
Cloud Infrastructure for Storing and Processing EEG and ERP
Experimental Data
Petr Je
ˇ
zek and Luk
´
a
ˇ
s Va
ˇ
reka
Department of Computer Science and Engineering, New Technologies for the Information Society,
Faculty of Applied Sciences, University of West Bohemia, Plze
ˇ
n, Czech Republic
Keywords:
EEG, ERP, Cloud, HDFS, Hadoop, Spark, Experiment, Infrastructure.
Abstract:
Current infrastructures for experimental data, results and computational tools make a shift from locally main-
tained solutions to remote cloud-based infrastructures. It brings a higher availability, sustainability and per-
formance. However, specifics of different research areas require development of customized solutions for
individual research domains. For example, electroencephalography and event-related potentials (EEG/ERP)
use specific devices, data formats and machine learning workflows. As a solution, a cloud-based system for the
EEG/ERP domain containing a distributed data storage, a signal processing method library and a client GUI
is presented. The signal processing method library is used for training of classifiers and classifying the data
in the cloud-based system controlled by the GUI. The presented system was tested using a machine learning
workflow based on the data stored in the system. In the workflow, various classifiers were trained and their
parameters stored into the system. Finally, testing data were classified using previously trained classifiers.
1 INTRODUCTION
Laboratories have been using locally maintained
databases and tools for experimental data storage and
processing. Experimental results have been pub-
lished and then locally stored or often even supposed
as no more needed and discarded. Nowadays be-
cause of increasing data storage capacity, fast Internet
connectivity, and cheaper hardware equipment, the
amount of experimental data is dramatically increas-
ing. With increasing amount of data, requirements
to data sustainability (Van Horn and van Pelt, 2008),
experiments reproducibility, and publishing also orig-
inal data not only experimental results (Abelard and
H
´
eloise, 2011) are increasing, too. These require-
ments make locally maintained infrastructures insuf-
ficient and obsolete. On one hand the computa-
tional performance is cheaper and easily available but
on the other hand the increasing number of devices
and technologies for data storing and processing re-
quire a higher demand on technical knowledge of re-
searchers.
A new view on data storage and management
makes a shift from locally maintained infrastruc-
tures to remote solutions usually referred to as cloud
computing. This approach defines a new deliv-
ery model Architecture-as-a-Service (AaaS) that sup-
poses a provider of a remote computational infrastruc-
ture who guarantees data integrity, backup and unin-
terrupted availability. Methods for data processing are
also installed in the remote infrastructure which en-
sures enough performance by scaling tasks over a dis-
tributed network of a computational cluster. This ap-
proach takes off the responsibilities from researchers
who can be focused on the laboratory work instead of
maintaining complex infrastructures.
Especially the domain of electroencephalography
(EEG) and event-related potentials (ERP) requires
various experts from signal processing, database or
hardware domains. Moreover, laboratory staff re-
sponsible for collecting experiments must be familiar
with all the systems in a complex chain of data col-
lecting, storing, processing and finally, presenting re-
sults. In the cloud computing approach the responsi-
bilities can be either partly dedicated to a paid service
or split out among individual experts not necessarily
located in the same laboratory.
We present a complex cloud-based infrastructure
for storing and processing experimental data. This in-
frastructure is validated on an ERP focused use-case
study. This infrastructure contains a data storage, a
library of signal processing methods, and a simple
GUI allowing users to easily control the whole sys-
tem. Section 2 presents similar concepts and eval-
274
Ježek, P. and Va
ˇ
reka, L.
Cloud Infrastructure for Storing and Processing EEG and ERP Experimental Data.
DOI: 10.5220/0007746502740281
In Proceedings of the 5th International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AWE 2019), pages 274-281
ISBN: 978-989-758-368-1
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

uates the approach. Section 3 briefly introduces the
ERP domain and introduces a technological stack for
the system. Section 4 describes implementation of the
system. Finally, Section 5 presents testing of the sys-
tem and Section 6 concludes the work.
2 RELATED WORK
There are some approaches that aim to move tradi-
tional desktop-based computing to cloud-based solu-
tions in medicine.
Cloudwave (Sahoo et al., 2013) is a platform that
provides paralleled algorithms for computing in a car-
diac domain. It supports real-time interaction with
large volumes of electrophysiological signals, and
features signal visualization and a querying function-
ality using an ontology-driven web-based interface.
The CARMEN e-science project (Watson et al.,
2008) is based on a grid computing concept that in-
tegrates heterogeneous resources across virtual orga-
nizations represented by registered laboratories. For
these laboratories a data/metadata storage, a data
analysis service, and a workflow enactment is ensured
but laboratories contribute by their own data and data
analysis methods.
Electrocardiographic (ECG) signal analysis us-
ing Linear Discriminant Analysis (LDA) and Sup-
port Vector Machines (SVM) framework is presented
in (Varatharajan et al., 2018). This framework pro-
vides a data storage for wearable sensors transferred
through a 4G network. Once data is transferred to the
data storage it is computed by LDA and SVM meth-
ods and results are transferred to a doctor’s computer
where he/she can see reports.
An IoT-based health-care architecture (Thota
et al., 2018) uses a fog computing approach for col-
lecting data from wireless sensors. The fog comput-
ing relies on the usage of so-called edge device com-
municating with wearable devices via a local network.
Then this edge-device is connected to the cloud solu-
tion via the Internet. Especially in this solution Rasp-
berry Pi is an edge-device that transfers data to the
cloud. Data in the cloud is accessible by health-care
providers.
3 METHODS
3.1 ERP Domain
Event-related potentials (ERP) (Sur and Sinha, 2009)
can be observed in the electroencephalographic
(EEG) signal as small voltages generated by the brain
as a reaction to stimuli. Many ERP experiments fol-
low an oddball paradigm (Garc
´
ıa-Larrea et al., 1992)
based on stimulation of tested subjects by two types
of auditory or visual stimuli. Oddball experiments
consist of a set of common (non-target) stimuli that
are infrequently interrupted by rare (target) stimuli.
The target stimuli are associated with the P300 wave-
form. Similar principle can also be applied to brain-
computer interfaces (BCIs) as originally proposed
with the P300 speller by (Farwell and Donchin, 1988)
in which the intended action of a user (such as a row
containing the letter that the user wishes to type) rep-
resents the target stimuli, and the task is to detect that
stimulus by means of machine learning (Hoffmann
et al., 2008).
The detection relies on extracting ERPs from the
ongoing EEG while minimizing noise. The common
procedure is based on epoch extraction and filtering
ERP epochs (trials). When aiming at P300 BCI clas-
sification, each extracted and processed ERP epoch is
evaluated by a binary classifier that is trained to sep-
arate between target and non-target trials. Finally, the
results of classification or parameters of the classifier
itself can be stored for further evaluation. (Hoffmann
et al., 2008)
A typical machine learning workflow in the ERP
domain can be organized as Fig. 1 depicts, and it can
consist of the following steps:
• Data reading. Either from filesystem (a binary
file specific for the EEG amplifier or a universal
open file format such as EDF (Kemp and Olivan,
2003)), or in real-time from stream such as Lab
Streaming Layer (LSL) (Kothe, 2014).
• Pre-processing commonly includes several sub-
steps. Subset selection of only relevant EEG
channels reduces data dimensionality. Band-pass
filtering attenuates undesired frequencies such as
line noise at 50 Hz. Epochs extraction splits con-
tinuous EEG into ERP epochs using stimuli mark-
ers. Each epoch has a pre-stimulus part (a time
window before the stimulus onset) and a post-
stimulus part (a time window after the stimulus
onset). Baseline correction is used to align the
ERP epoch around zero e.g. by subtracting aver-
age of the pre-stimulus part from the whole ERP
epoch. Many other methods can be used, e.g.
blind source separation methods such as Indepen-
dent Component Analysis (Luck, 2014).
• Feature extraction. Most common features to be
extracted are time point features and band power
features (Lotte et al., 2018). Discrete wavelet
transform, matching pursuit (Mallat and Zhang,
Cloud Infrastructure for Storing and Processing EEG and ERP Experimental Data
275

Data
reading
EEG data files
EEG on-line stream
Feature extraction
Preprocessing
Classification
Results visualization
Voltage [μV]
Time [ms]
Accuracy: 80 %
Precision: 78 %
Recall: 82 %
EEG channel
selection
Band-pass
filtering
Epoch
extraction
Baseline
correction
Figure 1: Simple machine learning workflow suitable for the P300 ERP detection.
1993) and other related methods for time fre-
quency analysis can also be applied. Feature se-
lection that usually aims at reducing data dimen-
sion by omitting redundant time points or band
power features (Lotte et al., 2018).
• Classification. Some methods used in the ERP
domains are e.g. Support Vector Machines
(SVM) (Subasi and Gursoy, 2010), decision
trees (Polat and G
¨
unes¸, 2007), logistic regression
(Subasi and Ercelebi, 2005), random forest (Frai-
wan et al., 2012), or neural networks (G
¨
uler et al.,
2005).
• Results visualization includes charts, tables or
single values.
3.2 System Requirements
The experimental workflow outlined in Section 3.1
defines requirements for the developed system. Data
must be collected in a robust and secured data stor-
age. Processing of data is a sequence of mostly lin-
early executed operations but e.g. classification that
uses deep learning methods is required to be paral-
leled. A cloud-based solution is suitable for classifier
training because it is usually the most time consum-
ing part of the workflow. However, the data is usually
collected on a local laboratory notebook connected ei-
ther to the Internet or at least to a local laboratory net-
work. Hence a secured data transfer from the labora-
tory notebook to the cloud solution must be ensured.
Data must be anonymized and the server located in-
side a protected network secured by firewalls.
3.3 Apache Hadoop and Spark
Commonly used open source technologies for data
storing and data processing in distributed environ-
ment include Apache Hadoop and Apache Spark.
Apache Hadoop (Apache Software Foundation, 2009)
is designed for performing massive, parallel jobs and
is de facto supposed as a place where data and com-
putational resources are shared and accessed. It is
an ecosystem of several technologies (Ishwarappa and
Anuradha, 2015). We used a Hadoop Distributed File
System (HDFS) that provides high-throughput access
to data stored in a distributed environment.
Hadoop provides a map-reduce implementation
for processing big data in the distributed environ-
ment (Ekanayake et al., 2008). However, Apache
Spark (Zaharia et al., 2016) reports significantly bet-
ter performance than the map-reduce implementation
(Gopalani and Arora, 2015). Moreover, an open-
source distributed machine learning library (MLlib)
(Meng et al., 2016) available for Apache Spark pro-
vides an implementation of various methods suitable
for training classifiers in the ERP domain.
ICT4AWE 2019 - 5th International Conference on Information and Communication Technologies for Ageing Well and e-Health
276

4 RESULTS
4.1 Server
The server operates HDFS for experimental data, a
data analysis package described in Section 4.2, and a
Spring boot (Webb et al., 2013) application that oper-
ates a REST API described in Section 4.3.
Once a processing of data is required, a job on the
server is created. The server handles all jobs in a job
manager. Once a job is done, a flag FINISHED is set
up and results are stored on the server. This approach
does not require to wait for every single job but allows
users to run any number of jobs in a row and see the
results once they are available.
ERP experiments (see Section: 3.1) require train-
ing of classifiers reusable for similar kinds of exper-
iments. Typically, same experiment is repeated with
various tested subjects. The server allows users to ei-
ther train a classifier on selected data or test a classi-
fier already trained and saved on the server.
The server is controlled by a GUI via the REST
API described in Section 4.4.
Because installing a server with all needed tools
is a difficult task and the server is supposed to be eas-
ily transferred to other environments, we used Docker
(Merkel, 2014). There are several Hadoop Docker im-
ages (e. g. (Vohra, 2015)). We used Cloudera Quick-
start (Taylor, 2010) that we extended by the tools we
implemented (described later in Sections 4.2 and 4.3).
A complete architecture is shown in Figure 2.
4.2 Methods Library
An aim of the methods library is providing the signal
processing tools needed for training and classifying
the ERP signal. We implemented three packages of
tools for (1) data reading and transformation, (2) fea-
tures extraction and (3) classification. The Apache
Spark API enables parallelism of the processes over
available nodes. This feature is useful especially for
deep learning methods. The methods library also con-
tains a pipeline builder responsible for running meth-
ods in a workflow as described in Section 3.1. The
Apache Spark distributes data and computational load
over nodes connected in the HDFS cluster and col-
lects results. Figure 3 shows all methods currently
implemented in the library.
4.3 REST API
The Apache Spark job is a single process configured
by Spark properties
1
. We provided a REST API for
1
https://spark.apache.org/docs/latest/configuration.html
controlling individual Spark jobs from a client com-
puter. The REST API methods are designed to allow
users to run any number of jobs they need, check their
status, and once they are finished, download results.
Available methods are:
• /jobs/submit/(id)?(qp) - to submit a job with an id.
Gp is a configuration via the Spark properties.
• /jobs/check/(id) - to check the status of a job with
an id
• /jobs/result/(id) - to get the result of a job with an
id
• /jobs/log/(id) - to get the log of a job with an id
• /jobs/cancel/id - to cancel a job with an id
• /jobs/configuration/(id) to get the configuration of
a job with an id
• /classifiers/list - to list all the saved classifiers
4.4 Client GUI
The graphical user interface is only a tool supposed
to be installed locally on a laboratory notebook. The
GUI implements a wizard through which the user can
upload a file or a complete folder, or delete existing
files or folders on the server. Once the data is up-
loaded the user can choose data for classifier train-
ing or testing. In the next step, the user lists avail-
able methods on the server, selects one and config-
ures its parameters. Finally, the REST API method
is called, data from the HDFS storage is submitted to
the Spark, and a Spark job is created and executed. A
print-screen of the GUI is in Figure 4. A complete se-
quence of steps from storing data to obtaining results
is in a sequence diagram in Figure 5. Communica-
tion between the client and the server is secured by
Kerberos (Neuman and Ts’o, 1994).
4.5 System Security
The server itself operates a daily updated Debian
Linux distribution. It is located on a secured univer-
sity network protected by the firewall. Communica-
tion with the server is secured by Kerberos and the
SSL channel. Moreover, all datasets are anonymized
before uploading. The datasets were anonymized by
using identification numbers and neither potential at-
tackers nor experimenters themselves would be able
to assign them to any real person. Since the server it-
self is secured and datasets are anonymized, raw EEG
data encryption was not necessary.
Cloud Infrastructure for Storing and Processing EEG and ERP Experimental Data
277
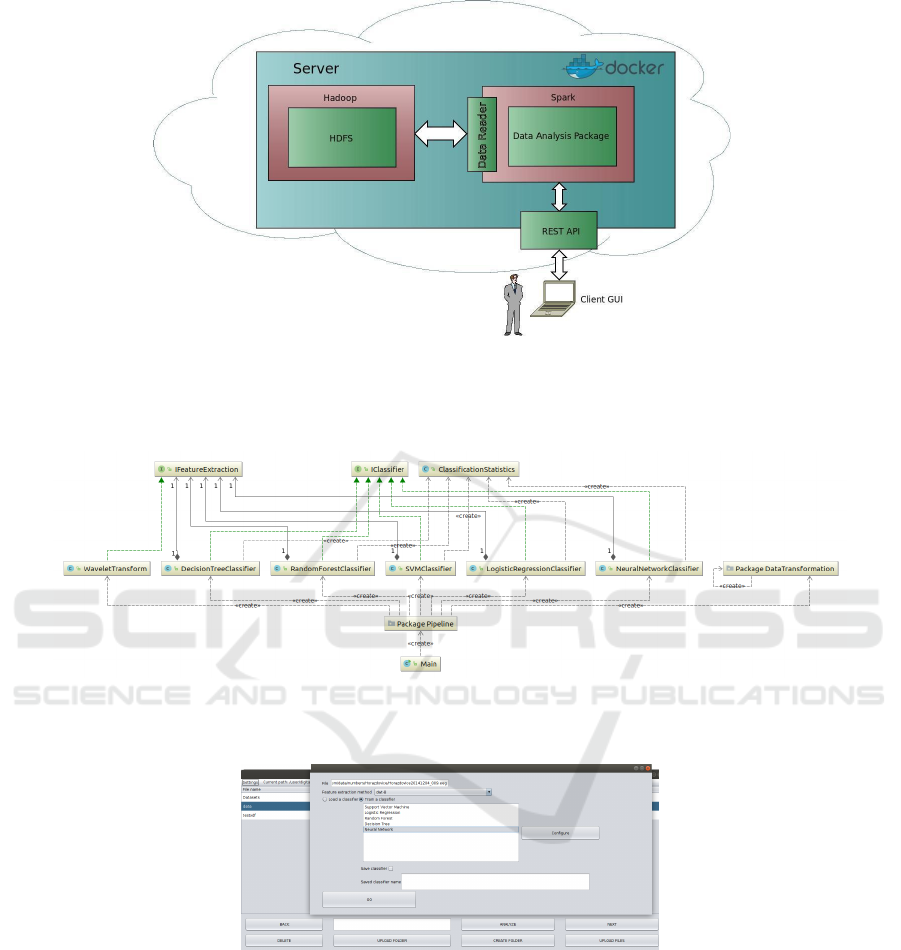
Figure 2: Architecture of the system. A cloud part of the architecture is a docker image containing on the left side: A Hadoop
distributed file system (HDFS) implemented in the Cloudera platform. On the right side: The remote server operating the data
analysis package. The server accesses data via a data reader and communicates with the client GUI program via the REST
API.
Figure 3: UML Diagram. Wavelet transformation implements the IFeatureExtraction interface. The IClassifier interface is
implemented by several classifiers including random forest, SVM, logistic regression and neural networks. A flow of the data
through the code is controlled by the Pipeline package.
Figure 4: Graphical User Interface
5 DISCUSSION
We have been collecting ERP experiments in a neu-
roinformatics laboratory at our department, in a hos-
pital university and also outside using portable de-
vices. Between autumn 2014 and spring 2015
Figure 2: Architecture of the system. A cloud part of the architecture is a docker image containing on the left side: A Hadoop
distributed file system (HDFS) implemented in the Cloudera platform. On the right side: The remote server operating the data
analysis package. The server accesses data via a data reader and communicates with the client GUI program via the REST
API.
Figure 3: UML Diagram. Wavelet transformation implements the IFeatureExtraction interface. The IClassifier interface is
implemented by several classifiers including random forest, SVM, logistic regression and neural networks. A flow of the data
through the code is controlled by the Pipeline package.
Figure 4: Graphical User Interface.
5 DISCUSSION
We have been collecting ERP experiments in a neu-
roinformatics laboratory at our department, in a hos-
pital university and also outside using portable de-
vices. Between autumn 2014 and spring 2015
we measured experiments at primary and secondary
schools for a science popularization event. Ten times
during that period, we attended various schools and
measured about 20 to 40 tested subjects per day. Later
in 2018, we attended the University hospital in Pilsen
about 10 times where we measured 2-3 tested subjects
per visit. The laboratory experiments from the same
time period consist of about 10 days of experiments
with about 20 tested experimental sessions per day.
The typical experiment we performed at schools is
a ’Guess the number’ experiment. The tested subject
is asked to choose a number from a range 1-9 (the tar-
get stimulus). He/she is then visually stimulated with
a monitor with randomly presented numbers from the
ICT4AWE 2019 - 5th International Conference on Information and Communication Technologies for Ageing Well and e-Health
278
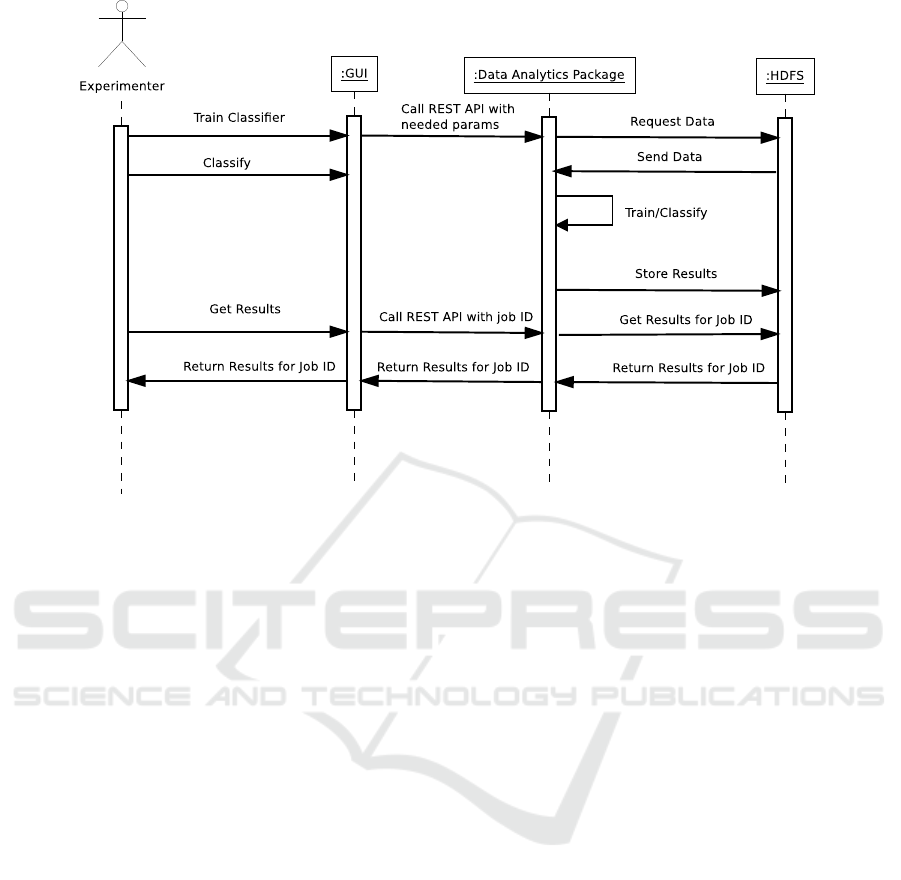
Figure 5: Sequence Diagram.
same range. The goal of the experimenter is to guess
the number based on observing the averaged wave-
forms. The P300 waveform is supposed to appear
following the target stimulus. Finally, we also asked
the participants to reveal the number thought and that
number was recorded as a part of metadata so that the
labels for classifier training were available. Details
about the ’Guess the number’ experiment as well as
the experimental data are described in (Moucek et al.,
2017).
We uploaded the datasets to the cloud infrastruc-
ture by the developed GUI. More than 400 experi-
ments have been stored (tens of gigabytes). Then
we prepared a bash script calling the REST API for
obtaining the datasets and running a simple machine
learning workflow. A part of the data was used for
training and another part for testing. For testing data,
classification accuracy was between 60-70 %. How-
ever, achieving high classification accuracy was not
the main aim of this paper. Classification accuracy is
strongly dependent on how the classifiers are config-
ured, how the testing/training data are selected, and
also on the quality of datasets themselves. The most
important was validation of the complete infrastruc-
ture. We validated that the system is suitably inte-
grated, the data is incorruptibly stored, and classifica-
tion gives reasonable results in reasonable time even
when tested on a single cluster. If more performance
is needed, a new node to the infrastructure can be
added without changing the presented system.
6 CONCLUSIONS
Increasing performance and availability of cloud so-
lutions open new possibilities for storing, processing
and visualization of experimental data. Cloud com-
puting concepts allow users to be more focused on
laboratory work without being worried about main-
taining complex data infrastructures. The usage of
cloud based approaches also relies on the researchers
willingness to entrust sensitive data to third-party
providers. The cloud solutions must be secured, re-
liable and easy-to-use to be accepted by researchers.
There are some existing cloud solution providers such
as Microsoft Azure, IBM Cloud or iCloud but they do
not provide sufficient means to satisfy specific needs
of the electrophysiology domain. Some existing solu-
tions were presented in Section 2. However, they are
not focused on the ERP domain and they are intended
to be used mainly in hospitals or in a clinician prac-
tice. However, there has been no solution for research
in laboratories.
We presented a system based on open source con-
cepts using Apache Hadoop and Spark. We used
HDFS for data storing and we implemented the meth-
ods for signal processing in the Spark context. The
methods are controlled by the REST API. We tested
this infrastructure by storing more than 400 datasets
into the distributed file system and by running simple
workflows aiming at training and testing classifiers.
The presented GUI provides a functionality for
Cloud Infrastructure for Storing and Processing EEG and ERP Experimental Data
279

data management and for controlling the available
methods. Moreover if a laboratory notebook operates
some open source tool for collecting data such as e.g.
Neurolab or LabRecorder it can be integrated with the
system using the REST API.
Our future work includes e.g. an implementation
of a workflow designer that is supposed to be a com-
fortable graphical tool for designing signal processing
workflows (as described in Section 3.1). Data running
through these workflows could be processed in the
presented system, too. There also exist micro devices
such as smart phones, smart watches or other wear-
able devices as heart belts, insulin pumps etc. Storing
data from these micro devices in the presented system
could be finally helpful in building complete Internet
of Things (IoT) infrastructures. Our future goal is also
extending the methods library to provide a larger col-
lection of methods.
ACKNOWLEDGEMENTS
The work was supported by the program INTER-
REG V-A (CZ-BY) Cross-border Cooperation Pro-
gram Czech Republic - Free State of Bavaria un-
der the project No. 85 Brain-driven computer assis-
tance system for people with limited mobility and the
project LO1506 of the Czech Ministry of Education,
Youth and Sports under the program NPU I. Also spe-
cial thanks go to Dorian Beganovic who implemented
most of the code as a part of the Google Summer of
Code 2017 project. Codes for the methods library are
available in (Jezek and Beganovic, 2019a), the client
GUI in (Jezek and Beganovic, 2019b) and the server
in (Jezek and Beganovic, 2019c).
REFERENCES
Abelard and H
´
eloise (2011). Why data and publications
belong together. D-lib magazine, 17(1/2).
Apache Software Foundation (2009). Apache hadoop.
http://hadoop. apache. org.
Ekanayake, J., Pallickara, S., and Fox, G. (2008). Mapre-
duce for data intensive scientific analyses. In eScience,
2008. eScience’08. IEEE Fourth International Confer-
ence on, pages 277–284. IEEE.
Farwell, L. A. and Donchin, E. (1988). Talking off the top of
your head: toward a mental prosthesis utilizing event-
related brain potentials. Electroencephalography and
Clinical Neurophysiology, 70(6):510–523.
Fraiwan, L., Lweesy, K., Khasawneh, N., Wenz, H., and
Dickhaus, H. (2012). Automated sleep stage iden-
tification system based on time–frequency analysis
of a single eeg channel and random forest classi-
fier. Computer methods and programs in biomedicine,
108(1):10–19.
Garc
´
ıa-Larrea, L., Lukaszewicz, A.-C., and Maugui
´
ere, F.
(1992). Revisiting the oddball paradigm. non-target
vs neutral stimuli and the evaluation of erp attentional
effects. Neuropsychologia, 30(8):723 – 741.
Gopalani, S. and Arora, R. (2015). Comparing apache spark
and map reduce with performance analysis using k-
means. International journal of computer applica-
tions, 113(1).
G
¨
uler, N. F.,
¨
Ubeyli, E. D., and G
¨
uler, I. (2005). Recur-
rent neural networks employing lyapunov exponents
for eeg signals classification. Expert systems with ap-
plications, 29(3):506–514.
Hoffmann, U., Vesin, J.-M., Ebrahimi, T., and Diserens, K.
(2008). An Efficient P300-based Brain-Computer In-
terface for Disabled Subjects. Journal of neuroscience
methods, 167(1):115–125.
Ishwarappa and Anuradha, J. (2015). A brief introduction
on big data 5vs characteristics and hadoop technology.
Procedia Computer Science, 48:319 – 324. Interna-
tional Conference on Computer, Communication and
Convergence (ICCC 2015).
Jezek, P. and Beganovic, D. (2019a). Data analysis pack-
age. https://github.com/NEUROINFORMATICS-
GROUP-FAV-KIV-ZCU/EEG DataAnalysisPackage.
Jezek, P. and Beganovic, D. (2019b). Data analysis pack-
age. https://github.com/NEUROINFORMATICS-
GROUP-FAV-KIV-ZCU/EEG ClientGUI.
Jezek, P. and Beganovic, D. (2019c). Data analysis pack-
age. https://github.com/NEUROINFORMATICS-
GROUP-FAV-KIV-ZCU/EEG RemoteServer.
Kemp, B. and Olivan, J. (2003). European data format
‘plus’(edf+), an edf alike standard format for the ex-
change of physiological data. Clinical Neurophysiol-
ogy, 114(9):1755–1761.
Kothe, C. (2014). Lab streaming layer (lsl). https://github.
com/sccn/labstreaminglayer. Accessed on October,
26:2015.
Lotte, F., Bougrain, L., Cichocki, A., Clerc, M., Congedo,
M., Rakotomamonjy, A., and Yger, F. (2018). A
Review of Classification Algorithms for EEG-based
Brain-Computer Interfaces: A 10-year Update. Jour-
nal of Neural Engineering, page 55.
Luck, S. (2014). An Introduction to the Event-Related Po-
tential Technique. A Bradford Book. MIT Press.
Mallat, S. and Zhang, Z. (1993). Matching pursuits with
time-frequency dictionaries. IEEE Transactions on
Signal Processing, 41:3397–3415.
Meng, X., Bradley, J., Yavuz, B., Sparks, E., Venkataraman,
S., Liu, D., Freeman, J., Tsai, D., Amde, M., Owen,
S., et al. (2016). Mllib: Machine learning in apache
spark. The Journal of Machine Learning Research,
17(1):1235–1241.
Merkel, D. (2014). Docker: lightweight linux containers for
consistent development and deployment. Linux Jour-
nal, 2014(239):2.
Moucek, R., Vareka, L., Prokop, T., Stebetak, J., and Bruha,
P. (2017). Event-related potential data from a guess
ICT4AWE 2019 - 5th International Conference on Information and Communication Technologies for Ageing Well and e-Health
280

the number brain-computer interface experiment on
school children. Scientific Data, pages 1–11.
Neuman, B. C. and Ts’o, T. (1994). Kerberos: An authenti-
cation service for computer networks. IEEE Commu-
nications magazine, 32(9):33–38.
Polat, K. and G
¨
unes¸, S. (2007). Classification of epilepti-
form eeg using a hybrid system based on decision tree
classifier and fast fourier transform. Applied Mathe-
matics and Computation, 187(2):1017–1026.
Sahoo, S. S., Jayapandian, C., Garg, G., Kaffashi, F.,
Chung, S., Bozorgi, A., Chen, C.-H., Loparo, K., Lha-
too, S. D., and Zhang, G.-Q. (2013). Heart beats in
the cloud: distributed analysis of electrophysiological
‘big data’using cloud computing for epilepsy clinical
research. Journal of the American Medical Informat-
ics Association, 21(2):263–271.
Subasi, A. and Ercelebi, E. (2005). Classification of eeg
signals using neural network and logistic regression.
Computer methods and programs in biomedicine,
78(2):87–99.
Subasi, A. and Gursoy, M. I. (2010). Eeg signal classifica-
tion using pca, ica, lda and support vector machines.
Expert systems with applications, 37(12):8659–8666.
Sur, S. and Sinha, V. K. (2009). Event-related poten-
tial: An overview. Ind Psychiatry J, 18(1):70–73.
21234168[pmid].
Taylor, R. C. (2010). An overview of the
hadoop/mapreduce/hbase framework and its cur-
rent applications in bioinformatics. In BMC
bioinformatics, volume 11, page S1. BioMed Central.
Thota, C., Sundarasekar, R., Manogaran, G., Varatharajan,
R., and Priyan, M. (2018). Centralized fog computing
security platform for iot and cloud in healthcare sys-
tem. In Exploring the convergence of big data and the
internet of things, pages 141–154. IGI Global.
Van Horn, J. and van Pelt, J. (2008). 1st incf workshop on
sustainability of neuroscience databases.
Varatharajan, R., Manogaran, G., and Priyan, M. (2018).
A big data classification approach using lda with
an enhanced svm method for ecg signals in cloud
computing. Multimedia Tools and Applications,
77(8):10195–10215.
Vohra, D. (2015). Pro Docker. Apress, Berkely, CA, USA,
1st edition.
Watson, P., Lord, P., Gibson, F., Periorellis, P., and Pitsilis,
G. (2008). Cloud computing for e-science with car-
men. In 2nd Iberian Grid Infrastructure Conference
Proceedings, pages 3–14. Citeseer.
Webb, P., Syer, D., Long, J., Nicoll, S., Winch, R., Wilkin-
son, A., Overdijk, M., Dupuis, C., and Deleuze, S.
(2013). Spring boot reference guide. Part IV. Spring
Boot features, 24.
Zaharia, M., Xin, R. S., Wendell, P., Das, T., Armbrust,
M., Dave, A., Meng, X., Rosen, J., Venkataraman, S.,
Franklin, M. J., Ghodsi, A., Gonzalez, J., Shenker, S.,
and Stoica, I. (2016). Apache spark: A unified engine
for big data processing. Commun. ACM, 59(11):56–
65.
Cloud Infrastructure for Storing and Processing EEG and ERP Experimental Data
281
