
Schema Matching with Frequent Changes on Semi-Structured Input
Files: A Machine Learning Approach on Biological Product Data
Oliver Schmidts
1
, Bodo Kraft
1
, Ines Siebigteroth
1
and Albert Zündorf
2
1
FH Aachen, University of Applied Sciences, Germany
2
University of Kassel, Germany
Keywords:
Schema Matching, Machine Learning, Classification, Natural Language Processing, Named Entity Recogni-
tion.
Abstract:
For small to medium sized enterprises matching schemas is still a time consuming manual task. Even ex-
pensive commercial solutions perform poorly, if the context is not suitable for the product. In this paper,
we provide an approach based on concept name learning from known transformations to discover correspon-
dences between two schemas. We solve schema matching as a classification task. Additionally, we provide
a named entity recognition approach to analyze, how the classification task relates to named entity recogni-
tion. Benchmarking against other machine learning models shows that when choosing a good learning model,
schema matching based on concept name similarity can outperform other approaches and complex algorithms
in terms of precision and F1-measure. Hence, our approach is able to build the foundation for improved
automation of complex data integration applications for small to medium sized enterprises.
1 INTRODUCTION
Operators of webshops usually use extract-transform-
load (ETL) workflows as an approach to fill their
backing data warehouse (DW). These workflows need
to be capable to handle a heterogeneous set of var-
ious data formats. Hence, one of the most frequent
performed activities in ETL workflows are schema
transformations (Vassiliadis et al., 2009). The task of
schema transformation is especially challenging for
small to medium sized enterprises (SMEs), since their
business model relies on their suppliers’ product data,
yet they do not possess enough market power to en-
force a standardized schema.
Data integration needs to be engineered depend-
ing on the data source, which is a time consuming
step. Schema matching is a tedious manual part of
this engineering process, that requires domain experts
depending on the complexity of data. Multiple indus-
trial grade tools (e.g. IBM InfoSphere Data Architect)
or open source tools (e.g. Talend Studio) aim to sim-
plify this process, but often these solutions perform
poorly depending on the context and domain. This
applies in particular to the field of biological product
data, where schemas of product data are not standard-
ized concerning language and naming labels.
Driven by a collaboration project with an SME to
automate the data integration process from receiving
data of product suppliers to webshop integration, we
focus on schema transformation of semi-structured
tabular data. Tabular data are characterized by the
separation of header and content, where the content
of each column refers to a single header. All data are
only available as string without further information on
context or relations between different columns.
This paper provides the following contributions:
• We propose a machine learning (ML) approach
to match tabular schemas, which contain minimal
textual metadata. This approach solves schema
matching as classification task by learning from
known concept transformations.
• We introduce a named entity recognition (NER)
approach to analyze, how the classification task
relates to NER.
• We evaluate the matching performance of differ-
ent ML models in a study on biological product
data.
The remainder of this paper is organized as fol-
lows: In Section 2 we provide background informa-
tion about the application domain and well under-
stood approaches for schema matching. In Section 3
and Section 4 we introduce learning approaches lever-
aging simple text processing and classification algo-
208
Schmidts, O., Kraft, B., Siebigteroth, I. and Zündorf, A.
Schema Matching with Frequent Changes on Semi-Structured Input Files: A Machine Learning Approach on Biological Product Data.
DOI: 10.5220/0007723602080215
In Proceedings of the 21st International Conference on Enterprise Information Systems (ICEIS 2019), pages 208-215
ISBN: 978-989-758-372-8
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
Daily
New
Weekly
Monthly
Mail
FTP
Cloud
...
Inbox System
txt
csv
xlsx
pdf
...
Document
Format
Product Information Inbox
Document Format
Unication to semi-
structured csv
Manual
ETL-System
Concept
Unication
Manual
Extract
Transform
Load
Website
Product Updates
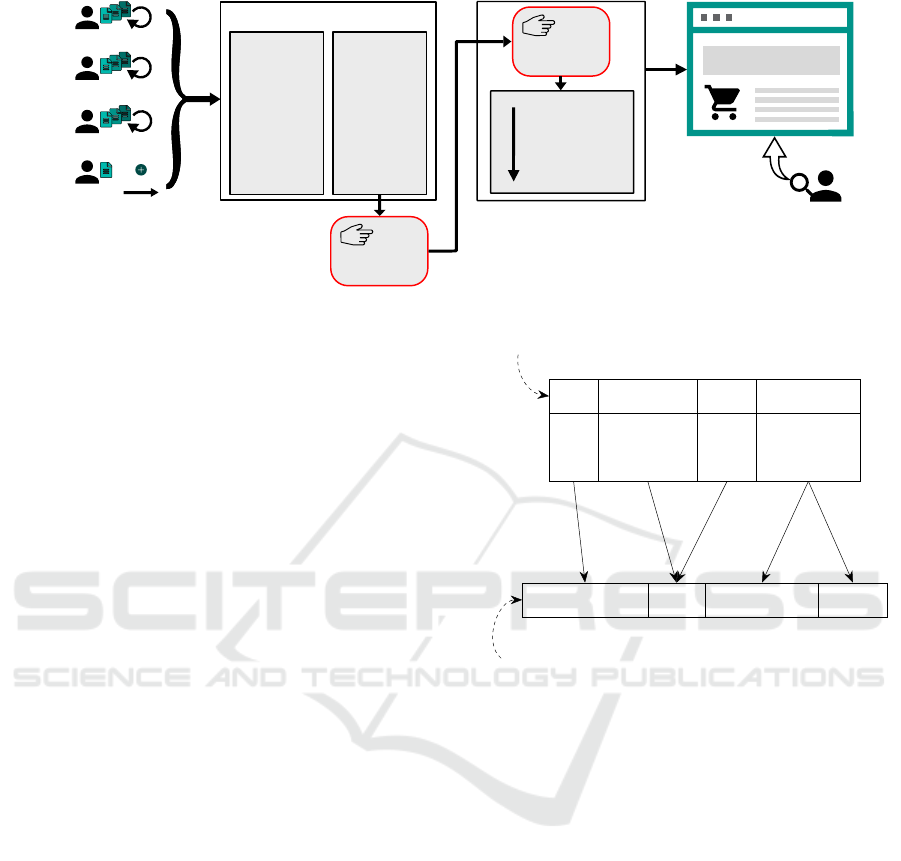
Figure 1: Simplified process of product data integration from suppliers to webshop.
rithms. Afterwards we evaluate and compare the
learning approaches to state-of-the-art approaches in
Section 5 and Section 6. Section 7 concludes this pa-
per in regards to the research questions.
2 BACKGROUND
Figure 1 illustrates a simplified process from prod-
uct data delivery to searchable product data. Update
cycles of product data differ in frequency (daily to
monthly) and it is not guaranteed, that a supplier uses
the same schema twice. Thus, an individual data inte-
gration workflow might be required every time a sup-
plier sends an update. Additional product suppliers
may require new individual workflows. Our collab-
orating SME has to deal with frequent product data
updates (up to 20 per month) for approximately two
million products by roughly 500 suppliers.
Product data may occur in any kind of format from
unstructured language based text to semi-structured
files (e. g. csv), and therefore require a manual unifi-
cation process (format and schema) before data can be
processed by an ETL pipeline. The conversion of text
based product information to semi-structured files is a
manual task, which may be supported by approaches
of natural language processing (NLP).
To match the schema of a tabular input file to a tar-
get schema, a combination of label and content infor-
mation to corresponding concepts of a target schema
may be utilized. Figure 2 shows the possible rela-
tions between input concepts and and target concepts.
We focus on terminological similarity between label
names of input and target concepts in 1:1 or n:1 rela-
tions. This issue definition is heavily motivated by the
biological product data domain of our SME collabora-
tion: n:1 relations emerge most frequently, while 1:n
or n:m relations emerge rarely.
Multiple grown approaches (Madhavan et al.,
Catalog
Special_Price ($)
List Price
Application tested
Catalog
Special_Price ($)
List Price
Application tested
T1: Input Schema
Supplier_Product_ID
Baseprice
Application_Advice
Application
T2: Target Schema
1:1 n:1 1:n
Relations
Input Concepts C
i
Target Concepts C
t
Figure 2: Schema matching of two tabular schemas with
different relations between corresponding concepts using
label names from the application domain.
2001; Do and Rahm, 2002; Melnik et al., 2002) fo-
cus on matching XML schemas. These schemas pro-
vide a tree structure and metadata (e. g. relation be-
tween elements, data types). Cupid (Madhavan et al.,
2001) represents a matching approach, which quan-
tifies similarity by combining structural information
and data type similarities. COMA (Do and Rahm,
2002) combines a set of different matchers, such as
ngrams, synonyms, name paths and data types, on dif-
ferent levels of a schema. Therefore, COMA is able to
match semi-structured schemas as well as structured
schemas. More recent approaches aim for schema
mediation (Saleem et al., 2008) or ontology merg-
ing (Raunich and Rahm, 2011).
Different approaches show, that name similar-
ity can be used to match two concepts (Do and
Rahm, 2002; Madhavan et al., 2005). Although these
approaches do not rely solely on name similarity,
they have proven, that name similarity is an impor-
tant parameter for matching schemas. Madhavan et
Schema Matching with Frequent Changes on Semi-Structured Input Files: A Machine Learning Approach on Biological Product Data
209
feature creation
from set of sim-
ilarity metrics
and target concepts
target
concept
DB
s
1
s
2
.
.
.
s
n
model creation
from concept
transformations
trained
model
concept
transfor-
mation DB
use
feature vector generation model training engine
machine learning
algorithm
model from training
c
1
i
c
2
i
.
.
.
c
n
i
matched
schema
estimation
use in production phase
use
ML production phase
input schema
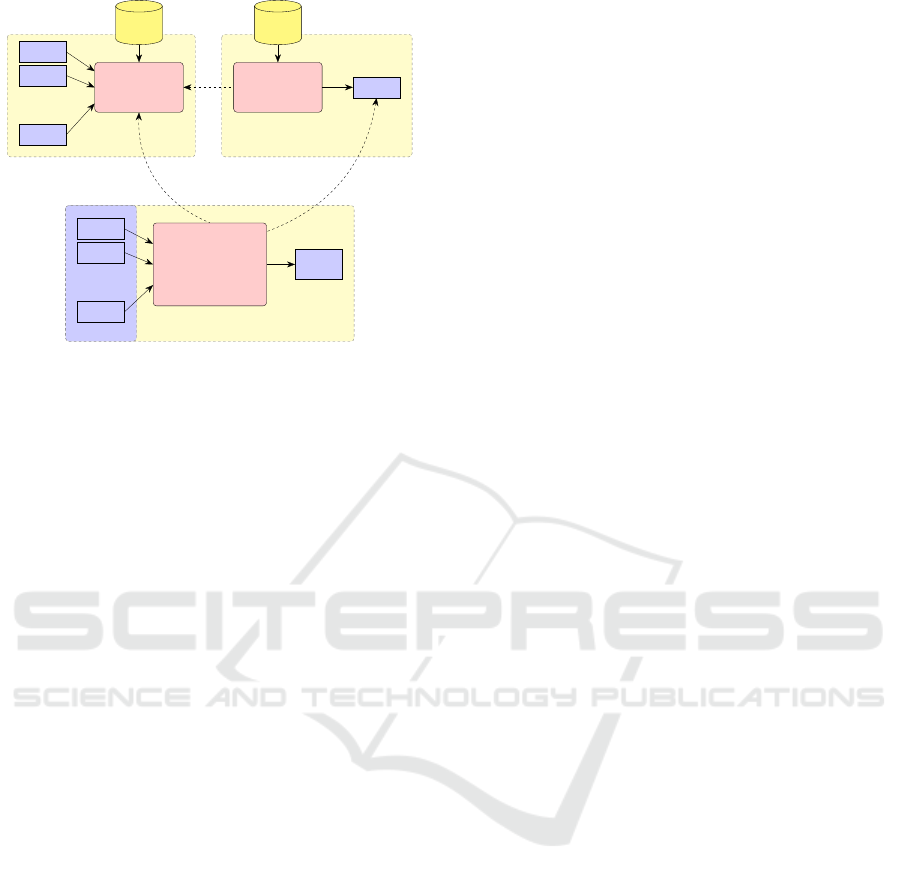
Figure 3: A ML system for supervised learning consisting
of three parts: feature vector generation, model training and
the system in production phase.
al. (Madhavan et al., 2005) proposed to use a name
learner based on classification methods as part of
their approach to avoid spelling errors or incomplete
names. This name learner tries to identify frequent
word roots to quantify similarity between concept
names.
Another option for determining the column label
similarity are token based or character based simi-
larity functions (Wang et al., 2011). Examples for
character based similarity functions are the minimum
number of single character edit operations required
to transform one string into another (Levenshtein-
Similarity) or cosine similarity. A more recent
approach by Zhang et al. (2014) extends classical
schema matching tools by crowdsourcing.
3 LEARNING APPROACH
In order to detect corresponding concepts in two
schemas, we try to estimate the similarity between
each input concept of the input schema C
i
= {c
1
i
..c
n
i
}
and each target concept from the set of all target con-
cepts C
t
= {c
1
t
..c
m
t
}. We build a ML model by lever-
aging lexical and phonetic similarity between input
and target concepts using known concept transforma-
tion provided as a ground truth, where target concepts
serve as classes to be distinguished.
A ML system for solving the classification prob-
lem requires three integral components (c. f. Fig-
ure 3): a feature generation engine, a training en-
gine, learning a matching model using those vectors,
and a classification engine, using the derived model
to match input schemas to target concepts.
3.1 Feature Creation
It is important to find a feature vector, that covers mul-
tiple aspects of string similarity, to receive good es-
timations from a learning model. We use functions
from the set of similarity functions Sim according to
the following Definition:
Definition 1 (Similarity Function).
Sim ={sim|sim : strings × strings → [0..1],
∀a ∈ strings : sim(a, a) = 1,
∀a,b ∈ strings : sim(a, b) = sim(b, a)}
A similarity function takes two strings and deter-
mines the similarity of these, where zero represents
no similarity and one is an exact match. How-
ever, the computed similarities may cause ambigu-
ous mappings. To reduce this issue, we use a set
of similarity functions S
sim
= (s
1
, . . . , s
n
), s
i
∈ Sim
composed of lexical similarity (Cosine-Similarity,
Greedy-Tiling (Wise, 1993), Levenshtein-Similarity,
Longest-Subsequence, Monge-Elkan (Monge and
Elkan, 1997)) and phonetic similarity functions
(Metaphone (Philips, 2000)). Further similarity func-
tions such as structural measures (ngrams) or token
measures, could be added. Applying every function
in S to two strings a and b results in a feature vector
f
S
(a,b) = hs
1
(a,b) ... s
n
(a,b)i.
3.2 Training Data
As depicted in Figure 3, a database with known con-
cept transformations provides ground truth training
data for each classification approach. Training a clas-
sification model requires specific samples for every
class to recognize; thus our training data consists of
pairs of concept variations C
v
= {c
1
v
..c
n
v
} and the cor-
responding target concept. Given a pair of corre-
sponding concepts (c
i
v
,c
i
t
) ∈ G, where G is the ground
truth, training data can be generated by evaluating
f
S
(c
i
v
,c
i
t
) and labeling the resulting vector with the
target concept c
i
t
.
3.3 Classification Models
To evaluate the effectiveness of different classification
models, we distinguish learning models into binary
and multiclass classification.
Binary classification can only decide between two
classes: either the input concept c
i
corresponds to the
target concept c
i
t
or not. Due to this property, we
need to build an individual model for every target con-
cept and compare their estimates. Given a concept to
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
210

match, we estimate matches by evaluating all mod-
els. Every model returns a confidence for a target con-
cept and a confidence for "other". Discarding results
for "other" leads to a ranking among all trained tar-
get concepts, where the target concept with the high-
est confidence equals the combined prediction of all
models.
We use conservative stochastic kernel logistic re-
gression (CSKLR) (Zhang et al., 2012) as a binary
classifier, which obtains sparse solutions by conser-
vatively rejecting updates, based on a binomial distri-
bution of the error on each update. The model was
trained according to Section 3.2 with the restriction
that a positive example for one class is a negative one
for all other classes. Hence, the label is either the cor-
responding target concept or else "other".
An alternative approach is multiclass classifica-
tion. Instead of building a separate model for each tar-
get concept, we build one model for multiple classes.
To achieve this, we adapt the feature vector creation.
Given C
t
and an input concept c
i
i
, we de-
termine the multiclass feature vector f
M
S
(c
i
) =
h f
S
(c
i
i
,c
1
t
), f
S
(c
i
i
,c
2
t
), . . . , f
S
(c
i
i
,c
m
t
)i. Hence, we
compare an input concept to every possible target
concept. Using this multiclass approach leads to a
high number of features and classes, which restricts
the choice of classifiers. For this reason we chose the
extra randomized trees (ERT) classifier (Geurts et al.,
2006), which is an ensemble method, built on top of
Extra Tree. The randomness of trees provides high
variance, yet a low bias.
4 NAMED ENTITY BASED
CLASSIFICATION
Natural language processing (NLP) provides ap-
proaches deriving structured information from natu-
ral language texts. One technique from this field, the
named entity recognition (NER), is an approach for
extracting information such as persons, organizations,
locations, and others.
To build a NLP application, which detects named
entities, complex tools are required. Most NLP
tools use a supervised learning approach to derive
named entities from plain text. Therefore, an anno-
tated corpus containing a set of samples with cor-
responding named entities is required as a training
dataset (Schreiber et al., 2018). During the produc-
tion phase, a NER tool uses a derived model to detect
named entities in a natural language text.
Usually, the first tool to use in a NLP applica-
tion is text segmentation, where a sentence is splitted
into a sequence of tokens. Tokens are not limited to
words, but also include punctuation marks and special
characters. Based on the token structure, a NER tool
searches for named entities in each sentence.
We now describe a schema matching approach
based on NER and how a corpus with customized
named entities can be derived from the target concept
database (DB) as a result.
4.1 Corpus Preparation
The first step when applying NER approaches to a
schema matching task, is building an annotated cor-
pus, which serves as training data. Hence, we need to
convert the concept transformation to a natural lan-
guage document.
Every target concept from the DB represents a
named entity to be detected in a sentence, and thus
a NER approach requires concept variations to repre-
sent sentences and named entities at the same time.
To clarify this procedure, consider the following ex-
ample.
Given the input schema of Figure 2, Spe-
cial_Price ($) and List Price correspond to the same
target concept (Baseprice). Therefore, Baseprice
is a named entity NLP tools shall detect. Spe-
cial_Price ($) and List Price are variations of this
concept. By interpreting Special_Price ($) as a sen-
tence, NLP tools become applicable. Firstly, we apply
text segmentation to split the concept variations into
tokens (e. g. Special, _, Price, (, $ , ) ). Afterwards,
we annotate these tokens with the corresponding tar-
get concept as named entities.
The resulting document, contains a lot of short
sentences, which are segmented into tokens and an-
notated with named entities, which allows us to train
NER tools to match schemas.
4.2 NER Pipeline
The performance of NER tools varies depending on
the application domain. For example, in some do-
mains the NER tool of StanfordNLP
1
might work bet-
ter than the NER approach of OpenNLP
2
. To work
around this issue we use NLPf
3
(Schreiber et al.,
2018), a framework for creating custom NLP models
and pipelines. NLPf allows us to determine the best
performing combination (pipeline) of a range of NLP
tools and thus train a domain specific model.
We used this framework to derive a best perform-
ing pipeline for schema matching based on our an-
notated corpus through NLPf, considering the ap-
1
https://nlp.stanford.edu/
2
https://opennlp.apache.org/
3
https://gitlab.com/schrieveslaach/NLPf
Schema Matching with Frequent Changes on Semi-Structured Input Files: A Machine Learning Approach on Biological Product Data
211

proaches of OpenNLP and StanfordNLP for text seg-
mentation and NER.
5 EXPERIMENTAL STUDY
We evaluate different machine learning approaches in
an experimental study with real data gathered from
integration jobs of biological product data.
5.1 Model Setup
Naive Bayes, binary CSKLR and multiclass ERT use
the same set of similarity functions (S
sim
), as provided
in Section 3.1, to generate the features. Each learn-
ing algorithm uses the default parameter configura-
tion as suggested by the original authors. To get an
overview of which approach performs best on the bi-
ological product data, we include further approaches
to the evaluation.
• A pragmatic approach that calculates the average
similarity of S
sim
for each known concept varia-
tion is stored in the concept transformation DB.
The target concept corresponding to an input con-
cept, is estimated by the maximum average sim-
ilarity of all variations. This approach could be
implemented easily by any SME and should be
outperformed by learning approaches.
• COMA (Massmann et al., 2011) is a mature ap-
proach to match schemas and ontologies. How-
ever, it should be mentioned that COMA is not de-
signed to match schemas solely based on concept
names of tables. We used the default trigram con-
figuration, and provided appropriate synonyms
and abbreviations. Since we do not have relational
tables, COMA can not exploit further properties
(e. g. data types).
• We built an annotated corpus according to the pre-
vious section from the same concept transforma-
tion DB as other ML approaches. Every named
entity corresponds to a target concept. We evalu-
ate the derived NER model in this study. We were
particularly interested in seeing differences be-
tween the NER approach and classification based
on similarity metrics. We expected the similar-
ity function based ML approach to perform bet-
ter compared to NER, since input data are of low
quality and for some target concepts, training data
are limited.
5.2 Training Setup
Each learning approach (Naive Bayes, CSKLR, ERT)
uses the system depicted in Figure 3. They differ
solely by the algorithm utilized and whether they use
binary or multiclass feature vectors. All approaches
use the same ground truth to train their models. In our
evaluation scenario, training data consists of about
600 concept variations. The training dataset is imbal-
anced, meaning not every target concept has the same
number of variations. The original schema consisted
of over 100 concepts. However, we had to reduce the
schema to 53 concepts due to the lack of sufficient
training data by excluding concepts with less than 15
schema variations.
5.3 Evaluation Scenario
To analyze the performance of all approaches, we
gathered a gold standard of concept mappings from
manually executed integration jobs. During that pro-
cess we excluded duplicates and took care of not mix-
ing any training and test data. To ensure test data and
training data are disjoint, we removed concept varia-
tions from the gold standard, which also occurred in
the training data as well. The resulting gold standard
consists of over 1000 concept mappings from schema
transformations.
Since we aggregated our test data from real jobs,
our test data is imbalanced. We therefore provide pre-
cision and F1-measure to compare the performance
of the approaches in the domain of biological product
data.
6 STUDY RESULTS
Table 1 provides detailed information about average
precision, recall and F1-measure, which have been
achieved on the evaluation scenario by the various ap-
proaches.
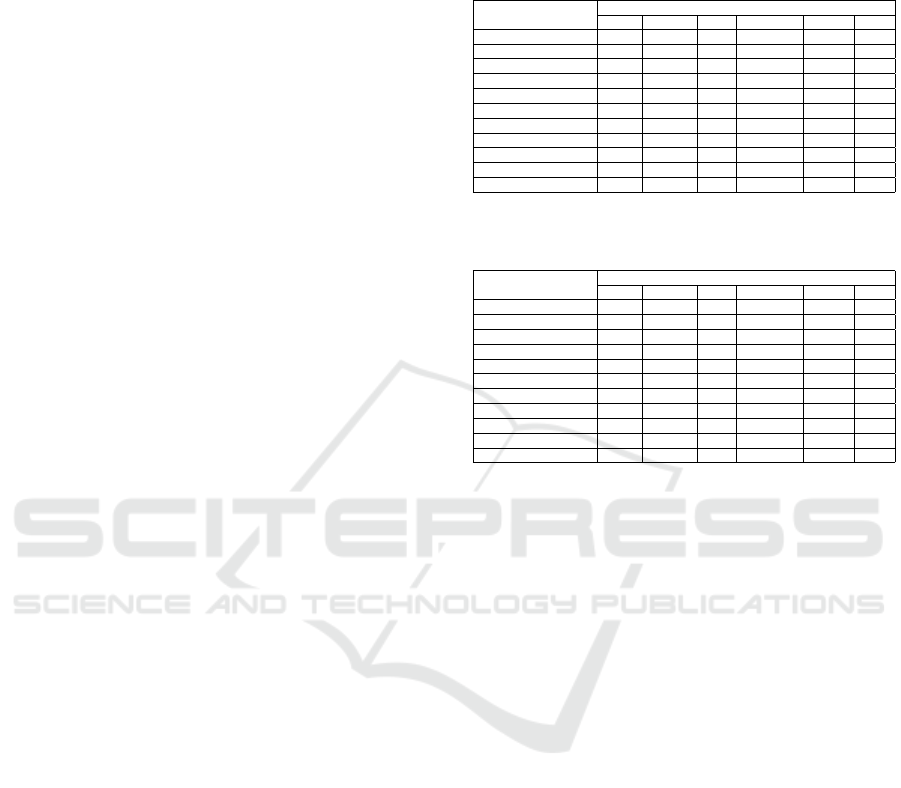
Table 1: Average results matching the test set over 53 target
concepts grouped by approach.
Approaches
Scores Bayes CSKLR ERT pragmatic COMA NER
Precision 0.45 0.71 0.69 0.66 0.60 0.45
Recall 0.45 0.49 0.51 0.58 0.53 0.09
F1-measure 0.38 0.53 0.55 0.58 0.49 0.13
Naive Bayes achieved an average precision and
recall of 0.45, while the average F1-measure only
achieved a score of 0.38. Therefore, Naive Bayes
achieves almost the lowest values for each measure.
Despite its poor performance Naive Bayes is reported
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
212
to be a standard name matcher, thus providing lots of
potential for improvement.
Both more advanced classification approaches,
CSKLR and ERT, achieve significantly higher aver-
age precision in our study. CSKLR reaches the high-
est precision of all approaches (0.71) closely followed
by the ERT approach (0.69), both outperforming the
pragmatic approach. However, results of the prag-
matic approach were better than expected. The per-
formance indicates, that choosing the maximum aver-
age value of multiple similarity functions is an effec-
tive approach for a small to medium sized enterprise
(SME) to start with. The average precision of 0.66
is close to the precision of ERT (0.69), and thus sig-
nificantly higher than the achieved precision of Naive
Bayes. Additionally, the pragmatic approach achieves
the highest recall and F1-measure of all evaluated ap-
proaches. Results show, that choosing a sophisticated
learning model, leads to significant higher precision
of concept matchings.
COMA achieves a better precision than the do-
main specialized Naive Bayes approach. However,
precision is lower than in more advanced models and
the pragmatic approach. This result can be explained
by the limitation of using simple similarity matchers
since no further information is available for COMA in
our study. Furthermore, COMA is not as specialized
to the application domain as learning approaches.
An overall noticeable result is, that recall for
CSKLR, ERT and COMA is almost similar (around
0.5). Recall of Naive Bayes is close to 0.5 as well. Re-
call might therefore be limited, due to the low quality
of data.
NER performs worst in every measure. While
reaching the precision of Naive Bayes, recall and F1-
measure are extremely low. We explain this result by
showing more details in the following section.
6.1 Detailed Results
CSKLR achieves satisfying results in terms of pre-
cision. 19 of 53 target classes are matched with
a precision of >0.9. Both, ERT and COMA reach
13/53, closely followed by the pragmatic approach
with 12/53. Naive Bayes only reaches a minimal pre-
cision of 0.9 in 9 of 53 cases. Surprisingly, the NER
approach is able to identify 18 of 53 target concepts
with a precision of 1.0. However, the model derived
by our annotated corpus seems to be extraordinary
sensitive. This is indicated by overall low F1-measure
for any target concept as depicted in Table 3.
Table 2 and Table 3 show more detailed match re-
sults in terms of precision and F1-measure on a repre-
sentative subset of 11 of 53 target labels for all evalu-
ated approaches.
Table 2: Precision details on a representative subset of 11
of 53 target labels.
Precision of Approaches
Target Concepts Bayes CSKLR ERT pragmatic COMA NER
ProductUrlHtml 0.35 0.56 0.78 0.86 0.66 1.00
Epitope 0.50 1.00 1.00 0.66 1.00 0.00
Immunogen 0.35 0.50 0.40 0.40 0.75 1.00
handlingAdvice 0.18 1.00 0.50 0.50 0.67 0.50
productNameOriginal 0.60 0.69 0.63 0.51 0.50 1.00
Gene1 0.72 0.88 0.80 0.69 0.36 0.75
ProductUrlPdf 0.15 0.00 0.80 0.66 0.36 0.75
Isotype 0.91 0.75 0.80 1.00 0.91 0.00
Purity 1.00 1.00 1.00 0.66 0.67 1.00
Description 0.17 0.67 0.38 0.56 0.60 0.40
AssayPrecision 0.17 1.00 1.00 0.50 0.25 0.00
Table 3: F1-measure details on a representative subset of 11
of 53 target labels.
F1-measure of Approaches
Target Concepts Bayes CSKLR ERT pragmatic COMA NER
ProductUrlHtml 0.25 0.14 0.54 0.65 0.56 0.15
Epitope 0.48 0.63 0.20 0.60 0.53 0.00
Immunogen 0.50 0.43 0.27 0.27 0.81 0.18
handlingAdvice 0.29 0.80 0.40 0.57 0.66 0.40
productNameOriginal 0.49 0.51 0.58 0.57 0.42 0.09
Gene1 0.28 0.85 0.51 0.67 0.43 0.11
ProductUrlPdf 0.16 0.00 0.65 0.60 0.20 0.18
Isotype 0.91 0.85 0.76 0.73 0.87 0.00
Purity 0.50 0.80 0.80 0.67 0.67 0.50
Description 0.14 0.17 0.33 0.46 0.29 0.08
AssayPrecision 0.29 0.80 0.67 0.50 0.40 0.00
CSKLR achieves high precision for the major-
ity of target concepts. However, no input concept
could be matched with the target productUrlPdf by
CSKLR. productUrlPdf always seems to be missrec-
ognized as productUrlHtml. CSKLR achieves preci-
sion of 1.0 for four target concepts, but recall ranges
from 0.09 (productUrlHtml, Description) to 1.0 (as-
sayPrecision). This result is probably related to the
use of language. Anything can be interpreted as de-
scription, while assayPrecision is very domain spe-
cific with a small range of variations. The suboptimal
matching performance for Immunogen is explainable
by looking into the training data. Product suppliers
use similar terms for epitopes and immunogens. This
makes it hard for a learning approach to distinguish
between these terms.
ERT suffers the same recognition issues for epi-
topes and immunogens. However, ERT is able to bet-
ter distinguish between productUrlHtml and produc-
tUrlPdf. Results of ERT for other target concepts are
a little lower than results of CSKLR.
The results of COMA show a perfect precision for
Epitope like CSKLR and ERT. Nevertheless, COMA
clearly outperforms every other approach in differen-
tiating between Immunogen and Epitope.
The approach based on NER provides high preci-
sion, yet low F1-measure. If an input concept can be
matched to a target concept, NER achieves high pre-
cision. The overall precision of 0.45 (c. f. Table 1) re-
sults from the NER approach not beeing able to recog-
Schema Matching with Frequent Changes on Semi-Structured Input Files: A Machine Learning Approach on Biological Product Data
213

nize some concepts as named entities. Interestingly,
NER achieves high precision, where other classifiers
do not and vice versa except for Purity. However,
the low F1-measure indicates, that NER is unable to
generalize concepts, and thus only detects the most
obvious matchings. This theory also explains the rel-
atively high precision across the board.
6.2 Additional Experiments
We briefly mention results of smaller, to some extend
manual experiments with feature vector creation and
different approaches.
• We extended the feature vector by trigram simi-
larity. We found that models including trigrams
as features performed significantly worse than
models based on other similarity functions. For
all learning approaches, precision, recall and F1-
measure dropped by 0.03-0.05. Learning similar-
ity exclusively from trigram similarity lead to sig-
nificantly worse results. The default configuration
COMA suggests trigram matchers as well. We as-
sume, that our input data from real jobs lack suf-
ficient quality for trigrams to achieve satisfying
results. Preparing concepts, using specialized text
segmentation approaches and further preparation,
could potentially improve the performance. Fur-
ther methods are definitely required to detect false
labeling by data suppliers.
• CSKLR and ERT aim to match 1:1 or n:1 relations
between input and target labels. Nevertheless,
both approaches indicate 1:n relations through the
score for small n (<=3). 1:n relations are indicated
if the highest two or three scores are close to each
other and >0.3.
• Special characters related to currency like $ or
C
are repeatably matched to the Baseprice label by
ERT, though it has never been trained with it as
special synonym. Only some concept variations
contain these special characters.
• We did not have enough variation for every tar-
get concept. However, it is possible to see the
result "nothing matches" indicated in the scores
when this condition occurs, if the highest score of
a learning matcher is <0.1.
• Intentional typos can be recognized correctly by
the binary CSKLR matcher. ERT recognizes only
some of them. For example concentratoin is
matched to Application_Advice instead of concen-
tration.
• The NER model seems to be very sensitive to dif-
ferent data formats. A lot of concept variations
for Baseprice contain numbers (e. g. List Price
2018). A concept variation containing a number is
detected correctly as named entity. If the number
is removed, the NER model is not able to further
on match the concept variation to the correspond-
ing target concept. Hence, we need to improve
modeling further.
Overall we conclude, that schema matching based
on concept names can be improved by using sophis-
ticated ML approaches, trained from known label
transformations. The matching results for single tar-
get concepts are depending on the derived learning
model. However, the combination of multiple ap-
proaches in an ensemble method (e. g. majority vote)
may cover weaknesses and emphasize strengths of
single approaches.
7 CONCLUSION
In this paper we presented different ML approaches
to schema matching based on concept names. We
showed, that multiple character based similarity func-
tions can be applied to build a feature vector. Further,
we introduced a NER approach for schema matching
tasks, where target concepts are used as named enti-
ties.
We evaluated and compared all approaches in a
study, to determine the practicability in real world
scenarios in the biology domain. Furthermore, we
compared the results to two non learning approaches,
which includes a pragmatic approach and the mature
schema matching system COMA.
The result shows, that a schema matching system,
based solely on learning concept name similarity, is
able to reach the same range of precision as the ap-
proach of Madhavan et al. (2005), even for data of
low quality. However, precision and F1-measure need
further improvements. Matching performance varies
for single concepts, depending on the learning method
used. Moreover, the results indicate, that classifi-
cation approaches with sophisticated models outper-
form COMA with the standard trigram configuration
as well as the NER approach.
We provide two ideas: Firstly, we combine his-
torical information on concept transformations with
simple similarity metrics to learn from. This simplic-
ity allows customization to special needs of SMEs.
Therefore, they can experiment with learning meth-
ods that are well suited for their domain. Secondly,
we introduce a NER based approach, which is trained
on an annotated corpus of known labels.
Automation of schema matching in an industrial
environment requires a precision of over 0.9. Due
ICEIS 2019 - 21st International Conference on Enterprise Information Systems
214

to this requirement further improvement of match-
ing precision and recall is required as well. Other-
wise, the cost of integration errors and manual cor-
rection prevails any benefit. To achieve further im-
provements, different concept name matching sys-
tems could be combined by using a majority voting.
Since the approaches we provide in this paper
achieve a lower precision than required for full au-
tomation, we built a recommending engine to inte-
grate research prototypes into industrial applications,
according to the MEDIATION approach (Schreiber
et al., 2017; Schmidts et al., 2018), to assist manual
data integration tasks. Additionally, the approaches
are used in a quality assurance system to aug-
ment possible wrong matches in manually matched
schemas.
REFERENCES
Do, H.-H. and Rahm, E. (2002). COMA: A System
for Flexible Combination of Schema Matching Ap-
proaches. In Proceedings of the 28th International
Conference on Very Large Data Bases, VLDB ’02,
pages 610–621, Hong Kong, China. VLDB Endow-
ment.
Geurts, P., Ernst, D., and Wehenkel, L. (2006). Extremely
randomized trees. Machine Learning, 63(1):3–42.
Madhavan, J., Bernstein, P. A., and Rahm, E. (2001).
Generic schema matching with cupid. In vldb, vol-
ume 1, pages 49–58.
Madhavan, J. et al. (2005). Corpus-Based Schema Match-
ing. In 21st International Conference on Data En-
gineering (ICDE’05), pages 57–68, Tokyo, Japan.
IEEE.
Massmann, S. et al. (2011). Evolution of the COMA match
system. In Proceedings of the 6th International Con-
ference on Ontology Matching-Volume 814, pages 49–
60. CEUR-WS. org.
Melnik, S., Garcia-Molina, H., and Rahm, E. (2002). Sim-
ilarity flooding: a versatile graph matching algorithm
and its application to schema matching. In Proceed-
ings 18th International Conference on Data Engineer-
ing, pages 117–128, San Jose, CA, USA. IEEE Com-
put. Soc.
Monge, A. and Elkan, C. (1997). An Efficient Domain-
Independent Algorithm for Detecting Approximately
Duplicate Database Records.
Philips, L. (2000). The Double Metaphone Search Algo-
rithm. C/C++ Users J., 18(6):38–43.
Raunich, S. and Rahm, E. (2011). ATOM: Automatic
target-driven ontology merging. In 2011 IEEE 27th
International Conference on Data Engineering, pages
1276–1279.
Saleem, K., Bellahsene, Z., and Hunt, E. (2008).
PORSCHE: Performance ORiented SCHEma media-
tion. Information Systems, 33(7):637–657.
Schmidts, O. et al. (2018). Continuously Evaluated Re-
search Projects in Collaborative Decoupled Environ-
ments. In Proceedings of the 5th International Work-
shop on Software Engineering Research and Indus-
trial Practice, SER&IP ’18, pages 2–9, New York,
NY, USA. ACM.
Schreiber, M., Kraft, B., and Zündorf, A. (2017). Metrics
Driven Research Collaboration: Focusing on Com-
mon Project Goals Continuously. In 2017 IEEE/ACM
4th International Workshop on Software Engineering
Research and Industrial Practice (SER IP), pages 41–
47.
Schreiber, M., Kraft, B., and Zündorf, A. (2018). NLP Lean
Programming Framework: Developing NLP Applica-
tions More Effectively. In Proceedings of the 2018
Conference of the North American Chapter of the As-
sociation for Computational Linguistics: Demonstra-
tions, pages 1–5, New Orleans, Louisiana. Associa-
tion for Computational Linguistics.
Vassiliadis, P., Simitsis, A., and Baikousi, E. (2009). A Tax-
onomy of ETL Activities. In Proceedings of the ACM
Twelfth International Workshop on Data Warehousing
and OLAP, DOLAP ’09, pages 25–32, New York, NY,
USA. ACM.
Wang, J., Li, G., and Fe, J. (2011). Fast-join: An efficient
method for fuzzy token matching based string similar-
ity join. In 2011 IEEE 27th International Conference
on Data Engineering, pages 458–469.
Wise, M. J. (1993). String similarity via greedy string tiling
and running Karp-Rabin matching. Online Preprint,
Dec, 119.
Zhang, C. J. et al. (2014). CrowdMatcher: Crowd-assisted
Schema Matching. In Proceedings of the 2014 ACM
SIGMOD International Conference on Management
of Data, SIGMOD ’14, pages 721–724, New York,
NY, USA. ACM.
Zhang, L. et al. (2012). Efficient Online Learning for Large-
Scale Sparse Kernel Logistic Regression. In AAAI.
Schema Matching with Frequent Changes on Semi-Structured Input Files: A Machine Learning Approach on Biological Product Data
215
