
Semi-automatic Segmentation of MRI Brain Metastases Combining
Support Vector Machine and Morphological Operators
Gloria Gonella
1
, Elisabetta Binaghi
1
, Paola Nocera
2,3
and Cinzia Mordacchini
2
1
Department of Theorical and Applied Science, Insubria University, Varese, Italy
2
C. S. Health Physics, ASST dei Sette Laghi, Varese, Italy
3
Department of Physics, University of Milan, Milan, Italy
Keywords: MRI Brain Tumour Segmentation, Support Vector Machine, Morphological Operators.
Abstract: The objective of this study is to develop a semi-automatic, interactive segmentation strategy for efficient and
accurate brain metastases delineation on Post Gadolinium T1-weighted brain MRI images. Salient aspects of
the proposed solutions are the combined use of machine learning and image processing techniques, based on
Support Vector Machine and Morphological Operators respectively, to delineate pathological and healthy
tissues. The overall segmentation procedure is designed to operate on a clinical setting to reduce the workload
of health-care professionals but leaving to them full control of the process. The segmentation process was
validated for in-house collected image data obtained from radiation therapy studies. The results prove that the
allied use of SVM and Morphological Operators produces accurate segmentations, useful for their insertion
in clinical practice.
1 INTRODUCTION
Magnetic Resonance Imaging (MRI) segmentation has
a central role in the assessment of a wide spectrum of
brain pathologies, in clinical settings. It allows
identification and delineation of tissues, thanks to the
high spatial resolution and contrast of images, and due
to enhanced signal differentiation (Greenberg et al.,
1999).
In radiation therapy (RT), a precise and accurate
segmentation of MR brain metasteses is important to
the planning of best-case treatments. In this context
automated methods of MRI brain segmentation
represent a valuable improvement to rough manual
detection and delineation, by supporting human
operators with varying degrees of automation, in
tracing the boundaries of pathological tissues and by
automatically providing accurate quantitative
measures used in further stages (Kaus et al., 2001;
Withey and Koles, 2008; Charron et al., 2018; Sharp et
al., 2014). Even though fully automated segmentation
algorithms have the advantage of computing results in
less time and low effort, semi-automated, interactive
methods could be preferable in principle, allowing to
incorporate useful prior knowledge from the experts
and then, making the overall segmentation procedure
more accurate and controllable
(Joe et al., 1999;
Pedoia et al., 2015). In the last years many methods
have been developed for the automatic segmentation of
MRI brain tumors. The proposed techniques make use
of a single image or multispectral pattern and are
supervised or unsupervised (Gordillo et al., 2013;
Bauer et al., 2013).
Even though a large number of techniques have
been proposed in the literature, their application to
brain metastases has received a lot less attention so far.
Yan Liu et al., (2016) propose an automatic
segmentation strategy for metastatic brain tumour
delineation on contrast-enhanced T1-weighted (T1c)
MR image for stereotactic radiosurgery (SRS)
applications. The strategy combines several techniques
such as clustering and regional active contour
technique. A fully automated method is proposed by
Charron et al., (2018). In their study, an existing 3D
convolutional neural network (DeepMedic) is adapted
to detect and segment brain metastases on multimodal
MRI.
Despite the relevant results recently obtained, there
is a need for further studies to investigate novel
approaches able to provide robust solutions and fulfil
spatial accuracy and reproducibility requirements.
The objective of this study is to develop a semi-
automatic, interactive segmentation strategy for
efficient and accurate brain metastases delineation on
Gonella, G., Binaghi, E., Nocera, P. and Mordacchini, C.
Semi-automatic Segmentation of MRI Brain Metastases Combining Support Vector Machine and Morphological Operators.
DOI: 10.5220/0008019304570463
In Proceedings of the 11th International Joint Conference on Computational Intelligence (IJCCI 2019), pages 457-463
ISBN: 978-989-758-384-1
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
457

Post Gadolinium T1-weighted (T1c) brain MRI
images. The salient aspects of the solutions proposed
are the combined use of machine learning and image
processing techniques, based on Support Vector
Machine (SVM) (Vapnik, 1995) and Morphological
Operators (Gonzalez and Woods, 2018) respectively,
to delineate pathological and healthy tissues. The
overall segmentation procedure is designed to operate
in a clinical setting to reduce the workload of health-
care professionals but leaving to them full control of
the process. It is then conceived semi-automatic, but
requiring limited user interaction in an attempt to
facilitate the insertion in current clinical practice.
The segmentation process was validated for in-
house collected image data obtained from RT studies,
where manually segmented images are also provided
by a team of experts.
2 METHODS
The overall segmentation procedure is hierarchically
structured in three phases:
Volume-of-interest (VoI) specification
Supervised Classification based on SVM
Segmentation Refinement based on Morphological
Operators
2.1 VoI Specification
The underlying assumption is that segmentation when
limited to a significant sub-region, could have
performances significantly better in terms of speed and
accuracy than if the segmentation were applied to the
entire scene.
Figure 1: Source Slice of a T1c volumetric MR scan with
the corresponding VoI slice.
In this step, a user specifies a volume of interest (VoI)
by drawing a rectangular region on one slice of the
input volume and selecting first and last slices in such
a way that the entire pathological area is bounded
within the specified parallelepiped (see Figure 1).
2.2 Supervised Classification of
Pathological and Healthy Tissues
In the second phase, a supervised classification is
applied to the selected sub-image.
Among the variety of automated classifiers well-
suited for biomedical image segmentation, we choose
the SVM model (Vapnik, 1995; Suykens et al., 2002).
In our previous works, we deal with MRI brain
tumor segmentations using several methods selected
from states of the art classifiers in the field of MRI
segmentation. In particular, we investigated the use of
Fuzzy connectedness and Graph Cut for glial tumor
segmentation (Pedoia et al., 2015; Binaghi et al.,
2016) and SVM for meningioma and edema
segmentation (Binaghi et al., 2018). Fuzzy
Connectedness and Graph Cut methods are
interactive asking experts to provide accurate
initialization information. Results obtained by these
methods were accurate but strongly influenced by the
prior knowledge provided by the users or by ancillary
methods. In RT domain, where a large number of
images are needed to be handled, they can be
laborious and time-consuming. We have shown that
SVM allows complete delineation of meningioma
and edema tissues and accurate volume estimation by
processing both volumetric and non-volumetric
imagery in a few minutes, without requiring manual
selection of example voxels.
Performances obtained were good confirming the
results obtained in other studies (Bauer et al., 2011).
Proceeding from these results, in the present work
we have considered SVM a potentially valuable tool
for brain metastases segmentation in RT daily care. In
this preliminary study, the in-house collected image
dataset is limited and the SVM model could optimize
the balance between accuracy and demand of the
number of training data.
Multidimensional input patterns are composed of
Post Gadolinium T1-weighted (T1c) voxel intensities
and corresponding textural and contextual features
extracted from the MR scan.
SVM classifier performs a binary hard
categorization labelling voxels as Metastasis (M) and
Healthy tissue (H). Different types of kernels are
tested such as linear, quadratic, cubic, fine-medium-
coarse gaussian. Given the results obtained, we
configured the SVM as soft-margin least square (LS)
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
458

model with linear kernel. The trained SVM classifier
receives in input patterns, in the form of vectors of
measured features and assigns labels to
corresponding T1c MR elements.
Different sets of features have been proposed in
the literature, selected in the function of the MRI
channels used and the classifiers adopted (Gordillo et
al., 2013; Bauer et al., 2013). On the basis of our
experience, in addition to image intensities from the
T1c MR scan, we consider features describing
quantitatively neighbour relationships and texture
(Tuceryan and Jain, 1998). Contextual and textural
features have been analysed systematically in order to
determine the combination that is most appropriate
for the current classification task.
In particular, several configurations of the
segmentation procedure have been experimented
initially providing input only intensity values of
central voxel and of neighbour voxels.
Different neighbourhoods have been considered
including incrementally neighbours along voxel
faces, corners and edges up to a maximum of 26
voxels. In a second step an enlarged feature set has
been considered adding textural features to the best
neighbourhood configuration. The following set of
features has been finally selected:
intensities from T1c scan
first order texture features, mean, variance,
skewness, kurtosis and entropy
intensities in 26 neighbourhood voxels
The features have been normalized to have zero mean
and unit variance.
During the training phase, the SVM learns an
approximation for the true input–output relationship
based on a given training set of examples constituted
by N input-output pairs
, where x
i
is the feature vector of length equal to 32 and
is a supervised label denoting the membership
in the metastasis or healthy class.
Several strategies are conceived to build the
appropriate training set during the learning stage. All
proposed training sets have been analysed
systematically in the experimental evaluation phase
in order to determine the combination that is most
appropriate for the classification task (see Section 3).
2.2.1 SVM Classifier
To make the work self-contained we briefly outline
the basic concepts of SVM adopted in the proposed
segmentation strategy. SVM is a classification
algorithm based on kernel methods (Vapnik, 1995;
Schoelkopf and Smola, 2002) map the input patters
into a high dimensional feature space. Classes which
are non-linearly separable in the original space can be
linearly separated in the higher dimensional feature
space.
Let
be a supervised training set of
elements for a two-class classification problem, with
and
. Considering the
case of linearly separable data, the solution to the
classification problem consists in the construction of
the decision function:
(1)
(2)
that can correctly classify an input pattern x not
necessarily belonging to the training set.
SVM classifier defines the hyperplane that causes
the largest separation between the decision function
values for the “borderline” examples from the two
classes. Mathematically, this hyperplane can be found
by minimizing the cost function:
subject to
(3)
for y
(4)
for y
(5)
The extension to the nonlinear classification is based
on the function
in which the
non-liner operator is introduced.
In this case the SVM cost function to be
minimized is
subject to
(6)
with
i 1,2,....l
(7)
Suykens (Suykens et al., 2002) proposed a new
formulation of SVM by adding a LS term in the
original formulation of the cost function. This
modification significantly reduces the computational
complexity.
2.3 Segmentation Refinement based on
Morphological Operators
Recent studies propose the allied use of SVM with
post-processing and/or regularisation procedures to
ensure spatial consistency in classification results
(Bauer et al., 2011). In our context, after the
segmentation, if the tumour area presents necrosis
Semi-automatic Segmentation of MRI Brain Metastases Combining Support Vector Machine and Morphological Operators
459

and inhomogeneity, small holes within the tumour
mass may be classified as healthy tissues and several
isolated elements in the background area may be
classified as tumour. Our strategy includes a
procedure based on the use of Morphological
Operators to refine the segmented masks in an
attempt to reduce omission and commission errors
and making the segmented tumour area more
compact.
For each selected slice, Opening and Closing
Operators with spherical shapes are applied
consequently. The Opening Operator removes from
the binary input image all the connected components
that have a lower number of pixels than a set value
and outputs a new binary image. The Closing
Operator closes holes present in the image and returns
the closed binary image. Three different tests were
performed, using for all a disk-shape structuring
element, aimed to tune parameters values of the
Morphological Operators and decide the order of
application. In the first test, only the opening
morphological operator (Open) was applied by
varying the radius; in the second test, only the
morphological closing operator (Close) was applied,
varying the radius; in the third test, both operators
were applied in a different sequence. The best result
was the third test, where the opening morphological
operator with a radius of 5 was first applied and then
the closing operator with a radius equal to 10.
3 EXPERIMENTS
The segmentation method was experimented on a
dataset of 20 patients with a total of 25 pathologies to
be segmented. Data are composed of T1c volumetric
MR scans. Volumes are acquired using a 3D sequence
characterized by 0,9 mm isotropic voxels, the pixel
spacing of 0,47 mm and the slice thickness of 2,67
mm.
We developed case-specific, intra-case analysis
and inter-case analysis. In case-specific analysis both
training and test sets were obtained from the reference
masks of the same VoI. In inter-case analysis training
and test data are extracted from VoIs of different MR
scans.
Accuracy of segmentation results is assessed by
comparing the spatial distribution of the masks
obtained by the automated segmentation with that of
the masks obtained through a manual segmentation of
the T1c images. The agreement between reference
and automated maps is measured in terms of Dice
(DSC) (Dice, 1945), Precision (P) and Recall (R)
indexes (Olson and David, 2008). The DSC index has
been used broadly in the field of segmentation as a
measure of spatial overlap and P and R indexes allow
to measure under- and over-estimations (Bouix et al.
2007).
Several experiments have been conducted for both
intra- and inter-case analysis distinguished by the
criteria for selecting training and test samples from
the VoIs under study. Experiments and accuracy
assessments computed according to cross-validation
scheme are detailed below.
3.1 Experiments for Intra-case
Analysis
Experiment 1a: training and test data are extracted
from the reference masks of the same VoI (intra-case
analysis) and built by randomly selecting elements in
the proportion of 70% and 30% respectively. An
equal number of elements labelled M and H was
considered. The number of contour elements
belonging to class M was increased to facilitate
recognition.
Experiment 2a: training and test sets are obtained
as above, but by limiting the random selection within
a region of 8 pixels wide, built around the contour of
the tumour reference masks. The underlying
assumption for this strategy lies in the fact that
metastases have little extensions and a high level of
heterogeneity occurs in the internal part of the
pathology due to the presence of necrosis. In this
context, an accurate delineation can be achieved by
identifying the partial contour region, subsequently
filled by the support of Morphological Operators.
Table 1: Dice (DSC), Precision (P), Recall (R) values
obtained for Experiment 1a and Experiment 2a over all 25
cases under study.
Experiment 1a
Experiment 2a
DSC
Mean
0.808
0.878
Var
0.008
0.003
Min
0.549
0.757
Max
0.908
0.963
P
Mean
0.824
0.884
Var
0.006
0.003
Min
0.648
0.749
Max
0.927
0.963
R
Mean
0.796
0.873
Var
0.012
0.003
Min
0.476
0.764
Max
0.923
0.963
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
460
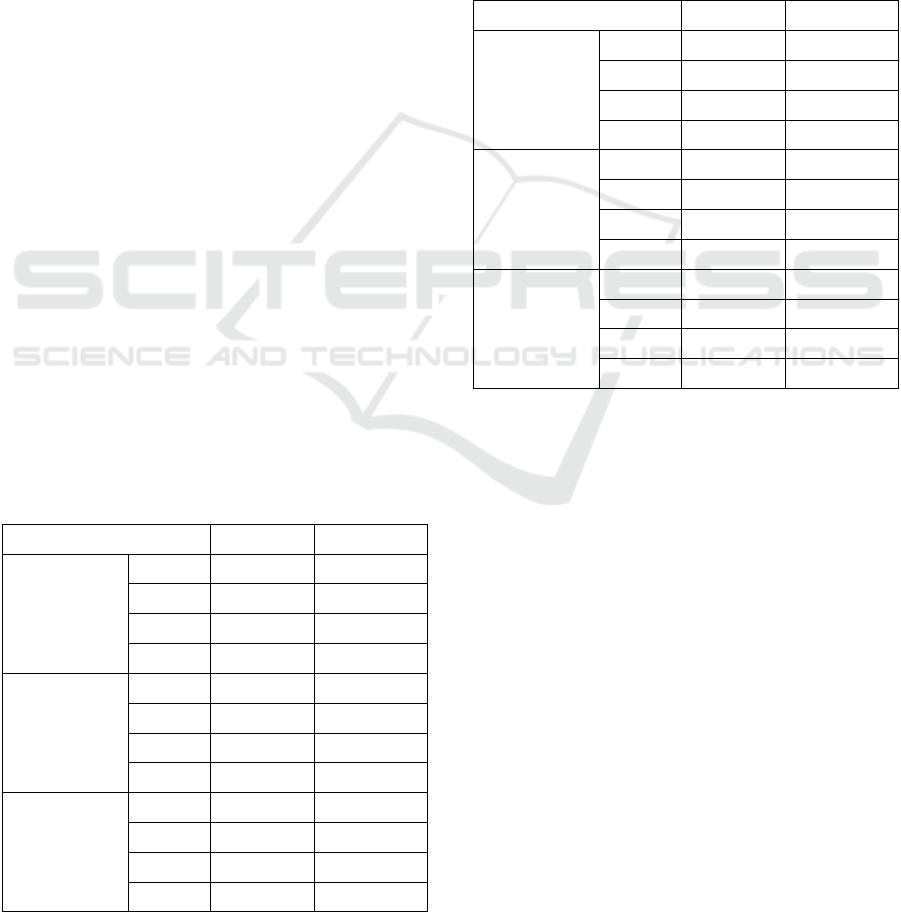
Table 1 shows the numerical results obtained for
Experiment 1a and Experiment 2a in terms of DSC, P
and R indexes. SVMs trained according to
Experiment 2a slightly prevail with a DSC value
computed over all 25 cases equal to 0.878. P and R
values highlight a significant reduction of omission
and commission errors.
3.2 Experiments for Inter-case
Analysis
Two types of experiments for inter-case analysis have
been conducted distinguished by an increasing level
of heterogeneity of the training set provided in input
to the SVM classifier. The random selection of
training elements was limited to contour regions of
tumour reference masks; the reason for this choice
lies in the fact that this strategy prevailed in the intra-
case analysis.
Experiment 1b: in this experiment, SVMs are
trained on data from one case and tested on all the
cases under study; in this way, we investigate the
generalisation power of the SVM when training data
present a minimum level of heterogeneity. 25 SVMs
are trained with training elements extracted from the
selected VoI, according to Experiment 2a and tested
on all the VoIs under study. Results obtained by the
best configuration when processing the 25 cases are
shown in Table 2. To isolate the contribution of
Morphological Operators within the overall
segmentation procedure, accuracy values obtained
with and without the use of them are computed.
Table 2: Dice (DSC), Precision (P), Recall (R) values
obtained by the segmentation procedure configured for
Experiment 1b with and without the use of Morphological
Operators (MO) and tested on the overall 25 cases under
study.
SVM
SVM+MO
DSC
Mean
0.701
0.693
Var
0.011
0.035
Min
0.462
0
Max
0.844
0.897
P
Mean
0.747
0.696
Var
0.026
0.047
Min
0.437
0
Max
0.997
0.997
R
Mean
0.737
0.769
Var
0.035
0.057
Min
0.410
0
Max
0.983
0.990
Experiment 2b: training elements are extracted
from a set of VoIs selected from cases well segmented
in intra-case analysis. Several configurations have
been considered obtaining 120 SVMs trained on
different sets of VoIs, according to the strategy
described in Experiment 2a, and tested on all the 25
VoI under study. The accuracy obtained with and
without the use of Morphological Operators are
computed as shown in Table 3.
Table 3: Dice (DSC), Precision (P), Recall (R) values
obtained by the segmentation procedure configured for
Experiment 2b with and without the use of Morphological
Operators (OM) and tested on the overall 25 cases under
study.
SVM
SVM+MO
DSC
Mean
0.653
0.660
Var
0.008
0.028
Min
0.390
0
Max
0.770
0.820
P
Mean
0.681
0.641
Var
0.017
0.025
Min
0.278
0
Max
0.968
0.881
R
Mean
0.710
0.762
Var
0.026
0.035
Min
0.482
0
Max
0.955
0.976
In general, results obtained in Experiment 1b are
better than those obtained in Experiment 2b. Looking
at values in Table 2 in more detail, we found that
performances obtained by the application of
Morphological Operators are worse on average.
However, when studying individual cases, we
have noticed that under-estimation and over-
estimation errors occur systematically when the
pathology occupies a very small volume (under the
100 elements) and is inserted in a highly
heterogeneous context. An example is illustrated in
Figure 2 where a slice (Slice 1) with a remarkable
small metastasis is shown. The refinement
accomplished by the Morphological Operators
deletes all the true positive elements identified by the
SVM classifier. On the contrary, the segmentation
masks of the larger pathological area in the slice
(Slice 2) shown in Figure 3, indicate that the
segmentation strategy benefits from the allied use of
SVM and Morphological Operators. Table 4 lists the
numerical results of the cases illustrated in Figure 2
and 3.
Semi-automatic Segmentation of MRI Brain Metastases Combining Support Vector Machine and Morphological Operators
461
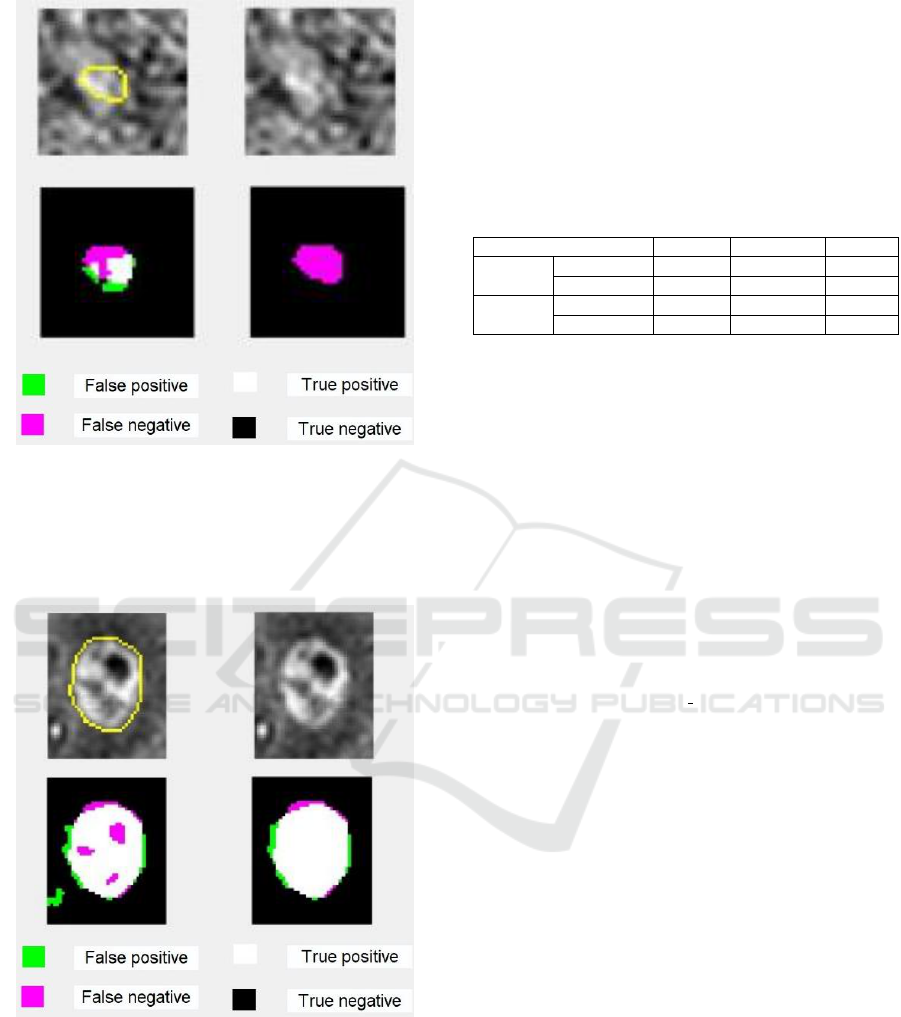
Figure 2: First row, from left to right: crop of a source slice
(Slice 1) of T1 MR Volume with superimposed the contour
of metastasis reference mask (dimension: 83 elements),
Slice of the corresponding VoI; second row from left to
right: Segmentation mask produced by SVM, refinement by
the Morphological Operators.
Figure 3: First row, from left to right: crop of a source slice
(Slice 2) of T1 MR Volume with superimposed the contour
of metastasis reference mask (dimension: 644 elements),
Slice of the corresponding VoI; second row from left to
right: Segmentation mask produced by SVM, refinement by
the Morphological Operators.
Automatic segmentations were evaluated
qualitatively through visual inspection. The complete
strategy including the combined use of SVM and
Morphological Operators have been judged
satisfactory. The limitations of the segmentation
procedure, inherent to specific cases, as illustrated
above, are considered acceptable and manageable
with interactive phases devoted to manual
refinements of the automated results.
Table 4: Dice (DSC), Precision (P), Recall (R) values
obtained by the segmentation procedure when processing
slices in Figure 2 and 3.
DSC
Precision
Recall
Slice 1
SVM
0.556
0.656
0.482
SVM+MO
0
0
0
Slice 2
SVM
0.885
0.898
0.873
SVM+MO
0.940
0.926
0.953
4 CONCLUSIONS
The objective of this study was to develop a semi-
automatic image segmentation strategy for
metastases segmentation in MR brain images. The
strategy was tested on a preliminary collected data
set. The results prove that the allied use of SVM and
Morphological Operators produces segmentation
sufficiently accurate for their insertion in clinical
practice. Future work contemplates the acquisition of
new data with which to perform a more significant
interpatient analysis and then to develop a more
robust evaluation. Moreover, the availability of a
wider set of data will allow developing a comparative
analysis with other promising segmentation
techniques, such as the Convolutional Neural
Network, the use of which is constrained to the
collection of huge data sets.
REFERENCES
Bauer, S., Nolte, L.P., Reyes, M., 2011. ‘Fully automatic
segmentation of brain tumor images using support
vector machine classification in combination with
hierarchical conditional random field regularization.’
In: Medical image computing and computer-assisted
intervention: MICCAI, Berlin, Springer, 14, 354-61.
Bauer, S., Wiest, R., Nolte, L.P., Reyes, M., 2013. ‘A
survey of MRI-based medical image analysis for brain
tumor studies.’ Physics in medicine and biology,
58(13), R97-R129.
Binaghi, E., Pedoia, V., Balbi, S., 2016. ‘Collection and
fuzzy estimation of truth labels in glial tumour
segmentation studies.’ Computer Methods in
Biomechanics and Biomedical Engineering: Imaging
and Visualization, 4 (3-4), 214-228.
Binaghi, E., Pedoia, V., Balbi, S., 2018. ‘Meningioma and
peritumoral edema segmentation of preoperative MRI
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
462

brain scans.’ Computer Methods in Biomechanics and
Biomedical Engineering: Imaging and Visualization, 6
(4), 362-370.
Bouix, S., Martin-Fernandez, M., Ungar, L., Nakamura, M.,
Koo, M.S., McCarley, W.R., Shenton, M., 2007. ‘On
Evaluating Brain Tissue Classifiers without a Ground
Truth.’ NeuroImage, 36, 1207-1224.
Charron, O., Lallement, A., Jarnet, D., Noblet, V., Clavier,
J.B., Meyer, P., 2018. ‘Automatic detection and
segmentation of brain metastases on multimodal MR
images with a deep convolutional neural network.’
Computers in Biology and Medicine.
Dice, L.R., 1945. ‘Measures of the amount of ecologic
association between species.’ Ecology, 26(3), 297-302.
Gonzalez, R., Woods, R., 2018. Digital Image Processing,
Pearson-Prentice Hall, 519-566.
Gordillo, N., Montseny, E, Sobrevilla, P., 2013, ‘State of
the art survey on MRI brain tumor segmentation.’ Magn
Reson Imaging, 31(8), 1426-38.
Greenberg, H., Chandler, W., Sandler, H., 1999. Brain
Tumors, Oxford University Press, Oxford.
Joe, B., Fukui, M., Meltzer, C., Huang, S.Q., Day, R.,
Greer, P., Bozik, M., 1999. ‘Brain Tumor Volume
Measurement: Comparison of Manual and
Semiautomated Methods1.’ Radiology, 212(3), 811-6.
Kaus, M., Simon, P., Warfield, K., Nabavi, A., Peter, M.,
Black, M., Jolesz, F., Kikinis, R., 2001. ‘Automated
segmentation of MRI of brain tumors.’ Radiology, 218,
586-591.
Liu, Y., Stojadinovic, S., Hrycushko, B., Wardak, Z., Lu,
W., Yan, Y., Jiang, S., Timmerman, R., Abdulrahman,
R., Nedzi, L., Gu, X., 2016. ‘Automatic metastatic brain
tumor segmentation for stereotactic radiosurgery
applications.’ Physics in medicine and biology, 61,
8440-8461.
Olson, D.D., David, L., 2008. Advanced Data Mining
Techniques, 1st Edition, Springer.
Pedoia, V., Balbi, S., Binaghi, E., 2015. ‘Fully Automatic
Brain Tumor Segmentation by using Competitive EM
and Graph Cut.’ In: International Conference on Image
Analysis and Processing, Genova, Italy.
Sharp, G., Fritscher, K., Pekar, V., Peroni, M., Shusharina,
N., Veeraraghavan, H., Yang, J., 2014. ‘Vision 20/20:
Perspectives on automated image segmentation for
radiotherapy.’ Medical physics, 41(5), 050902.
Schoelkopf, B., Smola, A., 2002. Learning with kernels:
support vector machines, regularization, optimization,
and beyond, MIT Press.
Suykens, J.A.K., Van Gestel, T., De Brabanter, J., De
Moor, B., Vandewalle, J., 2002. Least Squares Support
Vector Machines. World Scientific Publishing Co.,
Singapore.
Tuceryan M., Jain A., 1998. Texture analysis Inc. River
Edge, NJ: World ScientificPublishing.
Vapnik, V.N., 1995. The Nature of Statistical Learning
Theory, Springer-Verlag, New York.
Withey, D.J., Koles, Z.J., 2008. ‘A review of medical image
segmentation: Methods and available software.’
International Journal of Bioelectromagnetism, 10, 125-
14.
Semi-automatic Segmentation of MRI Brain Metastases Combining Support Vector Machine and Morphological Operators
463
