
Effective Frequent Motif Discovery for Long Time Series Classification:
A Study using Phonocardiogram
Hajar Alhijailan
1,2 a
and Frans Coenen
1 b
1
Department of Computer Science, University of Liverpool, Liverpool, U.K.
2
College of Computer and Information Sciences, King Saud University, Riyadh, Saudi Arabia
Keywords:
Time and Point Series Analysis, Frequent Motifs, Data Preprocessing, Classification, Phonocardiogram.
Abstract:
A mechanism for extracting frequent motifs from long time series is proposed, directed at classifying phono-
cardiograms. The approach features two preprocessing techniques: silent gap removal and a novel candidate
frequent motif discovery mechanism founded on the clustering of time series subsequences. These techniques
were combined into one process for extracting discriminative frequent motifs from single time series and then
to combine these to identify a global set of discriminative frequent motifs. The proposed approach compares
favourably with these existing approaches in terms of accuracy and has a significantly improved runtime.
1 INTRODUCTION
Time series analysis is directed at the extraction of
knowledge from temporally referenced data. The
usual application, and that of interest with respect to
this paper, is the construction of a classification model
for labelling (classifying) time series (Mueen et al.,
2009) . However, the time series of interest are typ-
ically too large to be considered in their entirety; for
example as a single feature vector. This issue can be
addressed by identifying motifs within the time se-
ries (Dau and Keogh, 2017) . A motif in this con-
text is some subsequence of points occurring within
a time series which is deemed to be representative of
the underlying class-label associated with the time se-
ries (Krejci et al., 2016). A representative motif can
be defined in various ways; that considered in this pa-
per is frequency of occurrence (Agarwal et al., 2015).
A number of motif discovery techniques have
been proposed (Gao et al., 2017; Dau and Keogh,
2017) . However, the discovery of good motifs in time
series remains computationally challenging; mainly
because of the large number of candidate motifs that
need to be considered. Schemes aimed at reducing
this complexity (Mueen et al., 2009) remain problem-
atic in that the classification accuracy tends to be ad-
versely affected, because of the nature of the various
proposed heuristics used to limit the time complexity.
a
https://orcid.org/0000-0002-4169-7911
b
https://orcid.org/0000-0003-1026-6649
A mechanism where by the motif discovery pro-
cess complexity can be reduced is by preprocessing
the data. A new technique, considered in detail in
this paper, is to prune the time series by removing
subsequences that are unlikely to be representative
of any class-label. Three categories of time series
subsequences for pruning can be identified: (i) sub-
sequences that exist in every time series and hence
not representative of any particular class, (ii) subse-
quences that appear so infrequently that they cannot
be deemed relevant and (iii) subsequences that appear
across two or more classes and thus cannot be usefully
employed to discriminate between classes.
The precise nature of the most appropriate pre-
processing technique to be adopted is very much de-
pendent on the application domain under considera-
tion. The application domain at which the work pre-
sented in this paper is directed at the classification
of canine Phonocardiograms (PCGs) according to a
variety of heart conditions. A PCG is a single vari-
ate time series, typically obtained using an electronic
stethoscope. The advantage offered, with respect to
computerised processing, is that the information con-
tained in a PCG is more than that can be distinguished
by the human ear or by visual inspection. The work
presented in this paper is thus directed at preprocess-
ing PCG data. Firstly by removing subsequences rep-
resenting “silent gaps”. Secondly by removing subse-
quences that cannot be frequent, using a novel tech-
nique involving clustering. Thirdly by discounting
motifs that are not good discriminators of a class.
266
Alhijailan, H. and Coenen, F.
Effective Frequent Motif Discovery for Long Time Series Classification: A Study using Phonocardiogram.
DOI: 10.5220/0008018902660273
In Proceedings of the 11th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2019), pages 266-273
ISBN: 978-989-758-382-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2 PREVIOUS WORK
The basic motif discovery technique is to determine
the frequency of occurrence of candidate motifs by
counting the number of similar subsequences that ex-
ist in the input data; in other words, by conducting
a large number of comparisons (Keogh and Pazzani,
2001). Efficiency gains can be made by preprocess-
ing/pruning the input data or using knowledge spe-
cific to the application domain to limit the number of
comparisons. Popular preprocessing techniques in the
context of time series include: Segmentation, Down-
sampling, Filtering, Decimation and De-noising.
An alternative pruning technique, applicable in
the context of certain time series applications, includ-
ing the PCG application considered in this paper, is
“silent gap” removal. This is a technique frequently
used with respect to applications that are founded on
audio data. Silent gap removal was first proposed
and adopted in the context of applications directed
at Voice Activity Detection (VAD) and speech recog-
nition (Sohn et al., 1999). The main motivation for
removing silent gaps in VAD and speech recognition
was that these subsequences were not likely to carry
any information (Yang et al., 2010); by removing
the silent gaps, the problem domain becomes more
tractable. Of course silent gap identification also al-
lows for the isolation of individual words, syllables
and sentences (Ram
´
ırez et al., 2004). To the best
knowledge of the authors, there is no work on silent
gap removal in the context of PCG data.
3 FORMALISM
This section presents a formalism for the work pre-
sented in this paper.
Definition 1, Time Series: A time/ point series P is a
sequence of x data values {p
1
, . . . , p
x
} associated
with a class-label c
i
taken from a set of classes
C = {c
1
, c
2
, . . . }. In the case of the training and
test data, this label is known. In the case of previ-
ously unseen data, this is what the classifier is in-
tended to predict. A collection of z labelled point
series is then given by T = {hP
1
, c
1
i, . . . , hP
z
, c
z
i}
where each P
i
is a point series and c
i
∈ C.
Definition 2, Pruned Time Series: A pruned time/
point series P
0
of P is a sequence of y data val-
ues P
0
= {p
1
, . . . , p
y
} such that y ≤ x, P
0
⊆ P.
Definition 3, Time Series Subsequence: A subse-
quence s of a time series P (P
0
) is any consecutive
set of points in the time series; s = {p
1
, . . . , p
ω
}
and ω is a pre-specified length of s. The set S is
the set of all possible subsequences, of length ω,
in P (P
0
); S = {s
1
, . . . , s
x−ω+1
}. The generation
of S is computationally expensive; given any
reasonably sized time series, there will x − ω + 1
(y − ω + 1) subsequences. This can be mitigated
against by generating only a limited number of
subsequences, S = {s
1
, . . . , s
max
}.
Definition 4, Motif: A motif m is a subsequence s
j
∈
S, where S is drawn from a point series P (P
0
),
that is representative of a class-label c
i
. One way
of determining whether a motif is representative
or not is to consider its frequency of occurrence.
For s
j
to be considered frequent, the set S must
include at least σ subsequences that are in some
sense all similar to s
j
, as define according to a
similarity threshold λ. The set of frequent motifs
generated from S is then given by M = {hm
1
, f
1
i,
hm
2
, f
2
i, . . . }, where f
i
is the frequency count.
Definition 5, Top k Motifs: The k most frequent mo-
tifs in a set M are the “Top k Motifs”. It is argued
that the Top k Motifs are the most representative
of the underlying class associated with S. The top
k motifs drawn from a set of frequent motifs M is
given by the set L = {hl
1
, c
1
i, . . . hl
k
, c
k
i}.
Definition 6, Motif Set: Given a collection of time
series T , the complete set of identified frequent
motifs is given by D =
S
i=z
i=1
L
i
, where z is the
number of records (examples) in T .
Definition 7, Pruned Motif Set: Each motif in D is
a good local representative of a class. This does
not necessarily mean that it is also a good global
representative. The set D therefore needs to be
pruned to give D
0
, a motif set that contains only
good global representatives. The complement of
D
0
in D is then the set D
s
, the set of motifs that are
not good global discriminators. A good represen-
tative motif is defined as one where either there
are no similar motifs in D or if there are similar
motifs in D, they are all linked to the same class;
D
0
and D
s
can be formally defined as follows:
• D
0
, D
s
⊂ D : D
0
∪ D
s
= D and D
0
∩ D
s
=
/
0.
• ∀ d
i
∈ D
0
@ d
j
∈ D : m
i
' m
j
∧ c
i
6= c
j
.
• ∀ d
i
∈ D
s
∃ d
j
∈ D : m
i
' m
j
∧ c
i
6= c
j
.
The set D
0
can then be used to build a classifica-
tion model of some kind.
4 FREQUENT MOTIF
DISCOVERY
The top-level process is given in Algorithm 1. The
input is a set of time series T , and a set of parame-
Effective Frequent Motif Discovery for Long Time Series Classification: A Study using Phonocardiogram
267

ters: (i) ω defining the size of the motifs, (ii) max to
limit the number of candidate frequent motifs, (iii) σ
to define the concept of frequency, (iv) λ to define the
concept of similarity and (v) k to limit the number of
frequent motifs selected. The output is a set D
0
of fre-
quent motifs. The algorithm operates by processing
each pair hP
i
, c
i
i in T in turn. The size of each time
series P
i
is first reduced (line 3) by removing “silent
gaps” so as to give a time series P
0
i
. This is then fur-
ther processed (line 4) to identify a set of sets candi-
date frequent motifs S
0
i
which is then used (lines 6 to
9) to discover the k frequent motifs, within S
0
i
, that are
good local discriminators of the class c
i
. In each case,
the motifs generated so far are collated into a set D
which, if not empty, is processed further (line 12) so
that only motifs that are good global class discrimina-
tors are retained; these are held in a set D
0
to be used
as the “data bank” in classification.
Algorithm 1: Frequent Motif Discovery.
Input: T , ω, max, σ, λ, k
Output: D
0
1: D ←
/
0, Set to hold “good” frequent motifs
2: for ∀hP
i
, c
i
i ∈ T do
3: P
0
i
← silentGapRemoval(P
i
)
4: S
0
i
← candidateFrequentMotifGen(P
0
i
, ω)
5: max =
max
|S
0
i
|
, k =
k
|S
0
i
|
6: for ∀S
0
i
j
∈ S
0
i
do
7: L ← localMotifDiscov(S
0
i
j
, max, σ, λ, k, c
i
)
8: D ← D ∪ L
9: end for
10: end for
11: if D 6=
/
0 then
12: D
0
← globalMotifSelection(D)
13: end if
14: return (D
0
)
The process, as shown in Algorithm 1, comprises
four subprocesses: (i) Silent gap removal, (ii) Can-
didate frequent motif generation, (iii) Frequent motif
discovery for local class discrimination and (iv) Fre-
quent motif selection for global class discrimination.
4.1 Silent Gap Removal
PCGs feature cycles (heartbeats) spaced by “silent
gaps”. These gaps, because they appear in all the time
series, cannot contribute to class discrimination and
therefore should be removed. The adopted mecha-
nism for removing silent gaps is founded on the Math-
Works mechanism
1
, which operates using a sliding
window, of a pre-specified length, to identify a col-
lection of non-overlapping subsequences S. For each
subsequence s
j
in S, two parameters are calculated:
1
https://uk.mathworks.com/matlabcentral/fileexchange/
28826-silence-removal-in-speech-signals
(i) the signal energy (e
j
) and (ii) the spectral centroid
(c
j
) which are then used for pruning.
The silent gap removal process is presented in Al-
gorithm 2. The input is a point series P and a win-
dow size w measured in milliseconds. The output
is a pruned point series P
0
. The process commences
(line 1) by segmenting P into a set of non-overlapping
subsequences S. The parameters e
j
and c
j
are then
computed for each subsequence s
j
in S and stored in
E and C respectively (lines 5 to 10). The thresholds,
t
e
and t
c
are then calculated; the calculation process is
described below. Then (lines 13 to 17) for each sub-
sequence s
j
in S, its parameters (e
j
and c
j
) must fulfil
the conditions to be appended to the end of P
0
, the set
of retained subsequences to be returned at the end.
The function findThreshold finds a threshold for
a collection of values. It starts by generating a his-
togram of the input values; this is then smoothed (H
0
).
The local maxima in H
0
are then identified and stored
in a set M. The threshold is then calculated according
to the number of maxima in M, which is then returned.
Algorithm 2: Silent Gap Removal.
Input: P, w
Output: P
0
1: S ← Set of subsequences of length w in P
2: P
0
←
/
0, Empty set to hold pruned point series P
3: E ←
/
0, Empty set to hold signal energy values
4: C ←
/
0, Empty set to hold spectral centroid values
5: for ∀s
j
∈ S do
6: e
j
← Signal energy calculated for s
j
7: E ← E ∪e
j
8: c
j
← Spectral centroid calculated for s
j
9: C ← C ∪ c
j
10: end for
11: t
e
← findThreshold(E)
12: t
c
← findThreshold(C)
13: for j = 1 to |E| do
14: if e
j
≥ t
e
and c
j
≥ t
c
then
15: P
0
← append(P
0
, s
j
)
16: end if
17: end for
18: return (P
0
)
4.2 Candidate Frequent Motif
Generation
Given a pruned point series P
0
, this can be pruned fur-
ther by removing subsequences that cannot be consid-
ered to be frequent so as to retain a set of candidate
frequent motifs S
0
(S
0
⊂ S), the complete set of sub-
sequence in P
0
of length ω. To do this, a novel algo-
rithm was proposed using a hypothetical motif r re-
ferred to as the “zero motif”. The similarity between
each subsequence s
i
= {s
i
1
, . . . , s
i
ω
}, s
i
∈ S, and r is
calculated using Euclidean Distance similarity. The
obtained similarity values were used to cluster the set
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
268

of subsequences S in P
0
into an ordered set of clusters,
CL = {CL
1
,CL
2
, . . . }, one cluster per unit distance
value, ordered according to size. The subsequences
in S contained the largest cluster, CL
1
, were retained;
however, if the difference between the size of CL
1
and
CL
2
was proportionally small, then CL
2
was also re-
tained. Whether one or two clusters are retained was
defined by a parameter θ, which was calculated using
Equation 1:
θ =
(
1 if
|CL
1
|−|CL
2
|
|S|
× 100 > α
2 otherwise
(1)
where, α is a user-defined percentage of the total
number of subsequences in S. α = 5 is suggested.
The candidate frequent motif generation subpro-
cess is given in Algorithm 3. The input is a pruned
time series P
0
and the window size ω. The output is
a set of sets of candidate frequent motifs S
0
; this may
consist of one or two sets. The algorithm commences
by defining the zero motif. Next, the set S is generated
and a corresponding set D. The set S is then processed
(lines 4 to 7) so as to populate D; there is a one-to-one
correspondence between S and D. The set S is then
clustered (line 8) according to D. The largest cluster
is retained (line 9) and the second largest might also
be retained depending on the θ value (lines 10 to 13).
The retained set of clusters S
0
is then returned.
Algorithm 3: Candidate Frequent Motif Generation.
Input: P
0
, ω
Output: S
0
1: r ← {r
j
: r
j
= 0, j = 1 to j = ω}
2: S ← Set of subsequences of length ω in P
0
3: D ←
/
0, Empty set to hold distance values
4: for ∀s
i
∈ S do
5: d ← Euclidean similarity between s
i
and r
6: D ← D ∪ d
7: end for
8: CL ← Ordered set of clusters obtained by clustering all s
i
∈
S according to d
i
∈ D
9: S
0
← CL
1
10: θ ← Parameter calculated using Equation 1
11: if θ = 2 then
12: S
0
← S
0
∪CL
2
13: end if
14: return (S
0
)
4.3 Frequent Motif Discovery for Local
Class Discrimination
The next subprocess, given a set of candidate frequent
motifs S
0
, is to identify the most frequent motifs that,
by definition, are deemed to be good local discrimi-
nators. However, even after the removal of silent gaps
and infrequent subsequences, the number of remain-
ing subsequences in S
0
is still likely to be large. It
is therefore proposed to limit the number of candi-
date frequent motifs considered using a user-defined
threshold max. The idea is to randomly select max
candidates from the set S
0
. The process is given in
Algorithm 4. The inputs are: a set of candidate fre-
quent motifs S
0
; the thresholds max, σ, λ and k; and
the class-label c associated with the given time series.
The output is a set L of identified frequent motifs. The
first step (lines 1 to 6) is to create a subset S
00
from S
0
comprised of only max subsequences. It is possible
that the value of max is greater than or equal to the
number of subsequences in S
0
in which case S
00
= S
0
.
Next, the frequency count f
i
for each subsequence s
i
in S
00
is calculated using Euclidean Distance and the λ
parameter for determining the similarity between sub-
sequences. Where the count is greater than or equal
to σ% of |S
0
|, the subsequence and count are stored
in M = {hm
1
, f
1
i, hm
2
, f
2
i, . . . } which is then ordered
according to frequency count (line 15). If k is greater
than the number of subsequences in M, k is adjusted
to |M| (lines 16 to 18). Next, (lines 19 to 21) the set L
of the k most frequently occurring motifs is generated.
The set L is then returned (line 22).
Algorithm 4: Local Frequent Motif Discovery.
Input: S
0
, max, σ, λ, k, c
Output: L
1: S
00
←
/
0
2: if max ≥ |S
0
| then
3: S
00
← S
0
4: else
5: S
00
← Set of max subsequences from S
0
6: end if
7: M ←
/
0, Empty set to hold motifs
8: for ∀s
i
∈ S
00
do
9: f
i
← The number of subsequences in S
0
that are similar
to s
i
according to the threshold λ
10: if f
i
≥
σ×|S
0
|
100
then
11: M ← M ∪ hs
i
, f
i
i
12: end if
13: end for
14: L ←
/
0, Empty set to hold top k frequent motifs
15: M ← The set M ordered by frequency
16: if k > |M| then
17: k ← |M|
18: end if
19: for i = 1 to k do
20: L ← L ∪hs
i
, ci (hs
i
, i ∈ M)
21: end for
22: return (L)
The silent gap removal, candidate frequent motif
generation and local frequent motif discovery subpro-
cesses are applied to each point series in the input set
T = {P
1
, P
2
, . . . } and a sequence of sets L generated,
{L
1
, L
2
, . . . }. These are collated into a set D.
Effective Frequent Motif Discovery for Long Time Series Classification: A Study using Phonocardiogram
269

4.4 Frequent Motif Selection for Global
Class Discrimination
Although the motifs in D were deemed to be good
local class discriminators, this did not automatically
mean that they were also good global class discrim-
inators; D may contain motif-class pairs that contra-
dict each other. Thus, the final step was to derive a
set D
0
⊂ D that comprised only good global class dis-
criminators. The process is presented in Algorithm 5.
The input is the set D. Each motif-class pair in D is
then compared with all other pairs in D and if no simi-
lar motif associated with a different class is found, the
motif-class pair is added to the set D
0
(line 11). Simi-
larity is again measured using Euclidean Distance and
the λ parameter. The dataset D
0
can then be used as a
Nearest Neighbour Classifier (NNC) “data bank”.
Algorithm 5: Global Motif Selection.
Input: D, λ
Output: D
0
1: D
0
←
/
0, Set to hold “good” frequent motifs
2: for ∀hm
i
, c
i
i ∈ D do
3: isGoodGlobalDisctimiator ← true
4: for ∀hm
j
, c
j
i ∈ D, i 6= j do
5: if c
i
6= c
j
and m
i
' m
j
then
6: isGoodGlobalDisctimiator ← false
7: break
8: end if
9: end for
10: if isGoodGlobalDisctimiator then
11: D
0
← D
0
∪ hm
i
, c
i
i
12: end if
13: end for
14: return (D
0
)
5 EVALUATION
For the evaluation, a dataset of canine PCGs was used
(described in Subsection 5.1). Five sets of experi-
ments were conducted. The first two were designed
to evaluate the operation of the first two subprocess.
The third set was designed to identify the most appro-
priate values for the parameters ω, max, λ and σ. The
fourth was directed at an investigation of the runtime
complexity of the proposed method, and the fifth at
the quality of the generated motifs.
5.1 Evaluation Data
The data used for the evaluation was a set of canine
PCGs encapsulated as WAVE files. It was collected
using an electronic stethoscope.The resulted point se-
ries were 72 series; the average length of a single se-
ries was 740, 550 points. Each point series had an
associated class-label selected from the class attribute
set {B
1
, B
2
,C,Control}. The first three are stages of
Mitral Valve disease that appear in the data collection.
5.2 The Silent Gap Removal Evaluation
Experiments were conducted to determine the most
appropriate value for w with respect to PCG signals.
To this end, a range of window sizes from w = 10
ms to w = 90 ms, increasing in steps of 10 ms, was
experimented with. It was found that a window size
of 10 ms produced the best result;this was the value
therefore used in this paper. Using w = 10, the input
time series was reduced by a little less than half as a
result of applying silent gap removal.
5.3 The Candidate Frequent Motif
Generation Evaluation
This subprocess takes as input a pruned point series
P
0
and a window size ω. Experiments were conducted
using ω = {100, 200, 300} to identify the most appro-
priate parameter settings. The results indicated that
when ω = 200, the point series size is reduced by ap-
proximately a further 45% (69% of the original size);
when ω = 100 and ω = 300, the point series size is
reduced by a further 27% (59% of the original size).
5.4 Parameter Setting for Frequent
Motifs Discovery
Recall that the process required five parameters: (i) ω:
desired frequent motif length expressed in terms of a
number of points, (ii) max: maximum number of mo-
tifs to be considered, (iii) λ: similarity threshold ex-
pressed in terms of a maximum distance between two
motifs, (iv) σ: frequency threshold expressed in terms
of a minimum percentage of possible motifs and (v) k:
maximum number of motifs to be selected (k < max).
The selected values for these parameters all affect
on the number of frequent motifs identified and con-
sequently the quality of any further utilisation of the
motifs. Clearly, the higher the σ value, the fewer the
number of motifs that would be identified because the
criteria for frequency would become stricter as σ in-
creased. Inversely, the higher the λ value, the greater
the number of motifs that would be identified because
the criteria for similarity would become less strict as
λ increased. It was anticipated that as ω increased,
the number of frequent motifs would decrease as there
would be fewer subsequences to choose candidate fre-
quent motifs from. The values for max and k would
also impact on the number of identified, and then se-
lected, candidate frequent motifs.
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
270

To identify the most appropriate parameter set-
tings, a range of values for ω and max were consid-
ered, {100, 200, 300} and {20, 40, 60} respectively.
The value for k was set to k =
max
2
although any value
less than max could be used. A range of values for
λ and σ was also experimented with, from 0.005 to
0.100 increasing in steps of 0.005. Both λ and σ
were considered to be more significant with respect
to the performance of the proposed approach and thus
a greater number of values was consider compared to
the range of values considered for ω and max. The
experiments indicated that there were clear “peaks” in
the number of motifs identified when ω = 100, while
in the case of ω = 200 and ω = 300 a “plateaux”
was reached before the number of motifs discovered
started to decrease as σ and λ increased. As expected,
the number of motifs discovered increases as max (k)
increased, and tended to decrease as ω decreased. The
best general setting for λ and σ, regardless of the
value of max or ω, was in the region of 0.025; al-
though it can be concluded that λ had more influence
on the number of motifs discovered than σ.
5.5 Runtime Evaluation
To determine the runtime complexity, nine sets of ex-
periments were conducted using ω = {100, 200, 300}
and max = {20, 40, 60}. The adopted values for both
λ and σ were 0.025; k =
max
2
was used. The runtime
results are presented in Table 1, these are average run-
time obtained by running each experiment ten times.
In the table, runtimes are presented in seconds for: (i)
Silent Gap Removal (SGR), (ii) Candidate Frequent
Motif Generation (CFMG), (iii) Local Frequent Mo-
tif Discovery (LFMD) and (iv) Global Frequent Motif
Selection (GFMS). The last column presents the aver-
age runtime to process a single PCG (the sum of the
values in the previous four columns divided by 72, the
number of records in the test dataset). From the table,
it can be clearly seen that the larger the ω value, the
less time that was required to generate a set of can-
didate frequent motifs because when using a large ω,
there were fewer subsequences to consider. However,
the runtime required for LFMD increased with ω be-
cause larger subsequences require more processing.
The runtime required for GFMS is negligible.
The total runtime required for the proposed pro-
cess to undertake the nine experiments was roughly
23 minutes. This compared very favourably with the
PCGseg technique reported in (Alhijailan et al., 2018)
which adopted a segmentation approach to reducing
the complexity of the motif generation and the MK al-
gorithm (Mueen et al., 2009) to extract motifs. In (Al-
hijailan et al., 2018), a runtime of 14 hours per experi-
ment was recorded (versus 2:59 minutes in this paper)
using the same dataset and similar hardware as used
in this paper. Using the best accuracy result, the pro-
posed process required only 0.46 second to mine one
record whereas PCGseg required 7 minutes.
Overall, the proposed algorithm reduced the gen-
eration time by a factor of 343 compared with
PCGseg, without adversely affecting the quality of
the motifs discovered as discussed further in Subsec-
tion 5.6 below. The algorithms were implemented us-
ing the Java object oriented programming language
and run on an iMac Pro (2017) computer with 8-
Cores, 3.2GHz Intel Xeon W CPU and 19MB RAM.
Table 1: Runtime for Frequent Motif Discovery process.
ω max SGR CFMG LFMD GFMS Avg.
100
20
4.01
8.40
20.36 0.02 0.46
40 40.57 0.09 0.74
60 60.90 0.18 1.02
200
20
6.82
62.84 0.01 1.02
40 126.11 0.06 1.90
60 190.63 0.14 2.80
300
20
6.02
132.44 0.00 1.98
40 264.22 0.01 3.81
60 398.44 0.04 5.67
5.6 Classification Accuracy
In the previous subsection, it was demonstrated that
the proposed algorithm speeds up the discovery pro-
cess to a matter of seconds per time series; much
faster than the runtimes presented in (Alhijailan et al.,
2018). However, any speed up in runtime must not
be offset against a loss in the quality of the iden-
tified motifs. The authors also wished to investi-
gate how the various parameters used by the pro-
posed approach might influence the quality of the
identified motifs. To quantify the quality of the mo-
tifs, a classification scenario was considered. The
idea was to use a subset of the selected motifs to
define a classification model and the remaining mo-
tifs to test the model. More specifically, the well-
known Nearest Neighbour Classification (NNC) ap-
proach (Dasarathy, 1991) was adopted because NNC
is frequently used in the context of time/point series
analysis (Wu and Chau, 2010). Two values for the
number of nearest neighbours to be identified were
used, k
NNC
= {1, 3}. The NNC bank comprised a set
of motifs, D = {hm
1
, c
1
i, hm
2
, c
2
i, . . . } where m
i
is a
frequent motif and c
i
is a class-label taken from a set
of classes C. The time series to be labelled, the query
time series, will then be processed, using the proposed
Frequent Motif Discovery algorithm, so that it is rep-
resented as a set of motifs, Q = {m
1
, m
2
, . . . } each of
which was to be matched with the motifs held in D.
Given that the proposed process will typically
Effective Frequent Motif Discovery for Long Time Series Classification: A Study using Phonocardiogram
271
identify more than one frequent motif in each time
series, |Q| classification labels will be identified; only
one is required, the most appropriate label. To select
it, three different methods were used: Shortest Dis-
tance (SD), Shortest Total Distances (STD) and High-
est Votes (HV). The SD simply chooses the class asso-
ciated with the most similar motif. The STD chooses
the class associated with the lowest accumulated dis-
tance; the total similarity distances is calculated for
each class and the class with the shortest total distance
selected. The HV chooses the most frequent class, the
class with the highest number of votes is selected. In
each case, if there is more than one winner, one of the
other class selection methods is applied. For the ex-
periments, the same parameter values as used in the
runtime experiments reported above was used here.
Ten-cross validation (TCV) was used throughout.
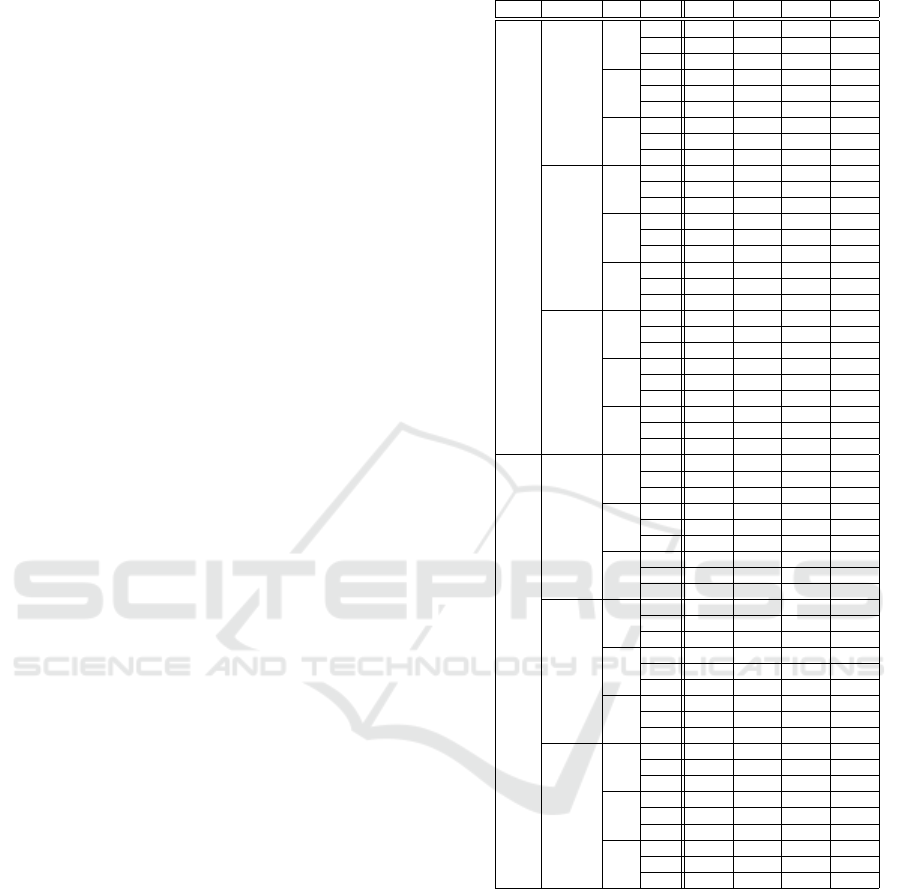
Average results are presented in Table 2; best results
highlighted in bold font. Inspection of the results indi-
cates that accuracy was around 70% regardless of the
k
NNC
value and class selection method used. It can
also be observed that there was little to choose be-
tween methods. Best accuracy, precision, recall and
f-score values were 73.0%, 0.386, 0.445 and 0.390,
this compares favourably with the results reported
in (Alhijailan et al., 2018) where the same dataset
and an alternative motif-based approach was used,
where accuracy, precision, recall and f-score values of
70.8%, 0.191, 0.312 and 0.218 were reported using
segmented data, and 71.9%, 0.218, 0.308 and 0.247
using unsegmented data. In other words, the proposed
algorithm identifies effective motifs.
The best accuracy was obtained using SD com-
bined with ω = 100 and max = 20, giving 73.0% and
72.2% for k
NNC
= 3 and k
NNC
= 1 respectively. For
SD and HV, the smaller the window size, the better the
accuracy while for STD the pattern for the accuracy
values seemed unclear. With respect to the max pa-
rameter, max = 20 produced the best result. It should
also be noted that the recorded standard deviation for
accuracy was good (in the region of 0.05.) Overall,
the combination of ω = 100 and max = 20, coupled
with SD, gave best result; a combination that was also
the fastest, 0.46 second to classify a single record.
Given the ω = 100 had produced the best results,
it was hypothesised that with an even smaller window
sizes, accuracy might increase. Hence, an extra exper-
iment was conducted using ω = 50 combined with the
best performing parameter settings and class selection
method (max = 20 with K
NNC
= 3 coupled with SD
class selection). However, it was found that accuracy
dropped to 70.2%. It was also hypothesised, given
that best results were obtained when max = 20, that
accuracy might be improved if a lower value for max
Table 2: Classification performance measures.
k
NNC
Method ω max Acc. Prec. Rec. F-S
1
SD
100
20 0.722 0.338 0.428 0.374
40 0.669 0.207 0.283 0.234
60 0.644 0.179 0.237 0.201
200
20 0.688 0.386 0.403 0.390
40 0.680 0.286 0.374 0.313
60 0.660 0.262 0.308 0.270
300
20 0.619 0.164 0.183 0.166
40 0.630 0.212 0.237 0.220
60 0.629 0.210 0.241 0.222
STD
100
20 0.660 0.266 0.312 0.283
40 0.611 0.199 0.208 0.197
60 0.611 0.208 0.245 0.217
200
20 0.633 0.226 0.253 0.227
40 0.652 0.268 0.333 0.281
60 0.673 0.373 0.345 0.355
300
20 0.598 0.112 0.145 0.122
40 0.670 0.289 0.324 0.298
60 0.581 0.137 0.116 0.122
HV
100
20 0.680 0.240 0.283 0.255
40 0.703 0.225 0.299 0.249
60 0.694 0.172 0.278 0.208
200
20 0.667 0.264 0.328 0.290
40 0.671 0.337 0.387 0.347
60 0.645 0.252 0.295 0.262
300
20 0.643 0.205 0.233 0.210
40 0.672 0.304 0.329 0.304
60 0.609 0.178 0.237 0.194
3
SD
100
20 0.730 0.341 0.445 0.381
40 0.676 0.201 0.287 0.231
60 0.658 0.185 0.254 0.211
200
20 0.687 0.360 0.412 0.380
40 0.665 0.274 0.358 0.300
60 0.686 0.320 0.378 0.331
300
20 0.626 0.176 0.191 0.178
40 0.622 0.174 0.212 0.189
60 0.599 0.147 0.195 0.159
STD
100
20 0.660 0.270 0.333 0.287
40 0.613 0.166 0.220 0.184
60 0.625 0.225 0.262 0.227
200
20 0.653 0.291 0.370 0.316
40 0.637 0.272 0.304 0.279
60 0.645 0.267 0.349 0.289
300
20 0.605 0.114 0.154 0.127
40 0.685 0.331 0.362 0.338
60 0.574 0.114 0.129 0.112
HV
100
20 0.695 0.219 0.299 0.250
40 0.689 0.188 0.291 0.224
60 0.687 0.145 0.266 0.185
200
20 0.680 0.306 0.395 0.340
40 0.671 0.239 0.362 0.278
60 0.638 0.213 0.270 0.228
300
20 0.658 0.247 0.254 0.246
40 0.645 0.243 0.279 0.250
60 0.588 0.139 0.212 0.161
was considered. An extra experiment with max = 10
was therefore undertaken, with all other parameters as
before, but it was found that this significantly reduced
the accuracy to 59.6%.
5.7 Comparison of Pruning Techniques
Further experiments were conducted to determine the
affect on accuracy when either the silent gap re-
moval phase or the candidate frequent motif genera-
tion phase was omitted, and when both were omitted.
In the first case, line 3 in Algorithm 1 was removed
and the candidateFrequentMotifGen function (line 4)
called with P
i
instead of P
0
i
. In the second case, line 4
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
272

was replaced with:
S
0
i
← Set of subsequences in P
0
i
of length omega
In the third case, both lines 3 and 4 in Algorithm 1
were replaced with:
S
0
i
← Set of subsequences in P
i
of length omega
The results obtained using the best performing pa-
rameters are given in Table 3. The accuracy results
are average results obtained using TCV. Considering
silent gap removal (SGR) first, the first two rows in
the table, it can be seen that SGR has a slight adverse
effect on accuracy. It did improve runtime although
this is not obvious from this table because different ω
and max values produced the best results (recall that
low ω and max values result in efficiency gains be-
cause they entail less calculation). Candidate frequent
motif generation (CFMG), on the other hand, had a
positive effect on accuracy and resulted in significant
speed up (although again it should be noted that the
results reported in Table 3 were obtained using differ-
ent ω values). When the two are run together, as in the
case of the earlier experiments, accuracy was slightly
reduced, because of the negative effect of SGR, but
runtime is enhanced considerably.
Table 3: The best classification accuracy for finding fre-
quent motifs with different preprocessing techniques.
Preproc. Tech. Attributes Results
SGR CFMG
Meth-
ω max k
NNC
Acc.
Runtime
od (Sec.)
X X HV 300 20 3 0.721 8.23
X X HV 100 60 1 0.714 16.44
X X SD 300 20 3 0.736 3.69
X X SD 100 20 3 0.730 0.46
6 CONCLUSIONS
An approach to Frequent Motif Discovery, applica-
ble to PCG time series, has been proposed. The pro-
posed method addresses the challenge of finding dis-
criminative motifs in long time series by proposing
two pruning mechanisms: (i) silent gap removal and
(ii) candidate frequent motif generation. The moti-
vation for the first was that little useful information
could be extracted from “silent gaps”. The second
mechanism featured a novel way of clustering sub-
sequences, without comparing all subsequences with
all other subsequences, to identify the most frequently
occurring subsequences. The performance of the pro-
posed approaches was ascertained in the context of
runtime and the quality of the motifs identified; the
latter analysed in terms of a classification scenario.
The results indicated a classification accuracy compa-
rable with other motif-based approaches but offering
significant runtime advantages.
REFERENCES
Agarwal, P., Shroff, G., Saikia, S., and Khan, Z. (2015).
Efficiently discovering frequent motifs in large-scale
sensor data. In Proceedings of the Second ACM IKDD
Conference on Data Sciences (CoDS’15), pages 98–
103.
Alhijailan, H., Coenen, F., Dukes-McEwan, J., and Thiya-
galingam, J. (2018). Segmenting sound waves to
support phonocardiogram analysis: The pcgseg ap-
proach. In Geng, X. and Kang, B.-H., editors, PRICAI
2018: Trends in Artificial Intelligence, pages 100–
112, Cham. Springer International Publishing.
Dasarathy, B. V. (1991). Nearest Neighbor (NN) Norms:
NN Pattern Classification Techniques. IEEE Com-
puter Society Press tutorial. IEEE Computer Society
Press.
Dau, H. A. and Keogh, E. (2017). Matrix profile v: A
generic technique to incorporate domain knowledge
into motif discovery. In Proceedings of the 23rd
ACM SIGKDD International Conference on Knowl-
edge Discovery and Data Mining, KDD ’17, pages
125–134, NY, USA. ACM.
Gao, Y., Lin, J., and Rangwala, H. (2017). Iterative
grammar-based framework for discovering variable-
length time series motifs. In IEEE International Con-
ference on Data Mining, pages 111–116. IEEE.
Keogh, E. J. and Pazzani, M. J. (2001). Derivative dynamic
time warping. In Proceedings of the 2001 SIAM Inter-
national Conference on Data Mining, pages 1–11.
Krejci, A., Hupp, T. R., Lexa, M., Vojtesek, B., and
Muller, P. (2016). Hammock: a hidden markov model-
based peptide clustering algorithm to identify protein-
interaction consensus motifs in large datasets. Bioin-
formatics, 32(1):9–16.
Mueen, A., Keogh, E., Zhu, Q., Cash, S., and Westover, B.
(2009). Exact discovery of time series motifs. In Pro-
ceedings of the 2009 SIAM International Conference
on Data Mining, pages 473–484.
Ram
´
ırez, J., Segura, J., Ben
´
ıtez, C.,
´
Angel Torre, and Ru-
bio, A. (2004). Efficient voice activity detection al-
gorithms using long-term speech information. Speech
Communication, 42(3):271 – 287.
Sohn, J., Kim, N. S., and Sung, W. (1999). A statistical
model-based voice activity detection. IEEE Signal
Processing Letters, 6(1):1 – 3.
Wu, C. and Chau, K. (2010). Data-driven models for
monthly streamflow time series prediction. Engineer-
ing Applications of Artificial Intelligence, 23(8):1350
– 1367.
Yang, X., Tan, B., Ding, J., Zhang, J., and Gong, J.
(2010). Comparative study on voice activity detection
algorithm. In Proceedings of the 2010 International
Conference on Electrical and Control Engineering,
ICECE ’10, pages 599–602, Washington, DC, USA.
IEEE Computer Society.
Effective Frequent Motif Discovery for Long Time Series Classification: A Study using Phonocardiogram
273
