
Use of Ontologies in Chemical Kinetic Database CHEMCONNECT
Edward S. Blurock
a
Blurock Consulting AB, Lund, Sweden
Keywords: Ontology Use Case, Database System, Chemical Kinetics, Experimental Data, Chemical Modelling.
Abstract: CHEMCONNECT is an ontology cloud-based repository of experimental, theoretical and computational data
for the experimental sciences domain that support the FAIR data principle, namely that data is findable,
accessible, interoperable and re-usable. The design also promotes the good scientific practices of
accountability, traceability and reproducibility. The key to meeting these design goals is the use of ontologies.
The primary goals of using ontologies include not only capturing a domain specific knowledge base (with
support of domain experts), but also to create a data/ontology driven software system for the data objects, data
entry, the database and the graphical interface. The impetus within (combustion research) domain, which is
the initial focus of CHEMCONNECT, of the knowledge base is formation and documentation of standard
data reporting practices. The ontology is a software technical implementation of practices within the
community. Storing and querying of specific instantiations of object data is done using a NOSQL database
(Google datastore). This initial design of CHEMCONNECT is modelled for the chemical kinetics and
combustion domain. Within this domain, the ontology defines templates of typical experimental devices
producing data, algorithms and protocols manipulating data and the data forms that are encountered in this
pipeline. These templates are then instantiated, with the aid of ontology driven cloud-based interface, to
specific objects within the database. The knowledge base is key to uniting data input in various forms
(including diverse labelling) to a common base for ease of search and comparison. The structure is not limited
to this domain and will be expanded in future collaborative work. CHEMCONNECT is currently implemented
with the Google App Engine at http:www.connectedsmartdata.info.
1 INTRODUCTION
CHEMCONNECT is a cloud-based database and
repository of experimental and modelling data
primarily in the area of chemical kinetics. Currently,
the emphasis is on combustion data, but a more
general structure is maintained to allow expansion
into other scientific domains.
As the name implies, the purpose and design of
the software system CHEMCONNECT recognizes
that individual data points are not isolated but are
interconnected with a multitude of other data
representing its history, origin and dependencies and
how points are used and related to other data points.
The impetus for this philosophy is to promote the
good scientific practices of accountability,
traceability and reproducibility. In addition, the
database is designed to promote FAIR(GO FAIR
2016; Hagstrom 2014; Wilkinson et al. 2016)
a
https://orcid.org/0000-0001-9487-3141
principles, meaning that the data is Findable,
Accessible, Interoperable and Re-usable. The goal of
CHEMCONNECT is to provide a platform that
encourages these practices in a semi-automated way
so as to not to incur an increased work-load for the
researcher. The goal is to provide a natural workflow
of data entry for the researcher.
This work has been spurred by the movement,
especially within the combustion community which
is the initial emphasis of CHEMCONNECT, of
standardizing data reporting. CHEMCONNECT
implements these standardizations through its
knowledge base ontology.
1.1 Interconnectivity of Data and
Knowledge Base
CHEMCONNECT recognizes that all data is not
isolated and has a complex interconnection with data
of different forms and purposes. This is the central
240
Blurock, E.
Use of Ontologies in Chemical Kinetic Database CHEMCONNECT.
DOI: 10.5220/0008069502400247
In Proceedings of the 11th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2019), pages 240-247
ISBN: 978-989-758-382-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

concept in promoting good scientific principles of
accountability, who and what devices generated the
data, traceability, the origins of all data, including
external data sources, and reproducibility, all the
information and algorithms of the entire process is
available.
We start with a single data point. Most likely this
point has been included from a data files within a
repository with its associated information of who
entered the data, data of entry, data of data
production, origin of data, etc. The data point itself
represents some knowledge as to its context. This
context lies within a network of interconnected
concepts. The database ontological knowledge within
CHEMCONNECT provides this context.
If the data point is a ‘direct’ measurement, there is
a connection with all the associated information with
the device. First, there is the specific device found at
a specific place (institute, department, etc.), within a
specific organization (university, research center,
etc.), performed by a specific researcher (including
collaboration, supervisor, position, and other
information about the researcher). The device itself
has a description and can be viewed as a collection of
subsystems each of which has a purpose and its role
in producing the data point. Within the device there
could be the actual component which produced the
data with its specific properties, including accuracy,
reliability and dependence on other components in the
device. Part of the CHEMCONNECT database is a
device description. The specific device used to
produce the point is, of course, related to similar
devices with similar properties. The
CHEMCONNECT ontology knowledge base
provides templates of device descriptions and the
device’s relation and composition in relation to other
similar devices. In addition to the meta-data about the
device (parameterized description, abstract
description, references to publications, institutes,
researchers, etc., the device is viewed as a set of
interconnected subsystems and components.
Templates of these descriptions are found in the
database which also gives their role and purpose
within a larger scientific context.
Final data point results reported in publication are
seldom direct measurements. Usually, there is a flow
of data manipulations from the ‘raw’ data
measurement from the device to the final result
reported in a table in the publication. It is becoming
more critical within the scientific community,
especially for the chemical kinetics community, that
this data trace is included, particularly in error
analysis, for traceability, accountability and
reproducibility. For example, the computation of
propagation of errors can be done a variety of ways
and can range from the simple, which is usually done
by the primary data producer, to complex, which can
be done by researchers with data expertise.
The chain of data manipulations from ‘raw’
results to final published results is represented by a
protocol. The interconnectivity of data is further
promoted by each component in this chain. A
protocol essentially consists of the entire set of
algorithms, procedures, devices and intermediate data
produced from those algorithms. Within these
components are further connections to specific
organizations, researchers, publications, and other
external references. Within the CHEMCONNECT
knowledge base, templates for protocols are given,
meaning typical experimental procedures leading to
final results. Instantiation of a protocol into the
database is done by providing the specific
information regarding the specific experiment. This
instantiation supplements the general context
knowledge, within the broader knowledge base of
experimental procedures and devices.
Within the knowledge base of CHEMCONNECT,
templates for algorithms and their specific
implementation can be given to the database.
Algorithms can be range from simple algorithmic
calculation, to computer software. Within the
algorithm description would be further references
giving a broader context to the algorithm. An
‘algorithm’ can also be a specific experimental
procedure describing (with references) how the data
was produced. The ontological knowledge base
algorithms provide information about the role and
purpose of the algorithm within the large context of
data manipulation.
1.2 Structure of CHEMCONNECT
The general structure of CHEMCONNECT consists
of the interaction between these entities:
Knowledge Base: This is the heart of the
CHEMCONNECT system. It is an ontology
describing the data structures and domain
structures and concepts.
Repository: This is the data in the original
form of the researchers. These are the files that
are parsed and interpreted using the knowledge
base and stored in the database.
Database: This is the primary persistent
storage of individual pieces of interconnected
data. The database not only holds the data
itself, but also the data specifications and
templates used to input and interpret data.
Use of Ontologies in Chemical Kinetic Database CHEMCONNECT
241

Cloud-based Interface: From a browser
interface, the data can be inputted, visualized,
compared and searched. The driving force of
the interface is the knowledge-base providing
for effective input, visualization and
comparison of data.
It is important to note that the role of the ontology
is to store ‘generic’ knowledge about structures
within CHEMCONNECT and the role of the database
is to store specific persistent instantiations of domain
data. Queries about specific domain objects is done
through the database. Queries using the ontology are
about the character and make-up of the entities used
by CHEMCONNECT.
The general workflow of using
CHEMCONNECT is in two stages. First, an initial
‘one-time’ setup phase setting up the organization,
researcher, device and protocols. During the
measurement phase, these references and protocols to
interpret the data are re-used. The philosophy is that
after the initial setup, the introduction of
measurements, including all their interpretations and
interconnections, is semi-automatic. In principle, the
data file is put into the repository and then parsed and
interpreted into the database.
A typical lab most likely has a limited number of
devices with a static configuration and a ‘fixed’
structure of the experimental data. This means that the
device description and the experimental protocols on
how to interpret the data are done once and then re-
used during the experimental phase.
The setting up of a protocol and data
specifications involves starting with a template within
the knowledge-base and making an instantiation
within the database. This instantiation, an entity
within the database, provides the exact interpretation
of data coming from the original raw input.
2 USE OF ONTOLOGIES
A primary design philosophy of CHEMCONNECT is
that the software system is knowledge-base driven.
The software components are fairly (emphasis on
‘fairly’) general and how they are pieced together is
determined by the knowledge base captured by an
ontology representation. The ontology knowledge
base is geared towards a particular focus group,
namely experimental and modelling researchers in
the field of chemical kinetics and combustion
research. However, in the design of
CHEMCONNECT a certain degree of generality is
maintained to expand out of this focus group, for
example, experimental research in general.
The ontology used in CHEMCONNECT is used in
several capacities:
Data Structures: These ‘general’ structures
have a one-to-one correspondence with data,
interface and persistent database structures.
Templates: These are generalized information
used to fill in domain information into the
general ‘data-structures’.
Concepts: This is the hierarchy of domain
specific concepts and classifications. The
concepts are also used to fill in domain
information in the templates.
Part of the design concept is to base
CHEMCONNECT ontology objects as much on
existing ontologies, both general and domain specific,
as possible.
2.1 Data Structures
The set of data structures is general enough to
accommodate the domain specific templates and
concepts. There are basically three levels of data
structures:
Catalog Structures: These are based on the
DCAT Catalog structure (Maali and Erickson
2014). These are the structures representing the
main data objects to represent the domain.
Record Structures: Base on the dcat:record
from the DCAT ontology, these are the records
of the catalog. Each record structure contains
several pieces of 'primitive' information.
Primitives: These are basically a single (string)
word primitives that make up the record.
For the current domain, the following types of catalog
objects were deemed sufficient:
Catalog Hierarchy: This is essentially a
directory structure virtually categorizing the
data. Sets of data within the same hierarchy
position are considered related.
Contact Data: It is important for
accountability to have a trace to individual
researchers and the organizations producing
the data.
Device and Component Data: Each device
and component is represented by a catalog
object. The device descriptions are a hierarchy
of subsystems and components.
Interpretation Data: These catalog objects
provide instructions on how to interpret data
object. For example, given a csv, spreadsheet
(in various formats), XML, yaml, etc. file with
a block (matrix) of data information. This
isolates and interprets the block as a matrix
object.
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
242
Observation/Protocol Specifications: These
catalog objects represent specifications of how
the data is to be interpreted relative to the
domain knowledge base.
Observations: This is the set of catalog objects
produced as a result of the observation
specifications with a protocol. Simply said, an
individual observation is a matrix of results,
where the columns are interpreted by the
observation specification (including which
knowledge base object is involved and the units
used) and each row is a set of actual
measurements.
2.1.1 Catalog Objects
The top data objects are the catalog objects
(dcat:catalog). Each catalog object consists a set of
records (dcat:record). Each catalog object has a set
standard records and a set of concept specific records.
In the ontology, a particular record can be one
instance of the record (restriction type of cardinality
of one) or there can be multiple instances of the same
record (restriction type some).
The other concept specific records contain the
defining information for that object. The standard
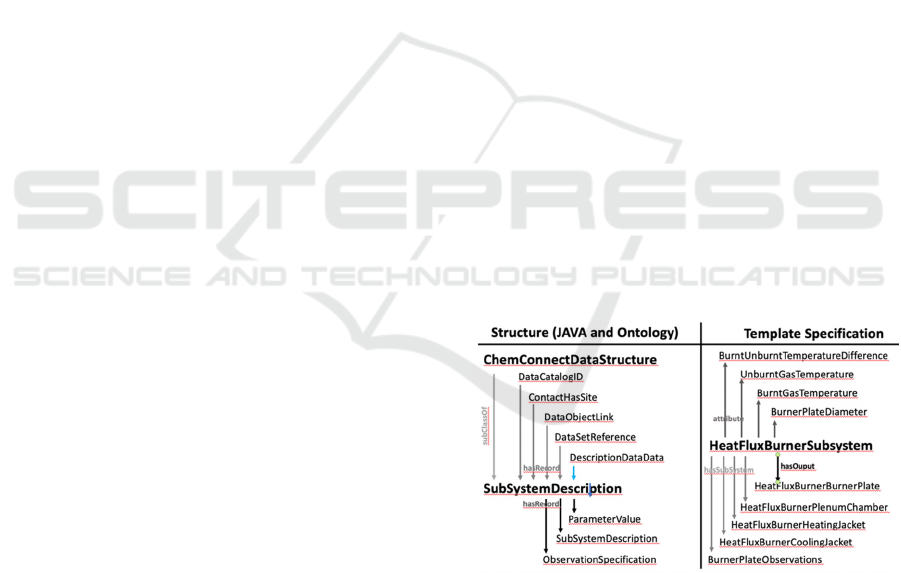
record information is (See structure part
ChemConnectDataStructure in Figure 1):
Catalog Hierarchy (DataCatalogID): The
position within the catalog hierarchy.
Description (DescriptionDataData): Title,
descriptions, ownership and concept
information.
References (DataSetReference): Publication
references
Data Links (DataObjectLink): Links to other
data objects with a corresponding link concept.
External Links (ContactHasSite): Links to
external sites with corresponding link concept.
2.2 Templates
Templates are more complex specifications, often
made of other specifications and concepts. Each
specification gives domain specific information on
how to fill in a particular catalog object (See Figure 1
for an example of a heat flux burner as a subsystem):
Parameter Specification (Attribute): The
template defining a parameter’s the essential
information, such as unit class (the actual class
is defined on instantiation), purpose, keyword,
standardized name, etc. A critical component
of the parameter ontology is the use of the
Quantities, Units, Dimensions and Data Types
Ontologies (QUDT 2018).
Observation Specification (hasOutput):
This is a template of a specific observation set
made up of a set of parameters. This
specification can be thought of defining which
parameter specifications define the columns of
a matrix of data. Based on the data cube
ontology (Cyganiak and Reynolds 2014).
Device Specification (hasSubSystem): This
defines a template for an experimental device.
The device is viewed as a hierarchy of
subsystems and components. Based on the SSN
(Haller et al. 2017) and the SOSA (Cox 2017).
Protocol Specification: A protocol is
essentially the specification that is needed to
define an experimental regime. An essential
part of the specification is used to interpret the
set of data input files making up the
experimental regime.
Within the interface, the template is used to generate
the interface in which the specific information for the
instantiation of the entity can be filled in. For
example, for a parameter, the template gives the
default information. In the interface, the actual units
used can be chosen. The template gives the unit class
(and maybe a suggestion for default units). There are
two levels of parameter specifications. For defining a
specific parameter, the value of the units and the
actual parameter value are given. But if the template
is used in, for example, defining the column of a
matrix, the value is no given.
Figure 1: The ontology describes the structure of all the data
objects. In this example, the structure describes a general
device (subsystem) structure. Within the ontology there is a
description of a typical device, such as a heat flux burner.
The template describes how the class structure should be
filled in for the device.
2.3 Concepts
Within CHEMCONNECT, keywords are arranged in
a hierarchy of Concepts (under the skos:Concept
Use of Ontologies in Chemical Kinetic Database CHEMCONNECT
243

ontological object). Most of the domain information
is stored as a concept. Within the hierarchy of
concepts there are several types. Those concepts
representing single word concepts, essentially a set of
standardized keywords are:
Keywords: These are concepts within a
hierarchy of concepts, with the hierarchy
giving them context and meaning.
Purpose: A standardized purpose keyword,
within a hierarchy.
Classifications: A concept keyword
representing a set of types of classes of objects.
The sub-classes of the classification are the
specific choices. Classifications are used, for
example, in the interface to produce a pull-
down list of choices.
Links: This is the concept that links two
entities, such as catalog objects. This concept
can have an extra property limiting the
structures it links to. This information, for
example, is used by the interface to produce a
list of choices from the database.
3 PARAMETERS
An important entity within a data repository is a
parameter. A parameter is simple representation that
condenses complex reality into a single value. One of
the challenges is data repository information is the
representing the ‘meaning’ and context of a particular
parameter. In scientific collaboration, unless there is
a conscience standardization, often there is a degree
of ambiguity data parameters, particularly in naming
the parameter (and the use of adjectives and
abbreviations) and in the actual units involved.
CHEMCONNECT’s knowledge base attempts,
through the use of ontologies, is to formalize these
concepts and reduce the ambiguity and increase the
comparability of parameters coming different
sources.
A parameter is used and defined on several levels
depending on how they are needed within the
respository.
A parameter specification defines the basic
information about a parameter, particularly the unit
type (unit class), uncertainty value type, a purpose, a
concept and whether it is an input (dimension) or an
output (measure). A specification does not involve
the actual value or even the specific units. This is
what is defined with a parameter template within the
ontology.
A value specification starts with the parameter
specification, but then the particular units from the
unit class are specified. An important aspect of the
parameter specification is the standardization of the
name that can be linked to from, for example, a
parameter name within a particular data file of a
researcher. A parameter specification can be thought
of as defining a column in a data matrix. Here, the
units of the values are specified and, if the matrix is
within a data file, a correspondence between the
parameter name in the file and the standardized name
in the ontology.
Within the description of a catalog object, there
can often be a parameterized description, a set of
attributes, of the object. Here, the parameter
specification is used and the units and the value of the
attribute is specified.
Similar to the attribute is the parameter value.
The only distinction here is that instead of the
parameter being a single attribute-value pair, it
represents a set of values, such as a column in the
matrix.
The CHEMCONNECT interface facilitates the
reading of a data file produced by a researcher so that
the data can be put on a standardized platform. The
key step is setting a correspondence between a
column of data, the set of data values, and a value
specification. The utility of this lies in the comparison
of data. The link to the standardized name within the
ontology links the two data sources for that
parameter. Knowing the particular units and the
conversion of units (primarily from the QUDT
ontology) allows direct numerical comparison. It
should be noted that the ‘extra’ work in setting up this
correspondence is done once for the particular format
of the researcher. For the most part, a given research
lab has a given device and a given data format that
they have used for years at a time.
4 SOFTWARE ENTITIES
The CHEMCONNECT systems consists of the
interplay between software entities of different types
defining the structure, its persistence, its knowledge
content (templates and instantiations), how it can be
visualized and its textual form (XML, yaml, etc.). A
single entity of CHEMCONNECT has several forms
serving different purposes. The purpose of this
section is to give a brief overview of the software
interactions with examples and key elements.
4.1 Ontology Object
One purpose of the CHEMCONNECT ontology is to
define data structures through the catalog, record and
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
244

primitive objects. This definition steers the
management of objects and information within the
JAVA software.
For example, the catalog object,
SubSystemDescripton
has (in addition to the
standard information for all catalog objects listed
previously) the following:
ParameterValue
(multiple): Parameterized
(attribute-value pairs) description of the
device.
ObservationSpecification
(multiple): A
specification of the observations that are made
from the subsystem.
SubsystemDescription
(multiple): The
subsystems of this subsystem (a device is
viewed as a hierarchy of subsystems).
In the ontology these are specified as a subclass
restriction with a
owl:onProperty
of
dcat:record
with
owl:someValuesFrom
, if
multiple, or
owl:onClass
if a singlet, which points
to the respective ontology record or catalog object. To
find the list of sub-objects of a catalog class a
SPARQL query is done. Within the JAVA software,
the SPARQL commands to access the ontology are
embedded in functions.
Figure 2: The ontology, shown on the left (from Prodigy)
gives the template for an observation. CHEMCONNECT
can use this template to create the interface to fill in values
for the observation elements.
4.2 Hierarchy of JAVA Classes
Corresponding to each catalog and record object
(primitives are usually just String classes) is a JAVA
class and there is a one-to-one correspondence to the
elements of the ontology definition. Each element in
the JAVA class is a string with the identifier of the
sub-object. If the object is a singlet, then the string is
the identifier of sub-object. If the object is multiple,
then the identifier points to a single
ChemConnectCompoundMultiple
object having a
reference to the multiple entities.
Each JAVA class corresponding to an ontology
catalog or record object is also persistent database
objects. CHEMCONNECT uses Google App Engine
(GAE) datastore objects (using Objectify API). This
is expressed with the
@Entity
and
@Index
annotations in the class definition.
A complete catalog object is actually a hierarchy
of JAVA catalog and record objects and is
represented in a tree of
DatabaseObjectHierarchyNode
objects. This
class holds the current object instantiation and the list
of
DatabaseObjectHierarchyNode
having the
sub-objects. When reading an object from persistent
storage (GAE datastore), the top object is read, put
into the top of the hierarchy and then using the
ontology description of the class and the identifiers
within the object, the sub-objects in the hierarchy are
filled in.
The hierarchy is made from individual classes as
java and the corresponding persistent objects in the
database. Associated with each object in the ontology
(corresponding to JAVA class) is an identifier. A
class is translated to a map using these identifiers. A
single class is translated to a map with the ontology
identifier pointing String object that is either an object
identifier if it another class or the class element value
if it is class information. The class hierarchy is a
hierarchy of these class maps. This mapping structure
facilitates the automated manipulation of the
hierarchy of class information and translation, for
example, to a text form such as YAML.
4.3 Domain Specific Information
The descriptions of the software objects in this
section up to now are general and the only connection
to domain information is what ‘general’ entities are
needed to describe the domain, which is this case is
experimental and modelling chemical information.
For example, the
SubSystemDescription
entity is
general enough to describe a large class of devices
and components. But there is no specific information
about the structure of a specific device. The primary
reason for this is to simplify the ‘hard-coded’
software and leave the ability to dynamically update
domain information as it is expanded in concert with
domain researchers. The domain researcher
concentrates on the ontology description of the
domain and the not the software technical details.
Under the
skos:Concept
hierarchy are
templates of typical domain instantiations of the
general catalog and record entities. For example, the
template needed for a
SubSystemDescription
entity fills in the four basis elements. The
Use of Ontologies in Chemical Kinetic Database CHEMCONNECT
245

subSystemType is the name of the subsystem
concept, for example dataset:HeatFluxBurner
(a device within the combustion chemical kinetics
domain). The particular device’s position in the
ontology skos:Concept hierarchy gives addition
context information about the device. For example,
the heat flux burner is in the hierarchy:
• dataset:DataTypeDevice
• dataset:CombustionInstrument
• dataset:FlameBurner
For analytical purposes, for example to
statistically compare the results of similar devices, it
is often useful to parameterize a description of the
device (or any catalog object for that matter). The
domain researchers, in analysing their particular
devices, often deduce a set of significant parameters
(range of operation, configuration, etc.) that should be
in the device description. These are presented as data
cube
cube:attribute sub-class restriction
owl:onClass
on a parameter within the ontology of
parameter specifications.
The device itself is viewed as a system of
subsystems. The top level being the overall device
description and the sub-levels describe important sub-
systems making up the device. The hierarchy can be
arbitrarily deep. Within the device ontology template
specification, is the set of subsystems making up the
device. Each subsystem is itself represents a template
instantiation of a
SubSystemDescription. This
is done through through a subclass restriction
owl:related and the ssn:hasSubSystem
property.
In a similar fashion, the set of observations, i.e.
the data that the subsystem can produce, is listed.
4.4 User Interface
Also associated with each catalog and record entity is
a user interface object which steers the look and feel
of the object presentation. The ontology and
particularly the templates defined object within the
ontology play an important part in the user interface.
The ontology plays a particularly important role
for the presentation of choices. For the specific choice
of a specific choice, the hierarchy of concepts found
below the top concept are presented as a pull-down
list (for example, for a classification) or a tree of
choices. For example, for the creation of a new
device, the concept hierarchy under the
dataset:DataTypeDevice is presented as a tree
menu. The nodes of the tree represent the device
templates to be presented. An end node is selected
and a new device is created using the device template
as an initial pattern.
Just as the objects themselves, the presentation
objects are set up as a hierarchy. For the most part,
the presentation object has a header object which
visualizes the direct information of the object. Under
this header is a tree of sub-objects as defined by the
particular object. If the object has elements that can
be changed, then those elements are presented as a
form.
CHEMCONNECT is cloud based as a Google
App Engine. The interface is built using Google Web
Tool Kit(Google GWT 2017) with GWT Material
Design(Google 2018) for the look and feel of the
presentation.
5 STANDARD ONTOLOGIES
Part of the design philosophy of the
CHEMCONNECT ontology was to build upon
available accepted ontologies from the W3C
community. The basic ontologies which have had a
particular influence are:
Dublin Core Terms(Dublin Core 2012): This
is the source of the basic terminology and the
basis of the other ontologies.
Simple Knowledge Organization System
(Miles and Bechhofer 2008): This is the basic
ontology for the concept terms and their inter-
relations.
Two ontologies serve as the basis for the contact
information:
Friend of a friend(Brickly and Miller 2014):
This is basic information about individuals.
Organization(Reynolds 2014): Organization
contact structure.
The ontology dealing with data structures are:
Data Catalog Vocabulary, DCAT (Maali
and Erickson 2014): This is the basic for the
category and record entities. This serves as the
basis for the basic data types and database
objects used within CHEMCONNECT.
Data Cube Vocabulary, cube (Cyganiak and
Reynolds 2014):
Several ontologies which contain domain knowledge:
Semantic Sensor Network, SSN (Haller et al.
2017):
Sensor Organization Sampling Actuator,
SOSA (Cox 2017):
Quantities, Units, Dimensions and Data
Types Ontologies, QUDT (QUDT 2018):
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
246

7 CONCLUSIONS
What has been described in this paper is the extensive
use of ontologies at several levels within the
CHEMCONNECT repository. The primary goal of
using ontologies is to decouple domain knowledge
from the software engine to produce a data/ontology
driven repository for experimental and modelling
information. The current version of
CHEMCONNECT is modelled on the domain
knowledge within the chemical kinetics and
combustion communities. An implementation of the
repository can be found at:
http:www.connectedsmartdata.info
and implemented on the Google Cloud Platform.
The templates for devices, protocols, algorithms, etc.
within CHEMCONNECT are derived in
collaboration of experts within the field. The
templates are implementations of best practice of data
reporting within the domain as established by domain
experts. CHEMCONNECT knowledge base is a
result of collaborations from the SMARTCATS
COST Action CM1303, Chemistry of Smart Energies
Carriers and Technologies.
ACKNOWLEDGEMENTS
The author would like to acknowledge the
cooperation of domain experts within the combustion
community that has been made possible through the
SMARTCATS COST Action CM1303, Chemistry of
Smart Energies Carriers and Technologies. In
particular, the members of the working task group
within the action for Standard definition for data
collection and mining toward a virtual chemistry of
Smart Energy Carriers, of which the author is the
group leader.
REFERENCES
Brickly, Dan, and Libby Miller. 2014. “FOAF Vocabulary
Specification.” January 14, 2014. http://xmlns.com/
foaf/spec/.
Cox, Simon. 2017. “SOSA Ontology - Spatial Data on the
Web Working Group.” April 18, 2017. https://www.
w3.org/2015/spatial/wiki/SOSA_Ontology.
Cyganiak, Richard, and Dave Reynolds. 2014. “The RDF
Data Cube Vocabulary.” January 16, 2014.
https://www.w3.org/TR/vocab-data-cube/.
Dublin Core. 2012. “Dublin Core Metadata Initiative.”
Dublin Core Metadata Initiative. June 12, 2012.
http://dublincore.org/.
GO FAIR. 2016. “FAIR Principles - GO FAIR.” 2016.
https://www.go-fair.org/fair-principles/.
Google. 2018. GWT Material Design (version 2.1.1).
Google. https://gwtmaterialdesign.github.io/gwt-
material-demo/.
Google GWT. 2017. Google Web Toolkit (GWT).
http://www.gwtproject.org/.
Hagstrom, Stephanie. 2014. “The FAIR Data Principles.”
FORCE11. September 3, 2014. https://www.
force11.org/group/fairgroup/fairprinciples.
Haller, Armin, Krysztof Janowicz, Simon Cox, Danh Le
Phuoc, Kerry Taylor, and Maxime Lefrancois. 2017.
“Semantic Sensor Network Ontology.” December 8,
2017. https://www.w3.org/TR/vocab-ssn/.
Maali, Fadi, and John Erickson. 2014. “Data Catalog
Vocabulary (DCAT).” January 16, 2014.
https://www.w3.org/TR/vocab-dcat/.
Miles, Alisair, and Sean Bechhofer. 2008. “SKOS Simple
Knowledge Organization System Namespace
Document 30 July 2008 ‘Last Call’ Edition.” August
20, 2008. https://www.w3.org/TR/2008/WD-skos-
reference-20080829/skos.html.
QUDT. 2018. “Quantities, Units, Dimensions and Data
Types Ontologies.” Quantities, Units, Dimensions and
Data Types Ontologies. December 16, 2018.
http://qudt.org/.
Reynolds, Dave. 2014. “The Organization Ontology.”
January 16, 2014. https://www.w3.org/TR/vocab-org/.
Wilkinson, Mark D., Michel Dumontier, IJsbrand Jan
Aalbersberg, Gabrielle Appleton, Myles Axton, Arie
Baak, Niklas Blomberg, et al. 2016. “The FAIR
Guiding Principles for Scientific Data Management and
Stewardship.” Scientific Data 3 (March): 160018.
https://doi.org/10.1038/sdata.2016.18.
Use of Ontologies in Chemical Kinetic Database CHEMCONNECT
247
