
Towards Automated Management and Analysis of Heterogeneous Data
within Cannabinoids Domain
Kenji Koga
1
, Maria Spichkova
2
and Nitin Mantri
2
1
iSelect, Cheltenham, Australia
2
School of Science, RMIT University, Melbourne, Australia
Keywords:
Software Engineering, Data Integration, Health Systems, Biomedicine, Bioinformatics.
Abstract:
Cannabinoid research requires the cooperation of experts from various field biochemistry and chemistry to
psychological and social sciences. The data that have to be managed and analysed are highly heterogeneous,
especially because they are provided by a very diverse range of sources. A number of approaches focused
on data collection and the corresponding analysis, restricting the scope to a sub-domain. Our goal is to
elaborate a solution that would allow for automated management and analysis of heterogeneous data within
the complete cannabinoids domain. The corresponding integration of diverse data sources would increase the
quality and preciseness of the analysis. In this paper, we introduce the core ideas of the proposed framework
as well as present the implemented prototype of a cannabinoids data platform.
1 INTRODUCTION
Since the beginning of the first phytocannabinoids
characterisation in the 20
th
century, see Grotenher-
men (2004), and the first studies using tetrahydro-
cannabinol and cannabidiol, we faced a boost in
research involving cannabinoids. With the use of
medicinal cannabis being legalised in a growing num-
ber of countries, several studies in different health ar-
eas have been conducted such as in inflammatory dis-
eases, see e.g., Hasenoehrl et al. (2017), neurologi-
cal disorders or related symptoms, see Solimini et al.
(2017); Pertwee (2012), cancer, see Pagano and Bor-
relli (2017); Naderi et al. (2018); Pastor et al. (2004);
Milano et al. (2017) and cardiovascular diseases, see
Mendizabal and Adler-Graschinsky (2007).
To conduct the cannabinoid research effectively
and efficiently, data from different sources have to
be considered. For example, a researcher interested
in finding the best treatment for a particular disease
has to analyse data on specific cannabinoid strains,
the treatment data and the corresponding effects that
patients or doctors have described. Since these data
are handled by different individuals and institutions,
which generally have their own data format, this task
requires the integration of multiple data sources that
are heterogeneous. Moreover, the diversity of the user
backgrounds requires the corresponding adjustments
of the system interface.
A number of approaches aimed to combine data
in areas such as pharmacology, see Gray et al. (2014);
Wishart et al. (2017); Samwald et al. (2011), and
health sciences, see Puppala et al. (2015); Reis et al.
(2018). Most of them applied some open standards
like OpenEHR
1
to present the collected data. This
solution is not applicable for the case of cannabinoids
research: we are dealing not with a single domain that
has already their data standards, but we have to collect
and integrate data from multiple heterogeneous sub-
domains. Thus, the challenges are not only in the in-
tegration of data collected from a single sub-domain,
e.g. health data records, but also in integration of mul-
tiple heterogeneous domains.
Contributions: In this work we propose a platform
for automated collection, management and analysis of
cannabinoids data. This platform will integrate data
from several cannabinoids data sub-domains in order
to provide means for higher quality research analy-
sis. We also present the implemented prototype of the
proposed platform.
Outline: The rest of this paper is organised as fol-
lows. Background and related work are discussed in
Section 2. Sections 3 and 4 introduce the core ideas
of the proposed cannabinoid data platform as well as
1
http://openehr.org/
Koga, K., Spichkova, M. and Mantri, N.
Towards Automated Management and Analysis of Heterogeneous Data within Cannabinoids Domain.
DOI: 10.5220/0007767405390546
In Proceedings of the 14th International Conference on Evaluation of Novel Approaches to Software Engineering (ENASE 2019), pages 539-546
ISBN: 978-989-758-375-9
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
539

its current implementation. Section 5 concludes the
paper and provides some future work directions.
2 BACKGROUND AND RELATED
WORK
In the cannabinoids domain, there are a number of
projects focusing on the acquisition of cannabinoid
data such as Strainprint
2
, SeedFinder
3
and Open
Cannabis Project
4
. The Strainprint project collects
personal data (user profile), data on strains, inges-
tion methods and dosage. Its core objective is to keep
track of the effectiveness of treatments. The other two
projects focus on collecting and sharing information
about cannabis strains, without any objective to inte-
grate these data with any other type of data to identify
the most effective treatment using cannabinoids.
Sawler et al. (2015) analysed the strain data classi-
fication using DNA samples which showed that strain
names do not represent genetically unique variety.
Samples with identical strain names were more ge-
netically similar to samples with different names than
to identical ones. This demonstrates another issue
that has to be covered when developing a system for
management and analysis of cannabinoids data: re-
searchers cannot rely on strain names only, the genetic
similarity has to be taken into account.
In genomics, Pharmacogenomics Knowledge-
base, see Whirl-Carrillo et al. (2012), and Public
Health Genomics Knowledge Base, see Yu et al.
(2016), are open Web-based knowledge bases that
collect, curate and provide information about human
genetics and population health. They focus on pro-
viding high-quality information to support medicine-
implementation projects and population health, re-
spectively. To achieve this, they periodically extract
data from, e.g. scientific publications, using manual,
natural language processing and Machine Learning
techniques. They differ from our approach because
they are not fully automated as well as they are fo-
cused on a single domain.
In what follows we discuss the approaches that do
not focus on cannabinoids domain, but present some
computer science concepts related to our research –
big data analytics in healthcare, cloud-based solutions
for health information systems, etc. Luo et al. (2016)
conducted a literature review on big data applica-
tion in biomedical research and health care, focusing
on the big data application in four major biomedical
2
http://strainprint.ca
3
https://en.seedfinder.eu
4
http://opencannabisproject.org
sub-disciplines: bioinformatics, clinical informatics,
imaging informatics, and public health informatics.
The authors identified 68 relevant papers, and their
study demonstrated “While big data holds significant
promise for improving health care, there are several
common challenges facing all the four fields in using
big data technology; the most significant problem is
the integration of various databases.” To provide an
effective and efficient solution for this problem is one
of the goals of the platform we propose.
Wang et al. (2015, 2018) presents a survey on 26
big data analytics cases in healthcare research field
and derived some of the best practices. This anal-
ysis resulted in an architecture with 5 logical layers
including data collection, data aggregation, analytics,
information exploration and data governance. In the
Data collection layer, they have all data sources col-
lection such as structured, semi-structured and un-
structured data. Data aggregation layer deals with
data extraction and transformation. In the Analytics
layer, they process and analyze data using, for exam-
ple, MapReduce and data mining. MapReduce is a
programming paradigm and an associated implemen-
tation for processing and generating large datasets,
see Dean and Ghemawat (2008). MapReduce can
be also seen as the core of Apache Hadoop
5
, an
open source platform for the distributed processing
of structured, semi- and unstructured data. In Infor-
mation exploration layer, Wang et al. (2015, 2018)
proposed to generate reports, alerts and notifications
outputs derived from the Analytics layer. In Data gov-
ernance layer, they propose to deal with ethical, legal,
and regulatory challenges managing all the life-cycle
of data, security, privacy and policies. In our work,
we derived data heterogeneity architectural layers fol-
lowing these best practices adapted to the cannabinoid
data domain.
Yusuf et al. (2015) and Spichkova et al. (2015)
presented a model of a cloud-based platform and
its open-source implementation, which allows re-
searchers to conduct experiments requiring complex
computations over big data. This platform might be
integrated within Analytics and Visualisation Layer
of the architecture we propose, see Section 3.
Calabrese and Cannataro (2015) reviewed main
cloud-based healthcare and biomedicine applications,
especially focusing on healthcare, biomedicine and
bioinformatics solutions. The authors summarised
core issues and problems related to the use of such
platforms for the storage and analysis of patients data.
Bahga and Madisetti (2015) developed and ex-
tended Cloud Health Information Systems Technol-
ogy Architecture, which allows clinical data inte-
5
https://hadoop.apache.org
ENASE 2019 - 14th International Conference on Evaluation of Novel Approaches to Software Engineering
540

gration, access and analytics. It achieves integra-
tion through mapping source-specific format to a do-
main model. It supports formats, such as Health
Level-7 messages and Clinical Document Architec-
ture and raw ASCII. Once data schema matches the
domain model, the framework proceeds with a par-
allel MapReduce aggregation and transformation task
and write the result to HDFS storage. Data access and
analytics can be performed through Hadoop ecosys-
tem components such as Pig
6
or Hive
7
providing
seamless access to all the data inside the cloud us-
ing HCatalog from Hadoop. Since we will deal with
different kinds of domain standards, we will have to
research which are the main standards available in the
cannabinoid data research area and provide equiva-
lence of semantics. However, we will take advantage
of a similar cloud infrastructure approach in order to
get all the benefits that it provides such as paralleliza-
tion of processing jobs, fault-tolerance and scalability.
eClims, see Savonnet et al. (2016), is another
example of an integration framework to deal with
data and schema variability in Biomedical Informa-
tion Systems (BIS). Since Biomedical research area
has to deal with the constant integration of increasing
number of databases and ontologies, they have cre-
ated eClims to facilitate the integration of new data
and extend data models at the same time assuring
quality using Databases and Semantic Web theory. In
this approach, the authors preferred a manual solution
for semantic analysis of collected data that were col-
lected from several providers, hence had a different
structure. The question of a full automation was still
open in that approach.
There are also several approaches to integrate het-
erogeneous pharmacology data such as the Life Sci-
ences Linked Open Data (LSLOD) cloud, but it stills
a difficult task to acquire relevant results. It is nec-
essary to combine knowledge generated from drugs,
physiological functions in biological systems and un-
derlying biological interactions. This demands ef-
forts on integrating multiple heterogeneous sources,
perform manual entity reconciliation and disambigua-
tion, which are non-trivial and non-scalable tasks.
Kamdar and Musen (2017) developed a Platform for
Linked Graph Analytics in Pharmacology to per-
form integration of four different data sources from
LSLOD cloud, using a data model, to abstract all the
relevant mechanisms of drug relations, query feder-
ation, where SPARQL
8
queries are performed in all
sources to generate k-partite network and probabilis-
tic model to discover associations between, for exam-
6
https://pig.apache.org
7
https://hive.apache.org
8
https://www.w3.org/TR/rdf-sparql-query
ple, drugs and adverse drug reactions. In our work,
we will take advantage of their infrastructure on how
to deal with Semantic Web Technologies, i.e., their
data model and query federation in order to perform
data integration.
3 PROPOSED ARCHITECTURE
In this section, we discuss the core aspects of our
Cannabinoids Data Platform (CDP) for the manage-
ment and analysis of cannabinoids data. Figure 1
presents a layered architecture of the CDP.
In the Analytics and Visualization Layer, all
the concepts regarding user interaction are handled.
Users should be able to interact with the CDP through
different kinds of interfaces. We analyze and provide
corresponding functionalities required for each user
type. Interactive and integrated visualizations are pro-
vide in different options for visualizations in charts
and tables. To implement an efficient search func-
tionality, data has to be prepared and indexed (in Data
Processing Layer). Like in the Data Capture Layer,
we have to consider here the specifics of Cannabi-
noids Domain to provide an easy-to-use interface that
allows to find, order/rank, match and analyse the col-
lected data.
As the functionalities provided within this layer
are mostly focused on researchers, it makes sense to
apply the technologies that are already used in the cor-
responding research community. Thus, to support the
analysis and visualisation of the large amount of col-
lected data, we are going to apply a research-oriented
cloud computing platform Chiminey that provides
user-friendly interface for the computation/ analysis
set up, as well as visualisation of the calculation re-
sults as 2D or 3D graphs, see Yusuf et al. (2015) and
Spichkova et al. (2015). Monitoring and alerts has to
be provided to notify users about updates in the data
they are interested (through a previous subscription),
for example, new treatment data or changes in exper-
iments.
In the Data Processing Layer, data are prepared
and provided in the repository of processed data. This
repository contains a number of data sets, each of
them is specialised in providing data for a specific
usage. This is achieved through prior data cleaning
and correction, where useless data is removed, i.e.,
data that is not useful for a specific data set context,
and corrected if there is a possibility to do so. Other
important steps are data classification, categorization
and indexing, as they provide a basis for better search
results as the data with similar features are kept to-
gether in an appropriate set and structured using Se-
Towards Automated Management and Analysis of Heterogeneous Data within Cannabinoids Domain
541
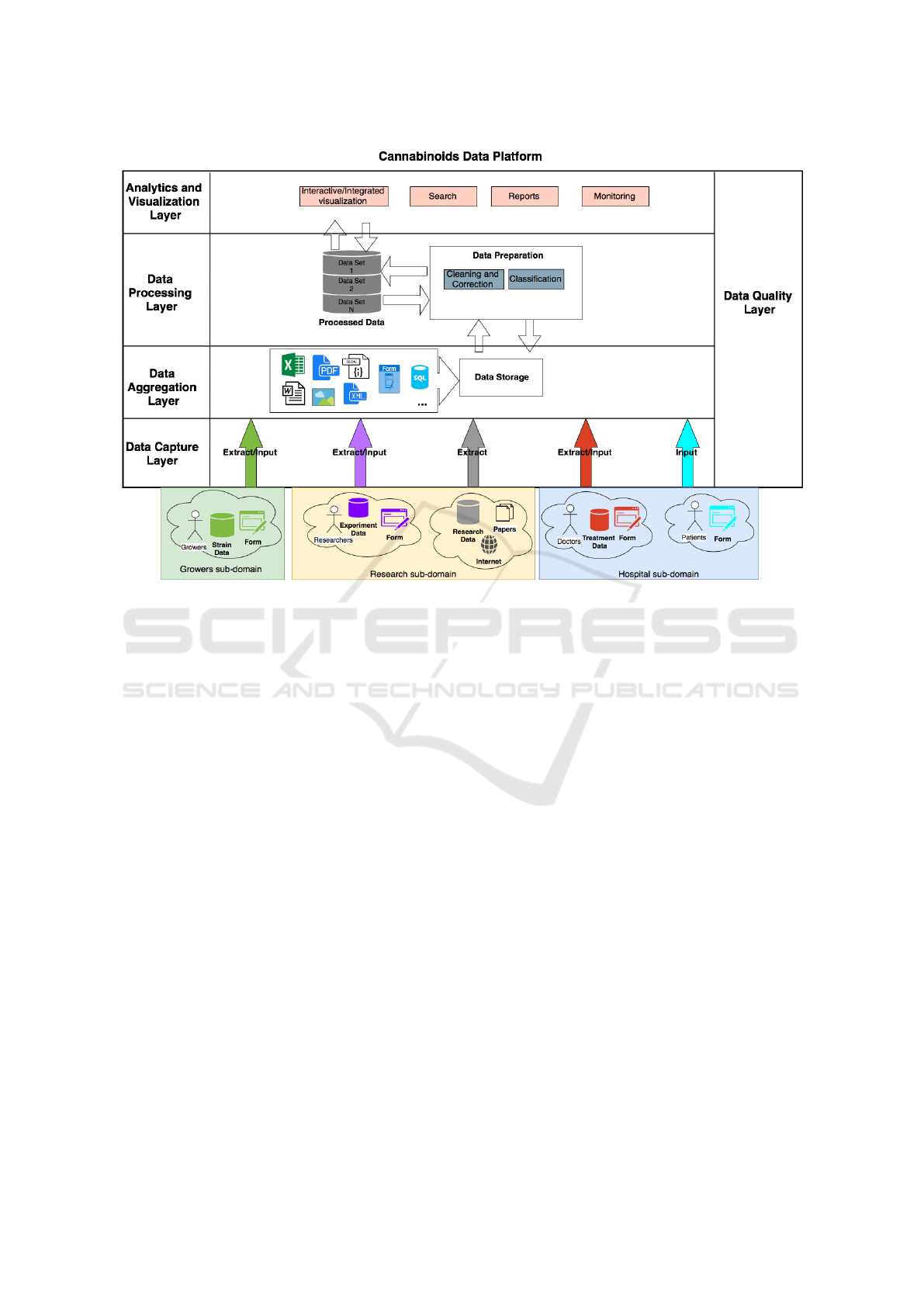
Figure 1: Architecture of the CDP.
mantic Web technologies such as presented in Kam-
dar and Musen (2017). The processing infrastructure
used here is similar to the one proposed by Bahga
and Madisetti (2015). In the Data Aggregation Layer,
data of any different format in the cannabinoids do-
main are aggregated and stored. We are consider-
ing storage of some of the unstructured (Excel, Word,
PDF and images), semi-structured (JSON and XML)
and structured (MySQL and PostgreSQL) data since
we do not have full knowledge of all available data
types in the cannabinoid domain.
In the Data Capture Layer, data is extracted from
research data bases as well as publications or manu-
ally fed in by the users (growers, researchers, doctors/
treatment coordinators, and patients). The users can
provide their data in several ways, for example up-
loading files or typing information in a Web form (in
the case of researchers, doctors / treatment coordina-
tors, and growers) or using a mobile application (in
the case of patients). The user interface for patients
should be as straightforward and simple as possible,
as some of the patients might have not much experi-
ence in using mobile applications.
Since each user of the platform can provide their
data using different mechanisms and interfaces, some
of the data might be unstructured, depending on the
sub-domain:
• The data collected within the hospital and growers
sub-domains is always structured, as all the users
within these sub-domains can provide the data
only using the corresponding forms / interfaces.
Thus, if we would limit the platform to these sub-
domains, a data warehouse solution woulds be
sufficient, see George et al. (2015).
• The vast part of the data collected within the re-
search sub-domain is unstructured and data ex-
traction becomes a challenge within this sub-
domain. This means that only the data lake so-
lution is applicable, see e.g., Soini et al. (2017);
Miloslavskaya and Tolstoy (2016); Fang (2015).
Also, the corresponding algorithms have to be devel-
oped to overcome this heterogeneity in data acquisi-
tion. A meta-format of the data should be applied,
so that all collected data can be represented within
this format providing a common basis for the further
analysis of data.
The Data Quality Layer is orthogonal to all the
other Layers and deals with the quality of data that
flows through these layers. Thus, it is dedicated to
syntactical and semantic data analysis and verifica-
tion, as well as quality assurance and usability as-
pects. One of important aspects is here also the track-
ing of data flows through all layers to provide means
to reproduce data processing conducted in our archi-
tecture.
ENASE 2019 - 14th International Conference on Evaluation of Novel Approaches to Software Engineering
542
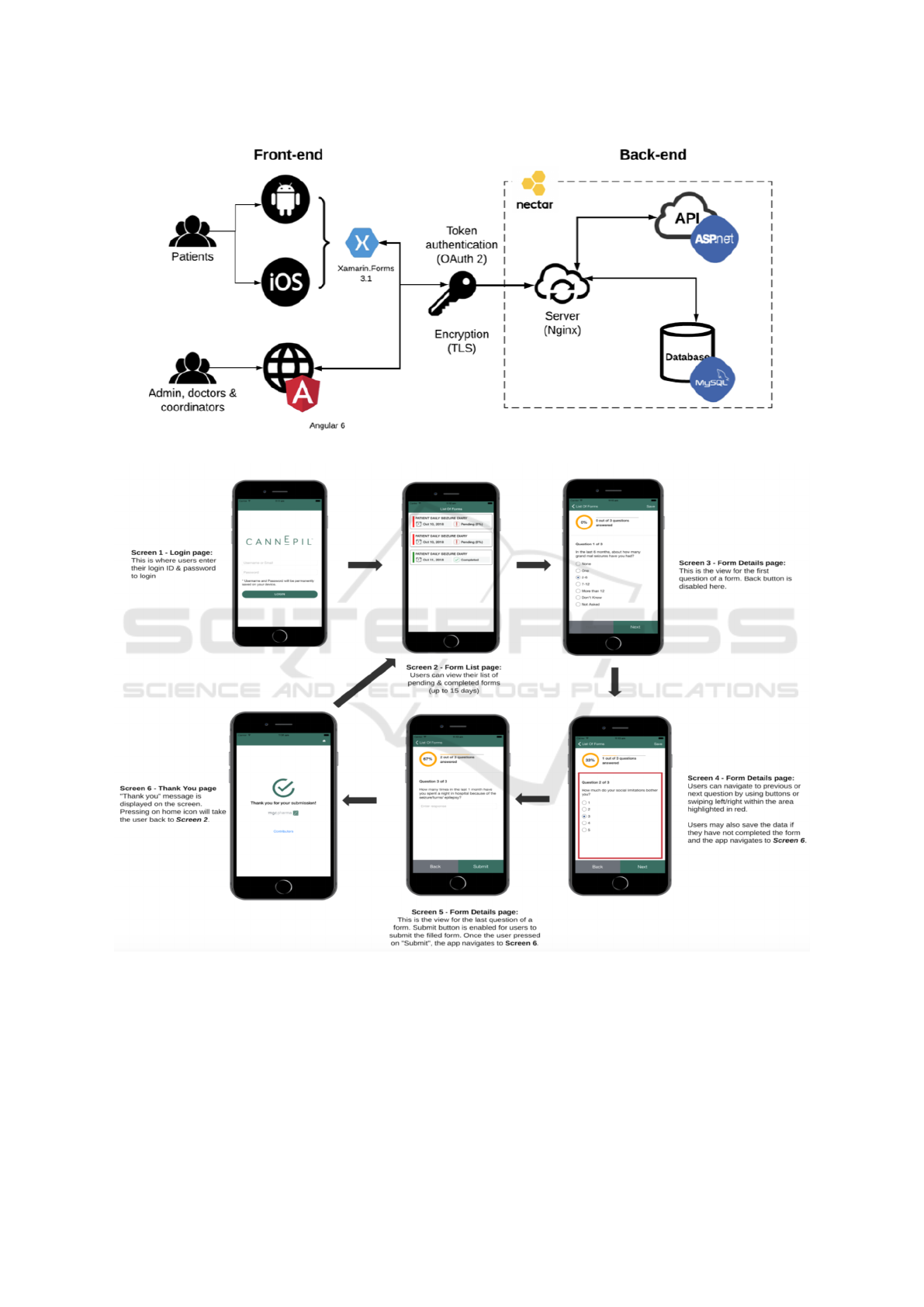
Figure 2: CDP: Architecture of the hospital sub-domain.
Figure 3: CDP: Flow Diagram for the hospital sub-domain (patient interface).
4 CANNABINOIDS DATA
PLATFORM
In this section, we introduce the current implemen-
tation of CDP. Currently, the prototype focuses on
the infrastructure required to collect data from the
users within the hospital and research sub-domains:
patients, doctors/ treatment coordinators and re-
searchers. Figure 2 presents the architecture of the
hospital sub-domain.
The prototype has two core interface components:
• mobile applications (both iOS and Android) de-
Towards Automated Management and Analysis of Heterogeneous Data within Cannabinoids Domain
543
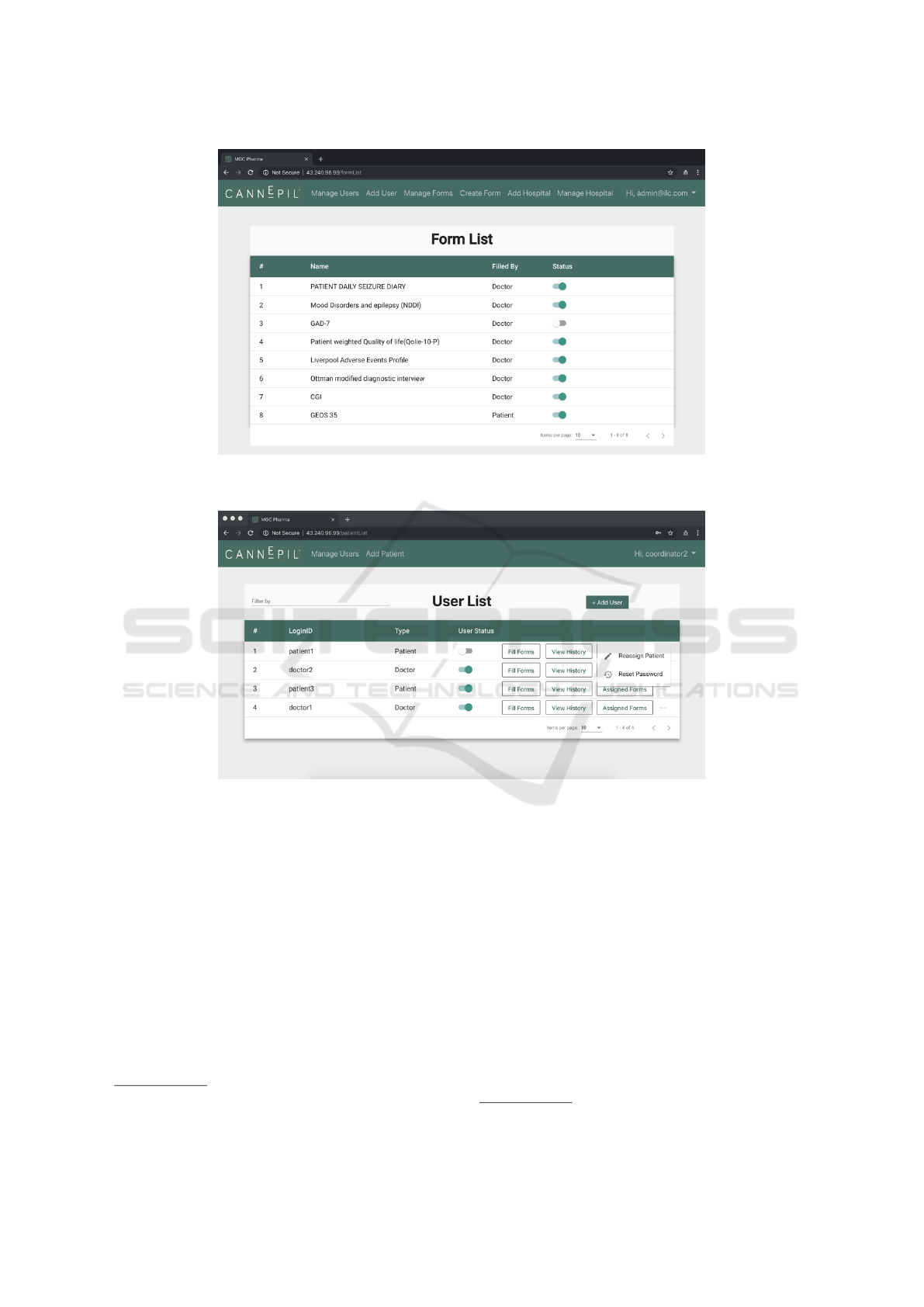
Figure 4: CDP: User interface for the hospital sub-domain (“Manage Forms” page, part of the functionality provided to doctor
/ treatment coordinator users).
Figure 5: CDP: User interface for the hospital sub-domain (“Manage Users” page, part of the functionality provided to doctor/
treatment coordinator users).
veloped for patients, see Figure 3; the applications
were built using Xamarin
9
, which provides cross-
platform compatibility between Android and iOS
platforms; Having a C#-shared codebase, devel-
opers can use Xamarin tools to write native An-
droid, iOS, and Windows apps with native user in-
terfaces and share code across multiple platforms.
• Web applications providing interfaces for doctors/
coordinators and researchers was developed us-
ing Angular 6, which is a TypeScript-based open-
source front-end web application platform
10
(see
Figure 4 for an example of the implemented Web
interfaces).
9
https://visualstudio.microsoft.com/xamarin
10
https://angular.io
Between the Front-end and Back-end, we imple-
mented an authentication system using OAuth2 with
refresh token grant type. In addition, the system is
using Transport Layer Security (TLS) cryptographic
protocols to provide communications security over a
computer network. In Back-end, MySQL Database is
used to store all resource data, and we implemented
an ASP.NET Web API project to communicate with
DB. The whole back-end including API (Application
Programming Interface) and data base are hosted in
Nectar Cloud
11
that provides free cloud services for
Australian Researchers.
Users have to register and log in before they are
provided a suitable interface for them. Patients can
11
https://nectar.org.au/research-cloud
ENASE 2019 - 14th International Conference on Evaluation of Novel Approaches to Software Engineering
544

add their treatment history including severity of con-
dition and effectiveness of particular formulations.
They can keep track of their treatment history to un-
derstand what works for their condition. Their as-
signed doctors have access to their treatment data to
customize the treatment.
Doctors can manage all their patient’s cases, allo-
cate to them corresponding questionnaires / forms to
fill out on regular basis to collect data on the progress
of the treatment and its effectiveness. Doctors can add
comments and annotations for an individual case, as
well as add/ remove treatments for the patients.
Researchers, users that have to request higher
privileges in the system because they can browse any
patient case to help their research. They can also
search for a comprehensive investigation of strain
data, which also includes added advanced search
functionalities on the system.
The standard functionality to deal with the user
profiles is also covered within the current version of
the prototype: all users can update their profile; pa-
tients and doctors can submit a request to become a
researcher with the CDP, where the researcher role
provides an access to anonymised data on treatments
and experiments being conducted.
5 CONCLUSIONS
In this paper, we presented the core ideas of the
Cannabinoids Data Platform, which goal is to in-
tegrate data from the complete cannabinoids do-
main, including research, hospital and growers sub-
domains. Dealing with multiple heterogeneous
sub-domains means additional challenges in col-
lecting and integrating heterogeneous and unstruc-
tured or semi-structured data from several sources,
as we cannot rely on a single data structure/ for-
mat or predefined data standards. However, a meta-
structure/format can be introduces to integrate the
data. We implemented a prototype of the platform,
which currently has two types of interfaces: iOS and
Android mobile applications developed for patients,
and Web applications developed for doctors/ treat-
ment coordinators and researchers. The user inter-
faces in different sub-domains also differ, as we have
to take int account not only the type of data we col-
lect from each sub-domain but also the preferences
and skills of the users within the sub-domain.
Future work will be focused on (1) extending the
prototype to cover the growers sub-domain, (2) im-
plementing the data extraction algorithm from the
data lake within the research sub-domain, and (3)
analysing the usability and efficiency aspects for man-
agement and analysis of collected cannabinoids data.
ACKNOWLEDGEMENTS
We would like to thank Nidhi Chawla, Rachita
Chugh, Rochelle Maria Gracias, Jitender Singh
Padda, Songyan Li, Minh Tuan Nguyen, for their con-
tributions to the implementation of the prototype plat-
form.
REFERENCES
Bahga, A. and Madisetti, V. K. (2015). Healthcare data
integration and informatics in the cloud. Computer,
48(2):50–57.
Calabrese, B. and Cannataro, M. (2015). Cloud computing
in healthcare and biomedicine. Scalable Computing:
Practice and Experience, 16(1):1–18.
Dean, J. and Ghemawat, S. (2008). Mapreduce: simplified
data processing on large clusters. Communications of
the ACM, 51(1):107–113.
Fang, H. (2015). Managing data lakes in big data era:
What’s a data lake and why has it became popular in
data management ecosystem. In Int. Conference on
Cyber Technology in Automation, Control, and Intel-
ligent Systems, pages 820–824. IEEE.
George, J., Kumar, V., and Kumar, S. (2015). Data ware-
house design considerations for a healthcare business
intelligence system. In World congress on engineer-
ing.
Gray, A. J., Groth, P., Loizou, A., Askjaer, S., Brenninkmei-
jer, C., Burger, K., Chichester, C., Evelo, C. T., Goble,
C., Harland, L., et al. (2014). Applying linked data
approaches to pharmacology: Architectural decisions
and implementation. Semantic Web, 5(2):101–113.
Grotenhermen, F. (2004). Clinical pharmacodynamics of
cannabinoids. Journal of Cannabis Therapeutics,
4(1).
Hasenoehrl, C., Storr, M., and Schicho, R. (2017). Cannabi-
noids for treating inflammatory bowel diseases: where
are we and where do we go? Expert Review of Gas-
troenterology & Hepatology, 11(4):329–337.
Kamdar, M. R. and Musen, M. A. (2017). Phlegra: Graph
analytics in pharmacology over the web of life sci-
ences linked open data. In Int. Conference on World
Wide Web, pages 321–329.
Luo, J., Wu, M., Gopukumar, D., and Zhao, Y. (2016).
Big data application in biomedical research and health
care: a literature review. Biomedical informatics in-
sights, 8:BII–S31559.
Mendizabal, V. and Adler-Graschinsky, E. (2007). Cannabi-
noids as therapeutic agents in cardiovascular disease:
a tale of passions and illusions. British journal of
pharmacology, 151(4):427–440.
Milano, W., Padricelli, U., and Capasso, A. (2017). Re-
cent advances in research and therapeutic application
of cannabinoids in cancer disease.
Towards Automated Management and Analysis of Heterogeneous Data within Cannabinoids Domain
545

Miloslavskaya, N. and Tolstoy, A. (2016). Big data, fast
data and data lake concepts. Procedia Computer Sci-
ence, 88:300–305.
Naderi, J., Dana, N., Javanmard, S. H., Amooheidari, A.,
Yahay, M., and Vaseghi, G. (2018). Effects of stan-
dardized cannabis sativa extract and ionizing radiation
in melanoma cells in vitro.
Pagano, E. and Borrelli, F. (2017). Targeting cannabi-
noid receptors in gastrointestinal cancers for thera-
peutic uses: current status and future perspectives.
Expert Review of Gastroenterology & Hepatology,
11(10):871–873.
Pastor, M. G., Garcia, C. S., and Roperh, I. G. (2004). Ther-
apy with cannabinoid compounds for the treatment of
brain tumors. US Patent App. 10/647,739.
Pertwee, R. G. (2012). Targeting the endocannabinoid sys-
tem with cannabinoid receptor agonists: pharmaco-
logical strategies and therapeutic possibilities. Philo-
sophical Transactions of the Royal Society of London
B: Biological Sciences, 367(1607):3353–3363.
Puppala, M., He, T., Chen, S., Ogunti, R., Yu, X., Li, F.,
Jackson, R., and Wong, S. T. C. (2015). Meteor:
An enterprise health informatics environment to sup-
port evidence-based medicine. IEEE Transactions on
Biomedical Engineering, 62(12):2776–2786.
Reis, L. F., Ferreira, D. G., Maranhao, P. A., Cruz-Correia,
R., and Vieira-Marques, P. (2018). Integration through
mapping — an openehr based approach for research
oriented integration of health information systems.
In Conf. on Information Systems and Technologies,
pages 1–5.
Samwald, M., Jentzsch, A., Bouton, C., Kallesøe, C. S.,
Willighagen, E., Hajagos, J., Marshall, M. S.,
Prud’hommeaux, E., Hassanzadeh, O., Pichler, E.,
et al. (2011). Linked open drug data for pharmaceuti-
cal research and development. Journal of cheminfor-
matics, 3(1):19.
Savonnet, M., Leclercq, ., and Naubourg, P. (2016). eclims:
An extensible and dynamic integration framework
for biomedical information systems. IEEE Journal
of Biomedical and Health Informatics, 20(6):1640–
1649.
Sawler, J., Stout, J. M., Gardner, K. M., Hudson, D., Vid-
mar, J., Butler, L., Page, J. E., and Myles, S. (2015).
The genetic structure of marijuana and hemp. PLOS
ONE, 10(8):1–9.
Soini, E., Hallinen, T., Kekoni, A., Kotimaa, J., Nyk
¨
anen,
M., Tirkkonen, J., and Tervahauta, M. (2017). Effi-
cient secondary use of representative social and health
care data in finland: Isaacus data lake, analytics and
knowledge management pre-production project. Value
in Health, 20(9).
Solimini, R., Rotolo, M. C., Pichini, S., and Pacifici, R.
(2017). Neurological disorders in medical use of
cannabis: an update. CNS & Neurological Disorders-
Drug Targets (Formerly Current Drug Targets-CNS &
Neurological Disorders), 16(5):527–533.
Spichkova, M., Thomas, I. E., Schmidt, H. W., Yusuf, I. I.,
Drumm, D. W., Androulakis, S., Opletal, G., and
Russo, S. P. (2015). Scalable and fault-tolerant cloud
computations: Modelling and implementation. In Int.
Conference on Parallel and Distributed Systems (IC-
PADS), pages 396–404. IEEE.
Wang, Y., Kung, L., and Byrd, T. A. (2018). Big data an-
alytics: Understanding its capabilities and potential
benefits for healthcare organizations. Technological
Forecasting and Social Change, 126:3 – 13.
Wang, Y., Kung, L., Ting, C., and Byrd, T. A. (2015). Be-
yond a technical perspective: Understanding big data
capabilities in health care. In 2015 48th Hawaii Inter-
national Conference on System Sciences, pages 3044–
3053.
Whirl-Carrillo, M., McDonagh, E. M., Hebert, J., Gong, L.,
Sangkuhl, K., Thorn, C., Altman, R. B., and Klein,
T. E. (2012). Pharmacogenomics knowledge for per-
sonalized medicine. Clinical Pharmacology & Ther-
apeutics, 92(4):414–417.
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu,
A., Grant, J. R., Sajed, T., Johnson, D., Li, C., Say-
eeda, Z., et al. (2017). Drugbank 5.0: a major update
to the drugbank database for 2018. Nucleic acids re-
search, 46(D1):D1074–D1082.
Yu, W., Gwinn, M., Dotson, W. D., Green, R. F., Clyne,
M., Wulf, A., Bowen, S., Kolor, K., and Khoury, M. J.
(2016). A knowledge base for tracking the impact of
genomics on population health. Genetics in Medicine,
18(12).
Yusuf, I. I., Thomas, I. E., Spichkova, M., Androulakis, S.,
Meyer, G. R., Drumm, D. W., Opletal, G., Russo, S. P.,
Buckle, A. M., and Schmidt, H. (2015). Chiminey:
Reliable computing and data management platform in
the cloud. In 37th International Conference on Soft-
ware Engineering (ICSE).
ENASE 2019 - 14th International Conference on Evaluation of Novel Approaches to Software Engineering
546
