How Complex is to Solve a Hard Problem with Accepting Splicing
Systems
Victor Mitrana
1,3 a
, Andrei P
˘
aun
2 b
and Mihaela P
˘
aun
3 c
1
Department of Information Systems, Polytechnic University of Madrid, Crta. de Valencia km. 7 - 28031 Madrid, Spain
2
Faculty of Mathematics and Computer Science, University of Bucharest, Str. Academiei 14, 010014 Bucharest, Romania
3
National Institute for Research and Development of Biological Sciences, Independentei Bd. 296, Bucharest, Romania
Keywords:
Splicing, Accepting Splicing System, Computational Complexity, Descriptional Complexity, 3-colorability
Problem.
Abstract:
We define a variant of accepting splicing system that can be used as a problem solver. A condition for halting
the computation on a given input as well as a condition for making a decision as soon as the computation has
stopped is considered. An algorithm based on this accepting splicing system that solves a well-known NP-
complete problem, namely the 3-colorability problem is presented. We discuss an efficient solution in terms
of running time and additional resources (axioms, supplementary symbols, number of splicing rules. More
precisely, for a given graph with n vertices and m edges, our solution runs in O(nm) time, and needs O(mn
2
)
other resources. Two variants of this algorithm of a reduced time complexity at an exponential increase of the
other resources are finally discussed.
1 INTRODUCTION
One of the basic phenomenon in genetic engineer-
ing is that by which genetic material is recombined.
This phenomenon, called splicing, allows to geneti-
cally modify a biological entity for different purposes
like: more resistant plants, organisms better adapted
to weather changes, production of hormones, etc. The
chemicals involved in the recombination of DNA se-
quences are two types of enzymes: restriction en-
zymes which cut the DNA at specific sites (called
recognition sites) yielding two fragments with the so-
called “sticky ends”, and ligase which rejoin frag-
ments with sticky ends. A computational model based
on an operation abstracted from the splicing operation
described above has been defined in (Head, 1987).
The model viewed as a language generating device
is called splicing system. Roughly speaking, the
two DNA molecules are represented by strings while
the restriction enzymes are represented by quadruples
of strings, called splicing rules, indicating the sites
where the two strings are to be cut. The compatibility
for rejoining is defined by the fact that two fragments
a
https://orcid.org/0000-0002-1457-8933
b
https://orcid.org/0000-0002-1644-8198
c
https://orcid.org/0000-0002-3342-9140
can be rejoined if they were obtained by applying the
same splicing rule. The splicing operation is illus-
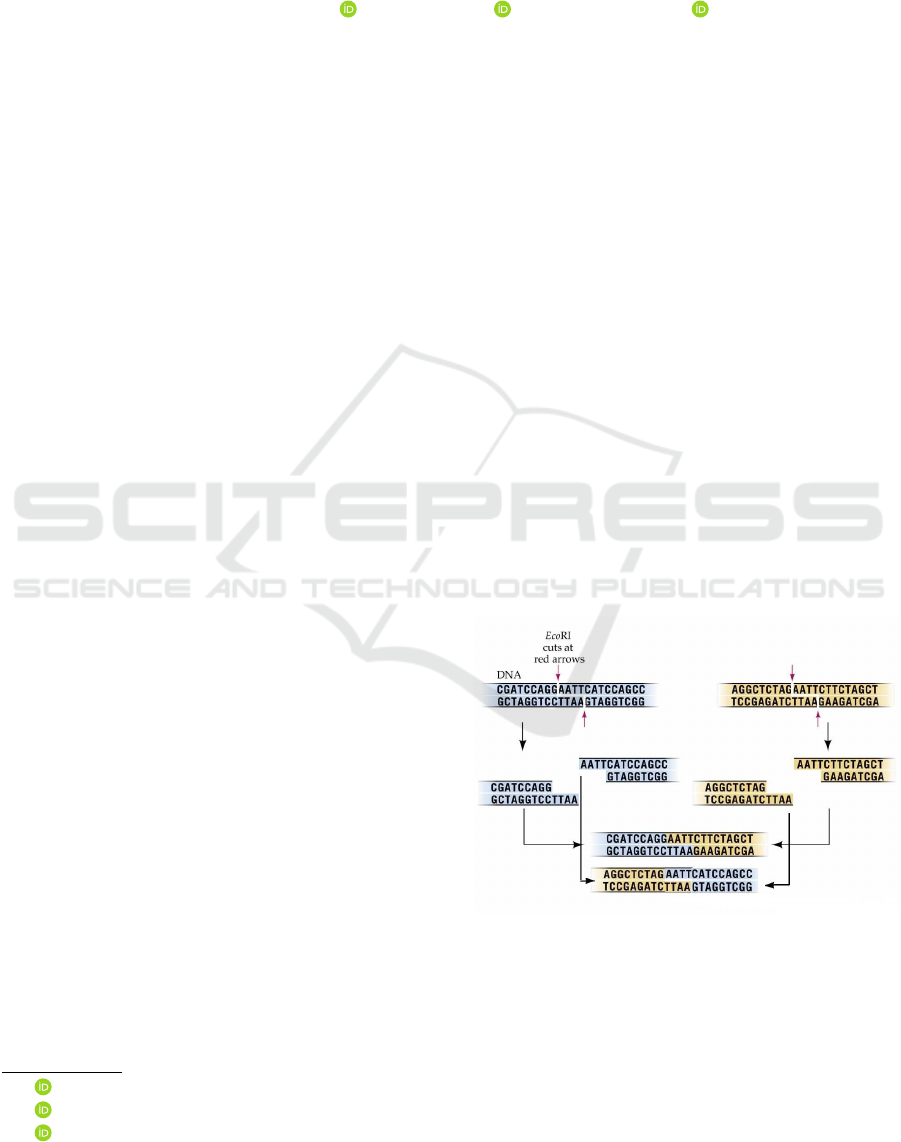
trated in Figure 1.
Figure 1: Splicing operation.
Most research in the area of splicing systems
has been focused on defining different types of such
systems and investigating their computational power
from a language generating point of view. For several
years, it was an open problem whether or not splic-
ing systems are more powerful than finite automata,
from a computational power point of view. The prob-
lem was first solved in (Culik and Harju, 1991), and
Mitrana, V., P
˘
aun, A. and P
˘
aun, M.
How Complex is to Solve a Hard Problem with Accepting Splicing Systems.
DOI: 10.5220/0007715900270035
In Proceedings of the 4th International Conference on Complexity, Future Information Systems and Risk (COMPLEXIS 2019), pages 27-35
ISBN: 978-989-758-366-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
27

later on, a constructive proof was proposed in (Pixton,
1996). In these papers it was proved that splicing sys-
tems are strictly weaker that finite automata. In order
to increase the computational power of these systems,
many variants have been defined and investigated; we
mention here just a few of them: distributed splicing
systems (Csuhaj-Varj
´
u et al. 1996)), extended splic-
ing systems (P
˘
aun et al. 1996), splicing systems with
multisets (Denninghoff and Gatterdam, 1989), splic-
ing systems with permitting and forbidding contexts
(Freund et al. 1999), splicing systems with a regular
set of rules (P
˘
aun, 1996), programmed and evolving
splicing systems (P
˘
aun et al. 1997). Under certain
circumstances, splicing systems are shown to be able
to simulate Turing machines, see (P
˘
aun et al. 1998).
This result suggests the possibility to consider splic-
ing systems as theoretical models of programmable
universal DNA computers based on the splicing op-
eration. Two comprehensive surveys can be found in
(Head et al. 1997) and (P
˘
aun et al. 1998).
Splicing systems working with circular strings are
inspired by a recombinant behavior of circular DNA
in plasamids. Two variants of circular splicing sys-
tems have been introduced in (Siromoney et al. 1992)
and (Pixton, 1995)
Networks with nodes hosting splicing processors
(NSP) have been considered in (Manea et al. 2007).
The NSP model resembles some features of another
distributed computing system, namely test tube dis-
tributed splicing systems introduced in (Csuhaj-Varj
´
u
et al. 1996)) and further investigated in (P
˘
aun, 1998).
A characterization of the complexity class NP as
the class of languages accepted by restricted NSPs
in polynomial time was proposed. Furthermore, a
similar characterization was proposed for the class
PSPACE as the class of languages accepted by re-
stricted NSPs with at most polynomial length of the
strings used in the derivation. In (Manea et al. 2006)
it was proved that NSPs (unrestricted, this time) of
constant size accept all recursively enumerable lan-
guages, and can solve all problems in NP in polyno-
mial time; also an universality result for NSPs was
proposed. In both cases the number of nodes needed
was 7. In (Loos et al. 2009) it is shown that compu-
tational completeness can be achieved by NSPs of 2
nodes. In the same paper a more involved construc-
tion, showing that NSPs of size 3 can simulate the
computations of a non-deterministic Turing machine
in parallel, is presented.
A new protocol of cooperation in networks of
splicing processors, namely the polarization, has been
introduced in (Bordihn et al. 2017) and further in-
vestigated in (Bordihn et al. 2018). The polariza-
tion protocol requires that each node has a polariza-
tion defined as a value in the set {+,0,−} (posi-
tive, neutral, negative) and the data have assigned a
value in the same set which is computed by a valu-
ation mapping. Now the communication is based on
the compatibility between the polarization of nodes
and the value of data. A quantitative generalization
of these networks, called networks of splicing pro-
cessors with evaluation sets have been introduced in
(G
´
omez-Canaval et al. 2016). In a network of splic-
ing processors with evaluation sets, unlike in all the
aforementioned cases, the valuation mapping returns
the exact value computed for data. The new model re-
fines the communication protocol based on polariza-
tion discussed above, in which each polarization may
be viewed as one of the intervals of integers (−∞, 0),
{0}, and (0,∞), to a more complex polarization based
on more intervals. The new model tries to resemble
the biological concept of the concentration gradient
in a solution. Now, the strategy of communication
between two nodes follows the compatibility between
their accepting values with respect to some predefined
evaluation sets and the values of the data computed
by a valuation mapping. More precisely, the values
of data have to be in the set of accepting values with
respect to some evaluation set of symbols. This new
communication protocol might be interpreted as the
movement of molecules or particles along a concen-
tration gradient between two areas.
Other fundamental algorithmic problems regard-
ing splicing systems, namely recognition and synthe-
sis are considered in a series of papers starting with
(Bonizzoni and Mauri, 2005). Recognition problem
refers to the design of an algorithm able to decide
whether or not a given regular language is a splicing
language, while the synthesis problem refers to the
possibility of effectively constructing a splicing sys-
tem that generates a given regular language.
A surprising application of splicing systems which
are used an automatic music composer is proposed in
(De Felice et al. 2017). This approach might be seen
as a possible bio-inspired strategy for automatic mu-
sic composition. The model in the aforementioned
work is tailored on 4-voice chorale-like music. Along
these lines, a new approach for both recognition and
automatic composition of styles for musical collec-
tives is designed in (De Prisco et al. 2017). Infor-
mally speaking, such a system exploits a machine
learning recognizer, based on one-class support vec-
tor machines and neural networks for style recogni-
tion, and a splicing composer, for music composition
(in the style of the whole collective).
Starting from some ideas considered in (Loos et
al. 2006), a novel look at splicing systems is pro-
posed in (Mitrana et al. 2010), namely these systems
COMPLEXIS 2019 - 4th International Conference on Complexity, Future Information Systems and Risk
28

are considered as accepting devices. More precisely,
one defines the concept of accepting splicing system.
The rough idea is to consider that an accepting splic-
ing system receives as input a string which enters, to-
gether with a given finite set of axioms, into an it-
erated splicing process until a string from a specific
finite sets of strings is produced. When such a string
is produced, the computational process halts. In this
paper, we consider the case when a decision can be
made as soon as the computation halts. To this aim,
we consider a two finite sets of strings, a set of halting
strings such that when a halting string is produced the
computation halts, and a set of accepting strings. For
making a decision, it suffices to check whether or not
at least one accepting string has been obtained.
The paper is organized as follows. In the next sec-
tion we present the basic definitions and the main con-
cept, that of an accepting splicing system viewed as a
problem solver. We define one computational com-
plexity measure , similar to the time complexity in
most models, which measures the number of splicing
steps necessary for a computation to halt. Then we
define a few descriptional complexity measures like:
the number of axioms, the number of supplementary
symbols, the number of rules, the length of the longest
splicing rule, and the number of halting or accepting
strings. In the third section, we consider a very well-
known NP-complete problem, that of 3-colorability,
and propose an algorithm based on accepting splicing
systems (an AccSS-algorithm) to solve this problem.
We give both informal explanations as well as formal
definitions on the way our algorithm works. After-
wards, we evaluate the computational and descrip-
tional complexity of our algorithm. Thus, we prove
that given a graph with n vertices and m edges, there is
an AccSS-algorithm that decides the 3-colorability for
this graph in O(nm) time and O(mn
2
) other resources
(axioms, symbols, splicing rules). Finally, we discuss
two variants of this algorithm that need a reduced time
complexity, but this is achieved at an exponential in-
crease of the other resources.
2 MAIN CONCEPT
We start by summarizing the notions used throughout
the paper. For all undefined notions the reader may
consult (Rozenberg and Salomaa, 1997). An alphabet
is a finite and nonempty set of symbols. The cardinal-
ity of a finite set A is written card(A). Any finite se-
quence of symbols from an alphabet V is called string
over V . The set of all strings over V is denoted by V
∗
and the empty string is denoted by λ. The length of
a string x is denoted by |x| while al ph(x) denotes the
minimal alphabet W such that x ∈ W
∗
.
A splicing rule over V is 4-tuple [(u
1
,u
2
);(u
3
,u
4
)],
with u
1
,u
2
,u
3
,u
4
∈ V
∗
. For a splicing rule r =
[(u
1
,u
2
);(u
3
,u
4
)] and a pair of strings x, y ∈ V
∗
, we
write
σ
r
(x,y) = {y
1
u
3
u
2
x
2
| x = x
1
u
1
u
2
x
2
,y = y
1
u
3
u
4
y
2
}
∪{x
1
u
1
u
4
y
2
| x = x
1
u
1
u
2
x
2
,y = y
1
u
3
u
4
y
2
}
for some x
1
,x
2
,y
1
,y
2
∈ V
∗
. The analogy with the
situation depicted in Figure 1 is immediate: the
two strings represent the two double stranded DNA
molecules, while the restriction enzyme is repre-
sented by the splicing rule. This definition is extended
to a set of splicing rules R and a set of strings A by
σ
R
(A) =
[
r∈R
[
w
1
,w
2
∈A
σ
r
(w
1
,w
2
).
When the set of splicing rules is clear, we omit the
subscript.
Note that this definition assumes that arbitrarily
many copies of all strings in A are available for splic-
ing. This is a natural assumption from the biological
point of view because every such string, encoding a
DNA molecule, can be duplicated sufficiently many
times by the PCR procedure, a well defined technique
for amplifying the genetic material (Rabinov, 1996).
We now recall the definition and terminology for
accepting splicing systems, following (Mitrana et al.
2010). It is worth noting that the definition presented
here differs from those in (Mitrana et al. 2010) and
(Arroyo et al. 2013) because we use here accepting
splicing systems as decision problem solvers. To this
aim, we need a condition for halting the computation
and a condition for making the decision. Furthermore,
we need that all systems halt on every input An ac-
cepting splicing system (AccSS for short) is a 8-tuple
Γ = (V,U,<,>, A, R, H, F),
where V is the input alphabet, U is the working al-
phabet, V ⊂ U, <,> are two symbols in U \V called
endmarkers, A ⊆ U
∗
is the set of initial strings (ax-
ioms), while R is a set of splicing rules over U. Fur-
thermore, H and F are finite sets of strings over U;
the elements of H are called halting strings and those
of F are called accepting strings.
Let Γ = (V,U, <, >, A,R,H,F) be an AccSS and a
string w ∈ V
∗
; we define the following iterated splic-
ing
τ
1
R
(A,w) = σ
R
(A ∪ {w}),
τ
i+1
R
(A,w) = σ
R
(τ
i
R
(A,w) ∪ A),i ≥ 1.
Again, the subscript R may be omitted when it is self-
understood. Note that if no splicing rule is applicable
to a string different than an axiom at some step, it
How Complex is to Solve a Hard Problem with Accepting Splicing Systems
29
disappears from the set of all available strings in the
next splicing step. A computation of Γ on w is the
sequence of sets (τ
i
R
(A,w))
i≥0
. Such a computation is
finite (we say that Γ halts on w ∈ V
∗
) if there exists
k ≥ 0 such that τ
k
R
(A,w)∩ H 6=
/
0. An input string w is
accepted by Γ if Γ halts on w as above and τ
k
R
(A,w) ∩
F 6=
/
0; otherwise, w is rejected. A string W is decided
by Γ if Γ halts on w. A set of strings L is decided by Γ
if Γ accepts all strings in L and rejects all strings that
do not belong to L. As we want to consider AccSS as
problem solvers, we assume that every AccSS in what
follows halts on every input string.
We now define the following computational com-
plexity measure for an AccSS Γ = (V,U,<,>,A,R,F)
that halts on every input:
Time
Γ
(w) = min{k | τ
k
R
(A,w) ∩ H 6=
/
0},
Time
Γ
(n) = max{Time
Γ
(w) | |w| = n}.
It is worth noting that a similar measure with a differ-
ent definition was introduced in (Loos and Ogihara,
2007) for generating splicing systems.
We also define the following descriptional complexity
measures for an AccSS Γ = (V,U, <,>,A,R,F) that
halts on every input:
Ax(Γ) = card(A),
Symb(Γ) = card(U \V),
NSplice(Γ) = card(R),
LSplice(Γ) = max{|u
1
u
2
u
3
u
4
| |
[(u
1
,u
2
);(u
3
,u
4
)] ∈ R},
Final(Γ) = max(card(H),card(F)).
We mention here another work considering some
descriptional complexity measures for generating
splicing systems, namely (Loos et al. 2008). In this
paper, the measures are the total length of the rules
and the size of the initial language. These measures
are related to the size of the minimal finite automata
accepting the same language.
We discuss below the potential of AccSS to solve
complex decidability problems. We now formally de-
fine what an AccSS-algorithm is. A decidability prob-
lem P is said to be solved in time O(t(n)) by AccSSs
if there exists a family G of AccSSs, which can be
constructed by an effective procedure, such that for
each instance x of size O(n) of the problem P , en-
coded by a string w, one can effectively construct an
AccSS Γ(x) ∈ G such that the computation of Γ(x )
on w halts in time O(t(n)) with at least an accepting
string obtained during the computation if and only if
the x is a true instance. This effective construction is
called an O(t(n)) time AccSS-algorithm for the con-
sidered problem.
3 AN AccSS -ALGORITHM FOR
THE 3-COLORABILITY
PROBLEM
Given a connected undirected graph without loops,
the 3-colorability problem is to decide whether or not
is it possible to color each vertex by using three colors
(say, red, blue, and green) such that no two adjacent
vertices are of the same color. For instance, the graph
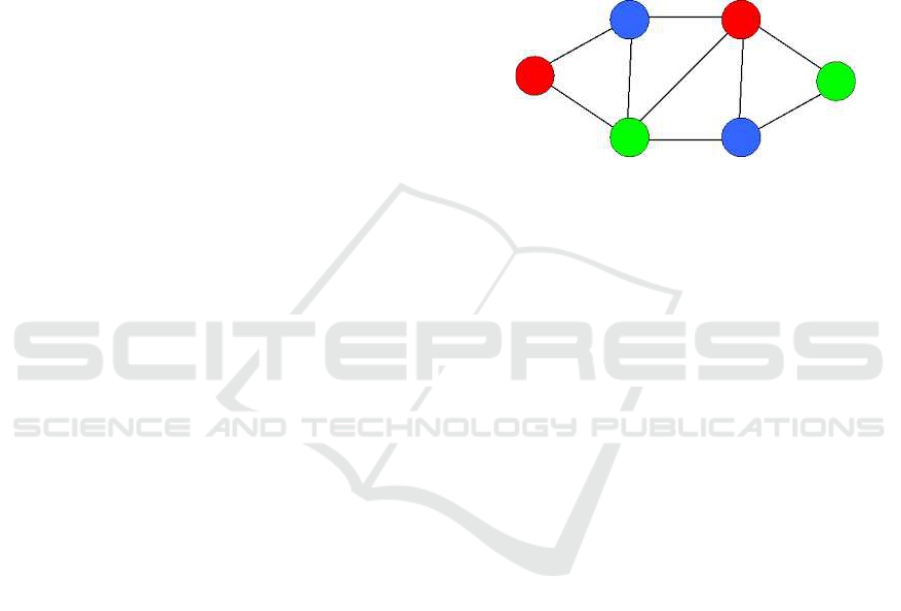
in Figure 2 is correctly colored.
Figure 2: Graph correctly colored with three colors.
The problem is known to be NP-complete (Garey
and Johnson, 1979). Let Y = (B, Q) be a graph with
set of vertices B = {1,2,...,n} and set of edges Q =
{e
1
,e
2
,..., e
m
}, where each e
k
is given in the form
e
k
= {k
1
,k
2
}, for some 1 ≤ k
1
6= k
2
≤ n.
We now present an AccSS-algorithm for the 3-
colorability problem and analyze its computational
and descriptional complexity.
3.1 AccSS-algorithm
The input string is < a
n
e
1
e
2
...e
m
>. The algorithm
has two main phases. In the first phase all strings en-
coding graph colorings, namely strings of the form
< x
1
x
2
...x
n
e
1
e
2
...e
m
>,
where x ∈ {r,b,g} are produced in parallel by splic-
ing. The meaning of this encoding is that the node i
is colored by red, blue, green if r
i
,b
i
,g
i
, respectively.
In the second phase, the graph colorings obtained in
the first phase are to be checked for correctness, that
is they are correct colorings. All the strings encoding
correct colorings will lead simultaneously to accept-
ing strings. If the graph cannot be correctly colored,
the computation will halt with no accepting string ob-
tained.
A complete formal solution is presented in the
next two sections. The general idea is that used by
most of the DNA-based algorithms: a first phase is
dedicated to the generation of solution space, and a
second phase which searches the correct solutions by
filtering the whole solution space. The parameters of
Γ are to be constructed dynamically as follows. We
give the splicing rules for each phase, hence the set
COMPLEXIS 2019 - 4th International Conference on Complexity, Future Information Systems and Risk
30

of splicing rules R of Γ is the set containing all these
rules. Similarly, the set of axioms A contains all the
axioms presented for each phase. The working alpha-
bet U is formed by all symbols that appear in the ax-
ioms and splicing rules listed for each phase.
3.1.1 First Phase
The first phase can be accomplished with the fol-
lowing splicing rules and axioms distributed in three
groups, each of them having subgroups:
1. [(<,a);(< C
1
,#)], and < C
1
# is an axiom.
2.1. [(†,C
i
a);(X
i
,Y
i
)],† ∈ {<} ∪ {b
i−1
,g
i−1
,r
i−1
}
and X
i
Y
i
is an axiom,1 ≤ i ≤ n.
2.2. [(X
i
C
i
a,†
1
);(X
0
i
†
2
C
i+1
,#)],†
1
∈ {a, e
1
},
†
2
∈ {b
i
,g
i
,r
i
},and X
0
i
†
2
C
i+1
# is an axiom,
1 ≤ i ≤ n − 1.
2.3. [(X
n
C
n
a,e
1
);(X
0
n
†,#)],† ∈ {b
n
,g
n
,r
n
},
and X
0
n
†# is an axiom.
2.4. [(<,Y
i
);(#,Y
0
i
)],and #Y
0
i
is an axiom,1 ≤ i ≤ n.
3.1. [(<,Y
0
1
);(X
0
1
,†C
2
a)],† ∈ {b
1
,g
1
,r
1
}.
3.2. [(†
1
,Y
0
i−1
);(X
0
i−1
,†
2
C
i
a)],†
1
∈ {b
i−2
,g
i−2
,r
i−2
},
†
2
∈ {b
i−1
,g
i−1
,r
i−1
},3 ≤ i ≤ n.
3.3. [(†
1
,Y
0
n
);(X
0
n
,†
2
e
1
)],†
1
∈ {b
n−1
,g
n−1
,r
n−1
},
†
2
∈ {b
n
,g
n
,r
n
}.
We explain how an arbitrary string
< x
1
x
2
...x
n
e
1
e
2
...e
m
>, with x ∈ {r,b,g}, is
produced from the input string w =< a
n
e
1
e
2
...e
m
>
by using the splicing rules listed above. The first
splicing rule that can be applied is the rule in
the group 1 which can be applied to the pair of
strings formed by the input string and the axiom
< C
1
#. This splicing step yields two new strings
< C
1
a
n
e
1
e
2
...e
m
> and < #. The string < # cannot
be involved in further splicing steps, hence it will
disappear in the next splicing step. Therefore, we
can say that the role of the first splicing step is to
introduce the symbol C
1
before the first occurrence
of a in the input string. The rough idea of the next
splicing steps is that symbols C
i
, 1 ≤ i ≤ n, scan
the input string from left to right following the next
procedure: each C
i
scans the symbol a next to it, re-
places this occurrence of a by a symbol in {b
i
,g
i
,r
i
},
moves to the right of this symbol, and changes itself
into either C
i+1
, if i < n or it is removed, if i = n.
Inductively, this happens as follows. Assume that the
current string is < x
1
x
2
...x
i−1
C
i
a
n−i+1
e
1
e
2
...e
m
>,
with x ∈ {r, b,g}, and i < n. A rule in the subgroup
2.1 is applied to the current string and the axiom
X
i
Y
i
. Two strings are obtained < x
1
x
2
...x
i−1
Y
i
and
X
i
C
i
a
n−i+1
e
1
e
2
...e
m
>. Now, a rule in the subgroup
2.2 is applied to X
i
C
i
a
n−i+1
e
1
e
2
...e
m
> and to
one of the axioms X
0
i
†C
i+1
#, where † ∈ {b
i
,g
i
,r
i
}.
Thus three new words are obtained simultaneously:
X
i
b
i
C
i+1
a
n−i
e
1
e
2
...e
m
>, X
i
g
i
C
i+1
a
n−i
e
1
e
2
...e
m
>,
and X
i
r
i
C
i+1
a
n−i
e
1
e
2
...e
m
>. In the same splicing
step, a rule in subgroup 2.4 is applied to an axiom #Y
0
i
and to < x
1
x
2
...x
i−1
Y
i
. As a result of this splicing the
string < x
1
x
2
...x
i−1
Y
0
i
is obtained. Now the process
of coloring the node i ends by applying a splicing rule
in the subgroup 3.2 to each of the three strings afore-
mentioned and to one of the strings < x
1
x
2
...x
i−1
Y
0
i
.
All the strings < x
1
x
2
...x
i
C
i+1
a
n−i
e
1
e
2
...e
m
> are
now produced.
It is worth mentioning that the rules in subgroups
2.3 and 3.3 replace the rules in 2.2 and 3.2, respec-
tively, when i = n in the above discussion. Now the
whole process described above is iterated until all
strings < x
1
x
2
...x
n
e
1
e
2
...e
m
>, with x ∈ {r, b,g} are
produced.
A short discussion is necessary here. First, it is
easy to note that all the other strings that are by-
products of the splicing rules applied in the process
discussed above will not be involved in further splic-
ing steps because they cannot be cut by any splicing
rule. Second, when applying a rule in the third group
the two strings that are joined do not necessarily come
from the same string cut with rules in the subgroup
2.1, but this fact does not introduce any illegal string,
that is all the strings obtained after applying rules in
the third group are still valid partial colorings of the
graph. Note that by partial coloring we mean that ev-
ery node of the graph is either colored with exactly
one color or it is not colored yet.
3.1.2 Second Phase
The second phase starts when the first
phase is finished, that is when all strings
< x
1
x
2
...x
n
e
1
e
2
...e
m
>, with x ∈ {r, b, g} are
produced. Informally, the second phase checks
whether every such string as above is a correct
coloring. This means that for each e
k
= {k
1
,k
2
},
x
k
1
6= x
k
2
holds. This is to be done by checking this
condition for e
1
, e
2
, ..., e
m
.
For a better understanding of this checking pro-
cess we need some preparatives. For every symbol
e
k
encoding the edge {k
1
,k
2
} we write e
k
,
b
e
k
, and
e
e
k
if one of the two nodes k
1
,k
2
is colored blue, green,
and red, respectively, while the other node is not col-
ored yet. Now the checking process is to be accom-
plished as follows. The prefix < x
1
x
2
...x
n
of each of
the strings < x
1
x
2
...x
n
e
1
e
2
...e
m
>, with x ∈ {r,b,g}
will be scanned by every symbol e
k
, 1 ≤ k ≤ m, from
right to left such that:
(i) If e
k
meets b
k
1
or b
k
2
to its left, then e
k
is replaced
How Complex is to Solve a Hard Problem with Accepting Splicing Systems
31

by e
k
.
(ii) If e
k
meets g
k
1
or g
k
2
to its left, then e
k
is replaced
by
b
e
k
.
(iii) If e
k
meets r
k
1
or r
k
2
to its left, then e
k
is replaced
by
e
e
k
.
(iv) If e
k
meets a symbol in {g
k
1
,r
k
1
,g
k
2
,r
k
2
} to its
left, then e
k
is deleted.
(v) If
b
e
k
meets a symbol in {b
k
1
,r
k
1
,b
k
2
,r
k
2
} to its
left, then
b
e
k
is deleted.
(vi) If
e
e
k
meets a symbol in {g
k
1
,b
k
1
,g
k
2
,b
k
2
} to its
left, then
e
e
k
is deleted.
(vii) In all the other cases, e
k
,
b
e
k
, and
e
e
k
are replaced
by a new symbol Q, which will continue to scan
the rest of the string until either the left endmarker
or another occurrence of Q is met.
(viii) If a symbol x
r
was scanned by all symbols e
k
, 1 ≤
k ≤ m, then it is deleted.
We give now the formal definitions of splicing
rules, distributed in groups and subgroups, and those
of axioms.
1. [(♣,x
i
Z
j
);(♦
i, j
,♥
i, j
)] and ♦
i, j
♥
i, j
is an axiom,
♣ ∈ {<, Q} ∪
{x
i−1
, if i > 1
/
0, if i = 1
Z
j
∈ {e
j
,e
j
,
b
e
j
,
e
e
j
},1 ≤ j ≤ m,1 ≤ i ≤ n.
2.1. [(♣,♥
i, j
);(#,♥
0
i, j
)] and #♥
0
i, j
is an axiom,
♣ ∈ {<, Q} ∪
{x
i−1
, if i > 1
/
0, if i = 1
1 ≤ j ≤ m,1 ≤ i ≤ n.
2.2. [(♦
i, j
x
i
Z
j
,x
i+1
);(♦
0
i, j
Y
j
x
i
,#)],
and ♦
0
i, j
Y
j
x
i
# is an axiom,x ∈ {b, g,r},
Z
j
∈ {e
j
,e
j
,
b
e
j
,
e
e
j
} and
• if i /∈ { j
1
, j
2
}, then Y
j
= Z
j
,
• if i ∈ { j
1
, j
2
}, and Z
j
= e
j
, then
Y
j
=
Z
j
, if x = b,
b
Z
j
, if x = g,
e
Z
j
, if x = r,
• if i ∈ { j
1
, j
2
}, then Y
j
= λ provided that
x ∈ {g,r} and Z
j
= e
j
,
x ∈ {b, r} and Z
j
=
b
e
j
,
x ∈ {b, g} and Z
j
=
e
e
j
,
•Y
j
= Q, in all the other cases,
1 ≤ j ≤ m,1 ≤ i ≤ n.
3. [(♣,♥
0
i, j
);(♦
0
i, j
,Z
j
x
i
P)],Z
j
∈ {Q, e
j
,e
j
,
b
e
j
,
e
e
j
,λ},
♣ ∈ {<, Q} ∪
{x
i−1
, if i > 1
/
0, if i = 1
P 6= #, 1 ≤ j ≤ m,1 ≤ i ≤ n.
In order to clarify the roles of these splicing rules
we show how they are used for fulfilling the con-
ditions (i)-(vi) and partly (vii) defined above. Let
< αx
i
Z
j
β, for some strings α,β, be an arbitrary string
available at this splicing step. We take the axiom
♦
i, j
♥
i, j
and apply a rule in group 1 to this pair of
strings. The two new strings are: α♥
i, j
and ♦
i, j
x
i
Z
j
β.
Now, in the same splicing step, α♥
i, j
is transformed
into α♥
0
i, j
by using a rule in subgroup 2.1, while
♦
i, j
x
i
Z
j
β is spliced by a rule in subgroup 2.2. As one
can easily see, all the conditions (i)-(vi) are consid-
ered in the definition of rules in subgroup 2.2. More-
over, the condition for introducing the new symbol Q
is also considered. The meaning for introducing Q is
that the coloring encoded by the string < αx
i
Z
j
β is
illegal for the edge e
j
.
For the second part of condition (vii) and condi-
tion (viii) we give only some informal explanations
without formally defining the rules, which can be eas-
ily defined by the reader. The condition (viii) assumes
that a symbol x
i
reached the right endmarker >. This
symbol can be easily deleted now by using one splic-
ing rule that cuts the string just before x
i
> and re-
places this segment by > only. As soon as a sym-
bol Q has still to scan symbols from right to left, this
can be easily done in three simple splicing steps using
rules similar to those in subgroups 2.1 (first splicing
step), 2.2 and 2.4 (second splicing step), and 3.2 (third
splicing step).
When no splicing rule can be applied anymore,
only strings of the form < Q
s
>, for some 1 ≤ s ≤ m
and possibly one string more, namely <>, have been
produced. By the aforementioned explanations, we
conclude that each string obtained in the first phase,
which does not encode a legal coloring, will eventu-
ally lead, at the end of the second phase, to a string
< Q
s
>, for some 1 ≤ s ≤ m. On the other hand,
each string obtained in the first phase, which does en-
code a legal coloring, will eventually lead, at the end
of the second phase, to <>. It is clear that, if <>
is obtained, it is produced before any of the strings
< Q
s
>. Now, it suffices to reduce, by splicing, all
the strings < Q
s
> to just one, namely < Q > and con-
sider the set of of halting strings H = {<>,< Q >}
and F = {<>}, and the construction of the AccSS-
algorithm is complete.
3.1.3 Evaluating Time Complexity
We first evaluate the time complexity of the AccSS
constructed in the Section 3. Insertion of C
1
takes one
step, each coloring of a node takes 3 steps, hence the
first phase takes a total number of 3n + 1 steps. Note
that each splicing step may be formed by a number of
individual splicings that are done in parallel. Again
we make use of the assumption that the each string
appears in a sufficient number of copies such that if
COMPLEXIS 2019 - 4th International Conference on Complexity, Future Information Systems and Risk
32

a splicing rule is applicable to a certain string, it can
effectively be used.
The second phase requires, in the worst case, that
the prefix < x
1
x
2
...x
n
to be scanned by every symbol
e
i
, 1 ≤ i ≤ m. Each such symbol can scan the prefix
in 3n splicing steps. As there are m such symbols,
the total number of steps is 3nm. Furthermore, the
number of steps necessary to delete the symbols x
i
as required by the condition (viii) is n. Finally, the
number of steps to reduce < Q
s
> to < Q > is at most
m.
We now conclude that the time complexity of our
algorithm is O(nm)
3.1.4 Evaluating Descriptional Complexity
We now evaluate the descriptional complexity mea-
sures defined in Section 2. In the first phase we have
the followings:
• The set of axioms used in the first phase is
A
1
= {C
1
#,X
n
b
n
#,X
n
g
n
#,X
n
r
n
#} ∪
{X
i
Y
i
| 1 ≤ i ≤ n} ∪ {X
0
i
†C
i+1
# |
† ∈ {b
i
,g
i
,r
i
},1 ≤ i ≤ n − 1} ∪
{#Y
0
i
| 1 ≤ i ≤ n}.
Therefore, the number of axioms is 5n + 1.
• We use the following set of symbols:
S
1
= {<,>,#} ∪ {C
i
,X
i
,Y
i
,X
0
i
,Y
0
i
| 1 ≤ i ≤ n} ∪
{b
i
,g
i
,r
i
| 1 ≤ i ≤ n}.
Therefore, the number of new symbols is 8n + 3.
• The number of splicing rules in each subgroup is
given in Table 1. Consequently, there are 16n − 5
Table 1: The number of splicing rules.
Subgroup Number of splicing rules
1 1
2.1 3n
2.2 3(n − 1)
2.3 3
2.4 n
3.1 3
3.2 9(n − 2)
3.3 9
splicing rules.
• The maximal length of a splicing rule is 8.
For the second phase an exact evaluation of all
measures is rather cumbersome, but we can evaluate
the order of these measures.
• The number of axioms used is at most 14n.
• The number of symbols used is 6nm + 4m + 2.
• The number of splicing rules is at most 5mn(n +
1) + 12mn + n + m
• The maximal length of a splicing rule is 8.
Finally, Final(Γ) = 2.
We conclude that
Ax(Γ) = O(n),
Symb(Γ) = O(mn),
NSplice(Γ) = O(mn
2
),
LSplice(Γ) = 8,
Final(Γ) = 2.
3.2 Two Variants
It is worth noting that the algorithm presented above
can be accelerated to O(n) time at an exponential cost
of rules and axioms (O(5
m
n)), and a linear cost of the
length of splicing rules. It can also be accelerated to
O(n) time at the same cost of rules, axioms and num-
ber of symbols but a constant length of the splicing
rules. We briefly discuss these variants in the sequel.
The second phase can be changed as follows. In-
stead of scanning the prefix from right to left with
each symbol e
i
, we can do this scanning process with
the whole segment e
1
e
2
...e
m
. The splicing rules for
doing this are:
[(♣,x
i
Z
1
Z
2
...Z
m
);(#,Y
1
Y
2
...Y
m
x
i
)],
where ♣ =
<, if i = 1
x
i−1
, if i > 1,
#Y
1
Y
2
...Y
m
x
i
is an an axiom,
Z
j
∈ {Q, e
j
,e
j
,
b
e
j
,
e
e
j
}, and
• if i /∈ { j
1
, j
2
}, then Y
j
= Z
j
,
• if i ∈ { j
1
, j
2
}, and Z
j
= e
j
, then
Y
j
=
Z
j
, if x = b,
b
Z
j
, if x = g,
e
Z
j
, if x = r,
• if i ∈ { j
1
, j
2
}, then Y
j
= λ provided that
x ∈ {g,r} and Z
j
= e
j
,
x ∈ {b, r} and Z
j
=
b
e
j
,
x ∈ {b, g} and Z
j
=
e
e
j
,
•Y
j
= Q, in all the other cases,
1 ≤ j ≤ m,1 ≤ i ≤ n.
As one can easily see, at every splicing step the
segment Z
1
Z
2
...Z
m
is updated with respect to the
value of x
i
and moves to the left. Therefore, after
n splicing steps either a segment consisting of some
occurrences of Q reaches the left endmarker, or the
string is <>.
As far as the descriptional complexity of this vari-
ant, we note that the number of splicing rules as well
How Complex is to Solve a Hard Problem with Accepting Splicing Systems
33

as that of axioms is O(5
m
n). Furthermore, the maxi-
mal length of a splicing rule is 2m + 4.
Another variant could consider symbols for en-
coding the segments, hence the number of symbols
explodes exponentially but the maximal length of a
splicing rule remains constant.
As a consequence of the constructions and com-
plexity evaluations above, we can conclude
Theorem 1. Let G be a graph with n vertices and m
edges, without loops.
1. There is an AccSS-algorithm that decides the 3-
colorability for G in O(nm) time and O(mn
2
)
other resources (axioms, symbols, splicing rules).
2. There is an AccSS-algorithm that decides the 3-
colorability for G in O(n) time, O(5
m
n) axioms
and splicing rules, and O(m) maximal length of
the splicing rules.
3. There is an AccSS-algorithm that decides the 3-
colorability for G in O(n) time, O(5
m
n) axioms,
symbols, and splicing rules, and a constant maxi-
mal length of splicing rules.
We also want to stress that the technique used in
the construction of the above AccSS-algorithm can
be employed for constructing similar algorithms for
other intractable problems.
4 CONCLUSIONS AND FURTHER
WORK
A new variant of accepting splicing system that can be
used as a problem solver was introduced. This model
may be used as a problem solver because the condi-
tion for halting the computation on a given input is
accompanied by a condition for making a decision as
soon as the computation has stopped. An algorithm
based on this accepting splicing system that solves
a well-known NP-complete problem, namely the 3-
colorability problem is presented. The time efficiency
of this solution is analyzed together with some de-
scriptional complexity measures for axioms, supple-
mentary symbols, number of splicing rules. More pre-
cisely, for a given graph with n vertices and m edges,
our solution runs in O(nm) time, and needs O(mn
2
)
other resources. Two variants of this algorithm of a
reduced time complexity at an exponential increase
of the other resources are also discussed.
We discuss here a few possible ways, that ap-
pear attractive to us, of continuing the work started
here. From a theoretical point of view, a complete in-
vestigation of the computational power of this model
seems to be of interest. This investigation could con-
tribute to a more complete picture of the study started
in (Mitrana et al. 2010). From a more practical
point of view, would like to consider possible soft-
ware implementations of this model that could be
similar to those reported in the literature for other
bio-inspired models. Along these lines, software sim-
ulators for this model using massively parallel plat-
forms for multi-core desktop computers, clusters of
computers and cloud resources similarly to the ap-
proached proposed in (G
´
omez-Canaval et al. 2015)
and (G
´
omez-Canaval et al. 2015) might lead to rel-
evant results. In (G
´
omez-Canaval et al. 2015), it
is shown that massively distributed platforms for big
data scenarios makes them potential candidates for
the development of ultra-scalable simulators able to
simulate different variants of bio-inspired models. In
particular, computing platforms by developing an en-
gine that uses Apache Giraph on top of the Hadoop
platform might be useful. The results of some experi-
ments with such simulators suggest that they might be
amenable to minimize the growth of processing data,
hence to be adapted for models like that discussed
here.
ACKNOWLEDGEMENTS
Work supported by a grant of the Romanian Na-
tional Authority for Scientific Research and Innova-
tion, project number POC P-37-257.
REFERENCES
Arroyo, F., Castellanos, J., Dassow, J., Mitrana, V., and
S
´
anchez-Couso, J.R. (2013). Accepting splicing sys-
tems with permitting and forbidding words. Acta Inf.
50 pp.1–14.
Bonizzoni, P. and Mauri, G. (2005). Regular splicing lan-
guages and subclasses. Theoret. Comput. Sci. 340 pp.
349–363.
Bordihn, H., Mitrana, V., P
˘
aun, A., P
˘
aun, M. (2017). Net-
works of polarized splicing processors. In Theory and
Practice of Natural Computing, TPNC 2017, LNCS
10687, Springer, Berlin, Heidelberg, pp. 165–177.
Bordihn, H., Mitrana, V., Negru, M.C., P
˘
aun, A., P
˘
aun, M.
(2018). Small networks of polarized splicing proces-
sors are universal. Natural Computing, 17 pp. 799–
809.
Csuhaj-Varj
´
u, E., Kari, L. and P
˘
aun, Gh. (1996). Test tube
distributed systems based on splicing. Computers and
AI 15 pp. 211–232.
Culik, K, and Harju, T. (1991). Splicing semigroups of
dominoes and DNA. Discrete Appl. Math. 31 pp. 261-
277.
Denninghoff, K.L. and Gatterdam, R.W. (1989). On the un-
decidability of splicing systems. Intern. J. Computer
Math. 27 pp. 133–145.
COMPLEXIS 2019 - 4th International Conference on Complexity, Future Information Systems and Risk
34

De Felice, C., De Prisco, R., Malandrino, D., Zaccagnino,
G., Zaccagnino, R., and Zizza, R. (2017). Splicing
music composition. Inf. Sci. 385 pp. 196–212.
Freund, R., Kari, L., and P
˘
aun, Gh. (1999). DNA com-
puting based on splicing. The existence of universal
computers. Theory of Computing Syst. 32 pp. 69–112.
Garey, M., and Johnson, D. (1979). Computers and
Intractability: A Guide to the Theory of NP-
Completeness. W. H. Freeman & Co. New York.
G
´
omez-Canaval, S., Mitrana, V., S
´
anchez-Couso, J.S.
(2016). Networks of splicing processors with evalu-
ation sets as optimization problems solvers. Informa-
tion Sciencs 369, 457–466.
G
´
omez-Canaval, S., Ortega, A., Orgaz, P. (2015). Dis-
tributed simulation of NEPs based on-demand cloud
elastic computation. In Advances in Computational
Intelligence, LNCS 9094, Springer, Berlin, Heidel-
berg, pp. 40–54.
G
´
omez-Canaval, S., Ordozgoiti, B., Mozo, A. (2015).
NPEPE: Massive natural computing engine for opti-
mally solving NP-complete problems in Big Data sce-
narios. In Communications in Computer and Infor-
mation Science 539, Springer, Berlin, Heidelberg, pp.
207–217.
Head, T. (1987). Formal language theory and DNA: an anal-
ysis of the generative capacity of specific recombinant
behaviours. Bull. Math. Biology 49 pp. 737–759.
Head, T., P
˘
aun, Gh., and Pixton, D. (1997). Language
theory and molecular genetics. Chapter 7, vol. 2, in
(Rozenberg and Salomaa, 1997).
Loos, R., Malcher, A., and Wotschke, D. (2008). De-
scriptional complexity of splicing systems. Intern. J.
Found. Comp. Sci. 19 pp. 813–826.
Loos, R. and Ogihara, M. (2007). Complexity theory for
splicing systems. Theor. Comput. Sci. 386 pp. 132–
150.
Loos, R., Martin-Vide, C., and Mitrana, V. (2006). Solv-
ing SAT and HPP with accepting splicing systems.
In Proc. 9th Parallel Problem Solving from Nature
(PPSN IX), LNCS 4193, Springer-Verlag, Berlin, pp.
771-777.
Loos, R., Manea, F., Mitrana, V. (2009). On small, reduced,
and fast universal accepting networks of splicing pro-
cessors. Theoretical Computer Science 410, pp. 406–
416.
Manea, F., Mart
´
ın-Vide, C., Mitrana, V. (2006). All NP-
problems can be solved in polynomial time by ac-
cepting networks of splicing processors of constant
size. In: DNA Computing. LNCS, vol. 4287, Springer,
Berlin, Heidelberg, pp. 47–57.
Manea, F., Mart
´
ın-Vide, C., Mitrana, V. (2007). Accepting
networks of splicing processors: Complexity results.
Theoretical Computer Science 371, pp. 72–82.
Mitrana, V., Petre, I., and Rogojin, V. (2010) Accepting
splicing systems. Theor. Comput. Sci. 411 pp. 2414–
2422.
P
˘
aun, Gh. (1996). Regular extended H systems are compu-
tationally universal. J. Automata, Languages, Combi-
natorics, 1 pp. 27–36.
P
˘
aun, G. (1998). Distributed architectures in DNA com-
puting based on splicing: Limiting the size of com-
ponents. In Unconventional Models of Computation,
Springer, Berlin, Heidelberg, pp 323–335.
P
˘
aun, Gh., Rozenberg, G., and Salomaa, A. (1996). Com-
puting by splicing. Theoret. Comput. Sci. 168 pp. 321-
336.
Paun, Gh., Rozenberg, G., and Salomaa, A. (1997). Com-
puting by splicing. Programmed and evolving splicing
systems. IEEE Intern. Conf. on Evolutionary Comput-
ing, Indianapolis, pp. 273–277.
Paun, Gh., Rozenberg, G., and Salomaa, A. (1998). DNA
Computing - New Computing Paradigms, Springer-
Verlag, Berlin.
Pixton, D. (1996). Regularity of Splicing Languages. Dis-
crete Appl. Math., 69 pp. 101-124.
Pixton, D. (1995). Linear and circular splicing systems. In
First International Symposium on Intelligence in Neu-
ral and Biological Systems. INBS’95, IEEE Herndon,
VA, USA, pp. 181–188.
De Prisco, R., Malandrino, D., Zaccagnino, G., Zaccagnino,
R., and Zizza, R. (2017). Splicing-inspired recog-
nition and composition of musical collectives styles.
In Theory and Practice of Natural Computing - 6th
International Conference, TPNC 2017, LNCS 10687,
Springer-Verlag, Berlin, pp. 219–231.
Rabinow, P. (1996). Making PCR: A Story of Biotechnology.
University of Chicago Press.
Rozenberg, G, and Salomaa, A. (eds.) (1997). Handbook of
Formal Languages, vol. I-III, Springer-Verlag, Berlin.
Siromoney, R., Suhramanian, K.G., and Rajkumar Dare, V.
(1992) Circular DNA and splicing systems. In In-
ternational Conference on Parallel Image Analysis,
ICPIA 1992, LNCS 654, Springer-Verlag, Berlin, pp.
260–273.
How Complex is to Solve a Hard Problem with Accepting Splicing Systems
35
