rhythmia Database (Goldberger et al., 2000b) at the
following scenarios: 1, 5, 10, 20, 30, 40, 46 full-
processed ECG signals. The obtained results are il-
lustrated in Fig. 8, evidencing a linear increase in the
number of RDF triples, and, therefore, in the use of
memory as we increase the number of full-processed
ECG signals (RDF repository).
5 CONCLUSION
In this work, we introduced MobileECG, a complete
cardiac activity monitoring and prediagnosis solu-
tion. The platform comprises everything from the
MobileECG: An Ubiquitous Heart Health Guardian
585

stage of signal acquisition, through biosensors, Ana-
log/Digital conversion through the Arduino, digital
signal transmission executed by a mobile applica-
tion, to the stage of ECG feature and data extrac-
tion, integrating and publishing, through Web Service
technology. Additionally, a collaborative database
is maintained to integrate and store ECG data com-
ing from heterogeneous sources and data on patients,
treatments, and drugs. The database is published us-
ing Linked Data standards. A case study was ana-
lyzed aiming at to demonstrate MobileECG proper-
ties. As future work, anonymizing patient data tech-
niques should be implemented, aiming at the protec-
tion of individual information. Moreover, machine
learning algorithms will be deployed for classifica-
tion/recognition of arrhythmia and other events.
ACKNOWLEDGEMENTS
Sources of funding: The authors acknowledge the
supports of the Brazilian Research Council, CNPq
(Grant n. 426002/2016-4), and Cear
´
a State Founda-
tion for the Support of Scientific and Technological
Development (BP3-0139-00284.01.00/18).
REFERENCES
Bond, R. R., Finlay, D. D., Nugent, C. D., and Moore, G.
(2011). A review of ECG storage formats. Int. journal
of medical informatics, 80(10):681–697.
da Silva, H. P., Carreiras, C., Lourenc¸o, A., Fred, A.,
das Neves, R. C., and Ferreira, R. (2015). Off-
the-person electrocardiography: performance assess-
ment and clinical correlation. Health and Technology,
4(4):309–318.
Daar, A. S., Singer, P. A., Persad, D. L., Pramming,
S. K., Matthews, D. R., Beaglehole, R., Bernstein,
A., Borysiewicz, L. K., Colagiuri, S., Ganguly, N.,
et al. (2007). Grand challenges in chronic non-
communicable diseases. Nature, 450(7169):494–496.
do Vale Madeiro, J. P., dos Santos, E. M. B. E., Cortez, P. C.,
da Silva Felix, J. H., and Schlindwein, F. S. (2017).
Evaluating gaussian and rayleigh-based mathematical
models for t and p-waves in ecg. IEEE Latin America
Transactions, 15(5):843–853.
Elhaj, F. A., Salim, N., Harris, A. R., Swee, T. T., and
Ahmed, T. (2016). Arrhythmia recognition and classi-
fication using combined linear and nonlinear features
of ecg signals. Computer methods and programs in
biomedicine, 127:52–63.
Goldberger, A. L., Amaral, L. A., Glass, L., Hausdorff,
J. M., Ivanov, P. C., Mark, R. G., Mietus, J. E., Moody,
G. B., Peng, C.-K., and Stanley, H. E. (2000a). Phys-
iobank, physiotoolkit, and physionet. Circulation,
101(23):e215–e220.
Goldberger, A. L., Amaral, L. A., Glass, L., Hausdorff,
J. M., Ivanov, P. C., Mark, R. G., Mietus, J. E., Moody,
G. B., Peng, C.-K., and Stanley, H. E. (2000b). Phys-
iobank, physiotoolkit, and physionet. Circulation,
101(23):e215–e220.
Gonc¸alves, B., Guizzardi, G., and Pereira Filho, J. G.
(2007). An electrocardiogram (ECG) domain ontol-
ogy. In Workshop on Ontologies and Metamodels for
Software and Data Engineering, Jo
˜
ao Pessoa, Brazil.
Gonc¸alves, B., Guizzardi, G., and Pereira Filho, J. G.
(2011). Using an ecg reference ontology for seman-
tic interoperability of ecg data. Journal of Biomedical
Informatics, 44(1):126–136.
Islam, M., Tangim, G., Ahammad, T., Khondokar, M., et al.
(2012). Study and analysis of ecg signal using matlab
&labview as effective tools. International journal of
Computer and Electrical engineering, 4(3):404.
Luz, E. J. d. S., Schwartz, W. R., C
´
amara-Ch
´
avez, G., and
Menotti, D. (2016). Ecg-based heartbeat classification
for arrhythmia detection: A survey. Computer meth-
ods and programs in biomedicine, 127:144–164.
Madeiro, J. P., Cortez, P. C., Marques, J. A., Seisdedos,
C. R., and Sobrinho, C. R. (2012). An innovative ap-
proach of QRS segmentation based on first-derivative,
hilbert and wavelet transforms. Medical engineering
& physics, 34(9):1236–1246.
Mandellos, G. J., Koukias, M. N., Styliadis, I. S., and Lym-
beropoulos, D. K. (2010). e-scp-ecg+ protocol: An
expansion on SCP-ECG protocol for health telemon-
itoring—pilot implementation. International journal
of telemedicine and applications, 2010:1.
Martis, R. J., Acharya, U. R., and Adeli, H. (2014). Current
methods in electrocardiogram characterization. Com-
puters in biology and medicine, 48:133–149.
Ngo, D. and Veeravalli, B. (2014). Applied semantic tech-
nologies in ecg interpretation and cardiovascular di-
agnosis. In Bioinformatics and Biomedicine (BIBM),
2014 IEEE Int. Conf. on, pages 17–24. IEEE.
Orwat, C., Graefe, A., and Faulwasser, T. (2008). Towards
pervasive computing in health care – a literature re-
view. BMC Medical Informatics and Decision Mak-
ing, 8(1):26.
Priyatna, F., Corcho, O., and Sequeda, J. (2014). Formal-
isation and experiences of r2rml-based sparql to sql
query translation using morph. In Proc. of the 23rd
Int. Conf. on World wide web, pages 479–490. ACM.
Venkatachalam, K., Herbrandson, J. E., and Asirvatham,
S. J. (2011). Signals and signal processing for the
electrophysiologist. Circulation: Arrhythmia and
Electrophysiology, 4(6):965–973.
Wen, C., Yeh, M.-F., Chang, K.-C., and Lee, R.-G. (2008).
Real-time ecg telemonitoring system design with mo-
bile phone platform. Measurement, 41(4):463–470.
Worringham, C., Rojek, A., and Stewart, I. (2011). Devel-
opment and feasibility of a smartphone, ecg and gps
based system for remotely monitoring exercise in car-
diac rehabilitation. PloS one, 6(2):e14669.
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
586
