
Chemical Named Entity Recognition with Deep Contextualized Neural
Embeddings
Zainab Awan
1 a
, Tim Kahlke
2 b
, Peter J. Ralph
2 c
and Paul J. Kennedy
1 d
1
School of Computer Science, University of Technology Sydney, Sydney, Australia
2
Climate Change Cluster, University of Technology Sydney, Sydney, Australia
Keywords:
Named Entity Recognition, Deep Learning, Word Representation, BiLSTM.
Abstract:
Chemical named entity recognition (ChemNER) is a preliminary step in chemical information extraction
pipelines. ChemNER has been approached using rule-based, dictionary-based, and feature-engineered based
machine learning, and more recently also deep learning based methods. Traditional word-embeddings, like
word2vec and Glove, are inherently problematic because they ignore the context in which an entity appears.
Contextualized embeddings called embedded language models (ELMo) have been recently introduced to rep-
resent contextual information of a word in its embedding space. In this work, we quantify the impact of
contextualized embeddings for ChemNER by using Bi-LSTM-CRF (bidirectional long short term memory net-
works - conditional random fields) networks. We benchmarked our approach using four well-known corpora
for chemical named entity recognition. Our results show that incorporation of ELMo results in statistically
significant improvements in F1 score in all of the tested datasets.
1 INTRODUCTION
The volume of biomedical literature is increasing at
an exponential rate (Khare et al., 2014) which makes
manual searching and reading slow and labour inten-
sive for researchers and database curators (Jelier et al.,
2005). Text mining tools are essential for automating
the literature curation workflow. Named entity recog-
nition (NER) is the first step towards literature cura-
tion which aims to locate and classify named entities
from unstructured texts into pre-defined categories
such as person, location or organization. Biomedical
NER aims at identifying biomedical entities such as
chemicals, genes, proteins and diseases from biomed-
ical text. Biomedical NER is more complicated than
general domain NER because of ambiguous terms
and lexical variations (Kim et al., 2005). Biomedical
NER helps in downstream relation extraction, event
extraction, and question-answering tasks for knowl-
edge base completion.
In this work, we focus only on the recogni-
tion of the chemical entities from biomedical lit-
a
https://orcid.org/0000-0002-5356-4227
b
https://orcid.org/0000-0002-9762-6573
c
https://orcid.org/0000-0002-3103-7346
d
https://orcid.org/0000-0001-7837-3171
erature. Chemical entity recognition from the lit-
erature helps scientists working in drug develop-
ment and discovery (Eltyeb and Salim, 2014) among
other areas. Traditionally, chemical named entity
recognition (ChemNER) has been performed by rule-
based, dictionary-based and machine-learning-based
approaches, mainly conditional random fields. All of
these methods have drawbacks, such as low precision
and recall, and labour-intensive feature engineering.
With the availability of word embeddings and neu-
ral networks, efforts have been made in the recent
past to build end-to-end deep learning-based Chem-
NER systems (Habibi et al., 2017; Giorgi and Bader,
2018; Crichton et al., 2017; Corbett and Boyle, 2018).
These methods rely on pre-trained word2vec embed-
dings. However, these embeddings do not take into
account the contextual information of the named en-
tities and the entities are mapped to the same vector
space irrespective of their context, which is problem-
atic.
In this work, we examine whether Bi-LSTM-CRF
(bidirectional long short term memory networks- con-
ditional random fields) network could lead to better
performance for ChemNER when the input represen-
tation includes contextual representations ELMo in
conjunction with static representation word2vec.
In this study we quantify the impact of ELMo rep-
Awan, Z., Kahlke, T., Ralph, P. and Kennedy, P.
Chemical Named Entity Recognition with Deep Contextualized Neural Embeddings.
DOI: 10.5220/0008163501350144
In Proceedings of the 11th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2019), pages 135-144
ISBN: 978-989-758-382-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
135

resentations for ChemNER. To the best of our knowl-
edge, this is the first application of ELMo to Chem-
NER, although it has been previously applied to gen-
eral domain English corpora (Peters et al., 2018).
The rest of this paper is organised as follows. Sub-
section 1.1 presents related work including research
gaps and motivation for this paper. Section 2 presents
network architectures used in this paper together with
the word embeddings and benchmark corpora evalu-
ated. In Section 3 we describe the experimental setup
and in Section 4 experimental results and discussion.
Finally, in Section 5 we conclude the paper.
1.1 Related Work
To validate our approach, we consider five deep
learning-based baseline methods for ChemNER.
Baselines use techniques such as transfer learning,
multi-task-learning, and ensemble-based methods. In
the following section, we will discuss each of the ap-
proaches.
1.1.1 Word2vec with Bi-LSTM-CRF
The Bi-LSTM-CRF architecture was proposed by
Lample et al. (2016) for NER for four different lan-
guages, English, German, Spanish and Dutch, with-
out relying on any language specific resources or
features. The model consists of forward and back-
ward LSTM layers and a conditional random field
(CRF) layer for classification. It also incorporates
word embeddings and, for out-of-vocabulary terms,
LSTM based character representations were also con-
catenated with word-embeddings for a richer repre-
sentation. This model reported state-of-the-art perfor-
mance for sequence tagging in a generic domain.
Later, this model was evaluated for biomedical
NER on 33 corpora for five biomedical entities by
Habibi et al. (2017). We believe that this was the
first application of Bi-LSTM-CRF to sequence tag-
ging task in the biomedical domain. For biomedical
NER, word2vec pre-trained embeddings trained on
Pubmed abstracts made available by Moen and Ana-
niadou (2013) were used in this study. The perfor-
mance reported for five biomedical entities over 33
corpora was at par with the best performing feature-
engineered based systems, without relying on any
syntactic features or lexicons. Later this architecture
was evaluated for transfer learning based approach
which we will discuss in the following section.
1.1.2 Transfer Learning for ChemNER
Transfer learning for ChemNER employs transfer of
weights from a source to target dataset using a pre-
existing neural network which is used for generic
NER called NeuroNER (Dernoncourt et al., 2017).
NeuroNER employs Bi-LSTM-CRF with LSTM-
based character embeddings and word2vec word em-
beddings, similar to the model proposed by Lample
et al. (2016). We discuss the Bi-LSTM-CRF architec-
ture in detail in subsection 2.1.
In summary, this approach does not initialize
model weights randomly for a gold standard corpus.
Firstly, the model is trained on a large silver stan-
dard corpora (Rebholz-Schuhmann et al., 2010) and
then weights of the layers are transferred to train on a
gold standard corpus (Giorgi and Bader, 2018). This
transfer of weights results in improved performance
compared to the model that was trained directly on
a gold standard corpus with random initialization of
weights. This method highlights the importance of
noisy silver standard corpora (i.e., those annotated by
text mining tools) that do not require human annota-
tion and can be easily generated with existing NER
tools. This method statistically significantly outper-
forms the method by Habibi et al. (2017).
1.1.3 Multi-task ChemNER
The multi-task ChemNER method (Crichton et al.,
2017) performs NER of biomedical entities using
three models: a single task model (Figure 1), a multi-
task multi-output model (Figure 2) and a dependant
multi-task model (Figure 3). Crichton and colleagues
show that multi-task settings do better than the single-
task model in terms of the F1-score. The single task
model is rather simple and inputs word2vec embed-
dings into a convolutional layer, whose output is then
fed into a fully-connected layer. The final output layer
is a dense layer with a softmax activation function.
Max-pooling layer was not used in this model as it
results in loss of positional information. In the multi-
task multi-output model all tasks share the input layer
and the convolutional layer. Each task (dataset) has
its own output layer.
The dependent multi-task model makes use of the
fact that some NLP tasks (in this case NER) can be
improved if they get information from other related
NLP tasks (called auxiliary tasks). For example, NER
can benefit from part-of-speech (POS) tagging. In the
dependent multi-task model, two single task models
are combined in such a way that a fully connected
layer of the main task receives input from another sin-
gle task model that performs POS tagging on the same
input. In this way, the supplementary information of
POS tags helps improve the performance of biomedi-
cal NER.
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
136
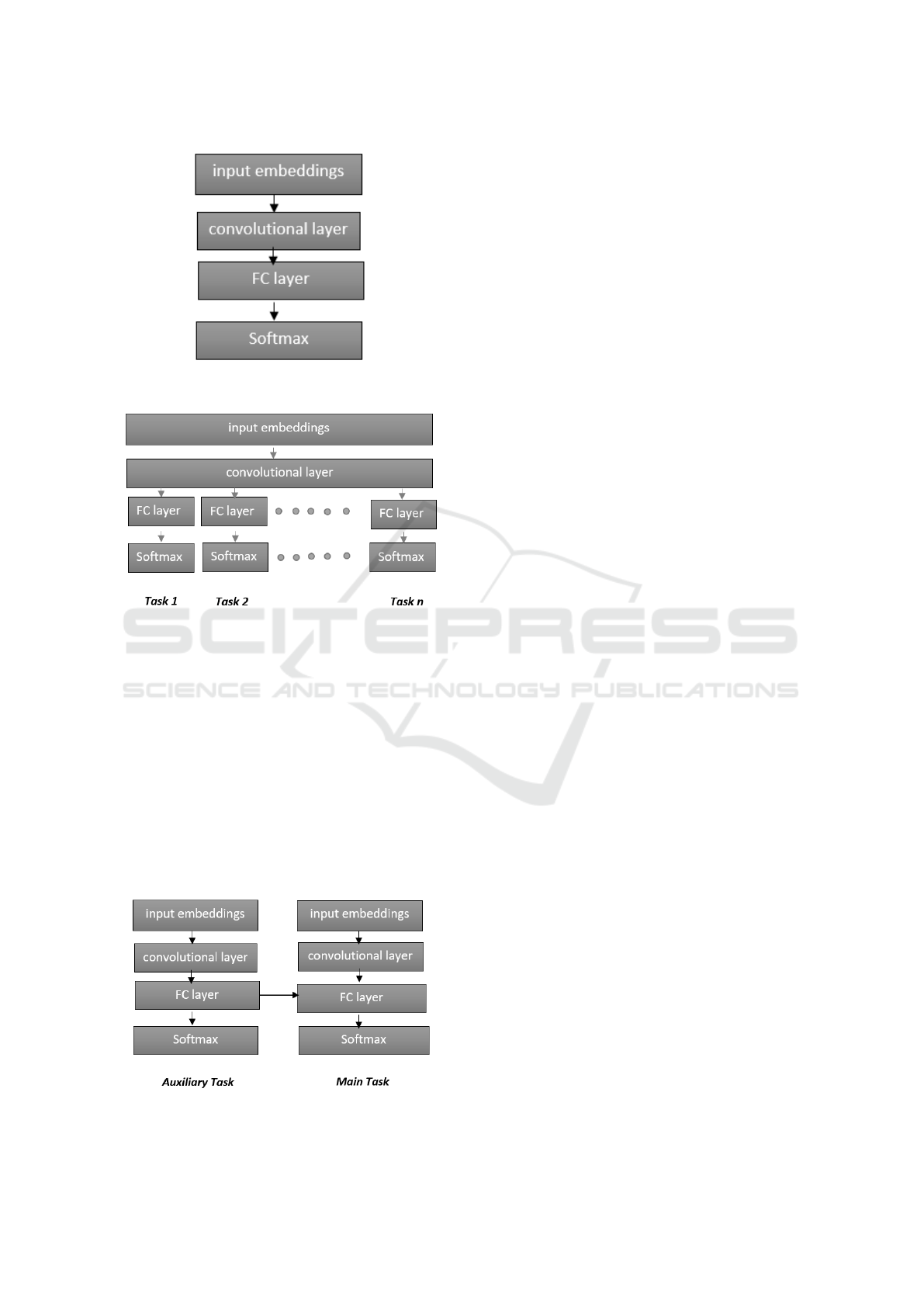
Figure 1: Single task model. FC stands for fully connected
layer.
Figure 2: Multi-task multi-output model where FC stands
for fully connected layer.
1.1.4 LSTMVoter
LSTMVoter (Hemati and Mehler, 2019) is a two-
stage method that uses five existing sequence tag-
ging tools including Stanford named entity recog-
nizer (Finkel et al., 2005), MarMot (M
¨
uller et al.,
2013), CRF++ (Kudo, 2010), MITIE (Geyer et al.,
2016) and Glample (Lample et al., 2016) in an initial
stage to label the sequences.
The outputs from stage one are transformed into
one-hot vectors with an attention layer on top of the
vectors. These one-hot vectors are then concatenated
Figure 3: Dependent multi-task model where FC stands for
fully connected layer.
with word2vec embeddings (Moen and Ananiadou,
2013) and a character level representation. A Bi-
LSTM with attention mechanism is used to represent
characters. The final representation is then fed into a
Bi-LSTM-CRF network for sequence tagging. This
method employs a Tree-structured Parzen Estimator
(TPE) (Bergstra et al., 2011) for hyperparameter opti-
mization.
1.1.5 Chemlistem
Chemlistem (Corbett and Boyle, 2018) is a combina-
tion of two approaches: a traditional and a minimal-
ist approach. The traditional approach uses a token-
based feature set with Glove word embeddings (Pen-
nington et al., 2014), trained on an in-house corpus of
patents, which is fed into LSTM layers.
The minimalist approach does not use any token
features and relies completely on character represen-
tations that are fed into the LSTM layers without even
using a tokeniser. The motivation behind the mini-
malist approach is that word segmentation in chemi-
cal texts is challenging and character representations
will allow a system to avoid tokenisation. Finally, the
results from the two systems are combined to get a fi-
nal prediction based on a scoring mechanism given in
Corbett and Boyle (2018).
1.1.6 Summary of Research Gaps
The methods discussed above rely on static word em-
beddings, which do not address the problem of pol-
ysemy. Polysemy is the capacity of a word to have
different meanings in different contexts. To overcome
this problem, ELMo representations model polysemy
by learning task-specific representations of words, en-
abling multiple representations based on context.
For instance, a word may be a chemical entity in
one case but a different biomedical entity in another
context. Context-dependency is particularly problem-
atic for abbreviations. For example, VHL could be a
gene, disease or a chemical depending on the context
in which it is used. To our knowledge, Bi-LSTM-
CRF has only been evaluated without ELMo repre-
sentations for ChemNER. In this study, we investigate
the impact of adding contextual representations to Bi-
LSTM-CRF and evaluate the performance using four
well-known corpora.
2 METHODS AND DATASETS
In this section, we describe the network architecture,
word embeddings, ELMo and the corpora used in this
study.
Chemical Named Entity Recognition with Deep Contextualized Neural Embeddings
137
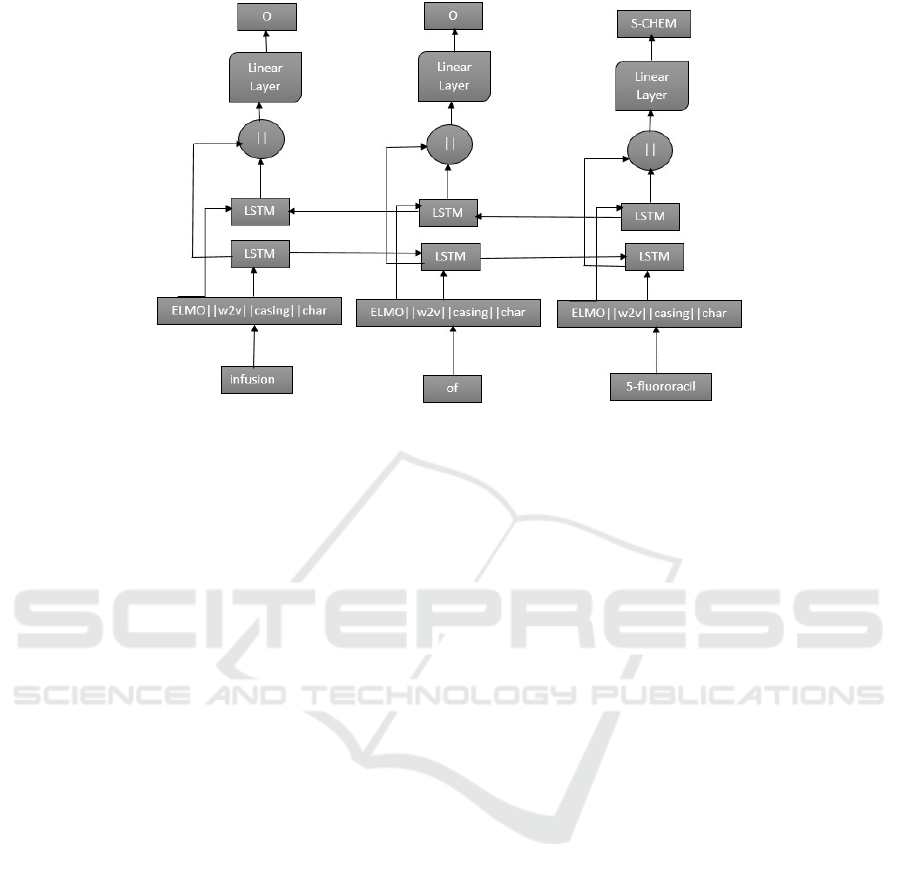
Figure 4: ELMo, word2vec, casing feature and character representations are concatenated together (k is the concatenation
operator) and fed into the Bi-LSTM-CRF network. The input sequence infusion of 5-fluororacil is labelled as “O, O, S-
CHEM”, where O means not an entity and S-CHEM means a single token chemical entity.
2.1 Neural Network Architecture
The architecture chosen was published by Reimers
and Gurevych (2017) and has previously been applied
to sequence labelling tasks (Huang et al., 2015; Ma
and Hovy, 2016; Chiu and Nichols, 2016). The net-
work is implemented in Keras 2.2.0 with Tensorflow
1.8.0 as a backend.
2.1.1 Bi-LSTM-CRF
Recurrent neural networks (RNNs) are a class of
neural networks that take a sequence of vectors
(x
1
, x
2
, . . . , x
t
) as input and return a new sequence
of vectors (h
1
, h
2
, . . . , h
t
). Theoretically, RNNs can
capture long-range dependencies in sequential data
but in practice they fail to do so due to the vanish-
ing gradient problem (Bengio et al., 1994; Hochre-
iter, 1998). LSTMs have been introduced to address
the issue with a memory-cell to retain long-range de-
pendencies. Memory cells in turn use several gates
to control the proportion of input kept in memory
and the proportion of input to forget (Hochreiter and
Schmidhuber, 1997). The following equations govern
LSTMs, where W and b are the weights and biases,
and σ are element-wise dot product and element-
wise sigmoid functions respectively. i
t
, o
t
are input
gate activation and output gate activation vectors, c
t
is a cell-state vector, and t indexes the time step.
i
t
= σ(W
xi
x
t
+ W
hi
h
t−1
+ W
ci
c
t−1
+ b
i
) (1)
c
t
= (1 − i
t
) c
t−1
+
i
t
tanh(W
xc
x
t
+ W
hc
h
t−1
+ b
c
) (2)
o
t
= σ(W
xo
x
t
+ W
ho
h
t−1
+ W
co
c
t
+ b
o
) (3)
h
t
= o
t
tanh(c
t
), (4)
For a given sentence (x
1
, x
2
, . . . , x
t
) containing t
words, each represented as a d-dimensional vector, an
LSTM computes a representation
−→
h
t
of the left con-
text of the sentence at every word t. A right context
←−
h
t
can also be added, which computes the representation
in the reverse direction. The left context network is
called a forward layer and the right context network is
called a backward layer. Both networks can be com-
bined to form a bidirectional LSTM represented as
h
t
= [
−→
h
t
;
←−
h
t
] (Graves and Schmidhuber, 2005). The
final representation is computed by concatenating the
forward and backward context vectors. A Bi-LSTM
runs over each sentence in a forward and backward
direction. The final outputs are concatenated together
and serve as the input to a CRF classifier.
A linear chain CRF (log-linear model) is used to
predict the probability distribution of tags of each
word in a complete sentence. Linear chain CRF can
also be referred to as a CRF. We have used a CRF
classifier instead of a softmax classifier because we
do not want to lose the sequential information (Laf-
ferty et al., 2001). Our final architecture is shown in
Figure 4.
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
138

2.2 Word Representation
In this section, we describe the word representations
word2vec, ELMo, casing feature and character repre-
sentation.
2.2.1 Word2vec
For the pre-trained word embeddings we have used
word2vec embeddings (Mikolov et al., 2013) trained
on Wikipedia-PubMed-PMC, 23 million Pubmed ab-
stracts, 700,000 full-text PMC articles and four mil-
lion Wikipedia pages. These embeddings were
trained and made publicly available by Moen and
Ananiadou (2013). We have used these embeddings
to be consistent with studies, Habibi et al. (2017)
and Giorgi and Bader (2018), that perform biomed-
ical NER using deep learning.
2.2.2 Embedded Language Models (ELMo)
ELMo is a linear combination of hidden states of Bi-
LSTM with character convolutions trained on a very
large corpus (Peters et al., 2018). In this case, we
have used the ELMo pre-trained on Pubmed and gen-
eral domain large corpora. To address the contextual
information loss in static word embeddings, ELMo
employs an approach where a word representation is
a function of the whole sentence in which it appears.
ELMo can integrate well into existing NLP applica-
tions such as question-answering, sentiment analysis
and NER (Peters et al., 2018).
ELMo has two main steps. Firstly, a three-
layer Bi-LSTM network is trained on a large unla-
belled corpus
1
in an unsupervised manner, which is
completely agnostic to ChemNER. Secondly, hidden
states of the network are taken, and their linear combi-
nation learned from the downstream task, in this case
ChemNER. Each task will learn its linear combina-
tion from the same weights. In this study, we have
used two pre-trained models of ELMo, one of them
pre-trained on 5.5B English words and the other pre-
trained on Pubmed articles.
2.2.3 Casing Feature
A casing feature (Reimers and Gurevych, 2017) is
a 7-bit one-hot vector that represents information
about a word. It is mainly numeric if more than
half of the characters are numeric, numeric if all
characters are numeric, all upper if all characters
are uppercase, all lower for all character if lower
case, initial upper if the first character is capital,
1
https://allennlp.org/elmo
contains digit if it has a digit, and the other label
is set to one when none of the rules could be applied.
2.2.4 Character Representation
Character-based convolutions are used for deriving
character representations based on the work of Ma
and Hovy (2016). Randomly initialized character em-
beddings are input to the convolutional layer followed
by a max-pooling layer, which provides a character
representation. Before feeding embeddings into the
convolutional layer, a dropout layer is also applied.
This is the same as that of Chiu and Nichols (2016),
with the only difference being the use of character
type features. In this work, we use a 50-dimensional
character representation.
2.3 Datasets
We have used four publicly available datasets for
ChemNER experiments as outlined in Table 1 which
shows the number of sentences used in the training,
test and validation sets.
BC5CDR. The Biocreative community chal-
lenge for chemical-disease relation extraction task
(BC5CDR) corpus was made available in a Biocre-
ative workshop (Li et al., 2016). The two subtasks
of BC5CDR are identifying chemical and disease
entities from Medline abstracts. The corpus has
1500 abstracts from Pubmed and chemical entities
are annotated by a team of indexers from Medical
Subject Headings (MeSH).
Annotations were done by two groups, and the
inter-annotator agreement was 96.05% for chemical
entities. The corpus has been split into training, test
and validation sets, where each set has 500 abstracts.
We have used this corpus in BIO (Beginning, Inside,
Outside) tagging scheme for ChemNER only.
BC4CHEMDNER. This dataset has been provided
by BioCreative community challenge IV for the de-
velopment and evaluation of tools for Chemical NER
(Krallinger et al., 2015). BC4CHEMDNER was used
for the recognition of chemical compounds and drugs
from Pubmed abstracts. The inter-annotator agree-
ment between human annotators is 91%. Ten thou-
sand abstracts were annotated by expert literature cu-
rators. We have downloaded training, validation and
test sets in the IOBES tagging scheme from Github
2
.
2
https://github.com/cambridgeltl/MTL-Bioinformatics-
2016/tree/master/data/BC4CHEMD
Chemical Named Entity Recognition with Deep Contextualized Neural Embeddings
139

Table 1: Gold standard corpora - number of sentences in
train, test and validation sets.
Data # Train # Test # Val
BC5CDR 9578 4686 1774
BC4CHEMDNER 30682 26364 30639
BioSemantics 15557 8840 3511
BCV.5CEMP 24145 9843 4773
Chemical Entity Mentions in Patents (CEMP)
Biocreative V.5. CEMP V.5 is based on Chem-
NER from patents. Twenty-one thousand patents
from medicinal chemistry were curated by experts
for annotation of chemical entities (P
´
erez-P
´
erez et al.,
2017). Patents are different from regular research ar-
ticles in that they use rather a complex language and
could contain up to 100 pages. This is why this task
focuses on the detection of chemical entities from
patents only.
Training, development and test sets each con-
tained seven thousand patents. Gold labels of the test
set used for evaluation are not made publicly avail-
able. We therefore first combine all fourteen thou-
sand patents and then split into the train, validation
and test sets in the ratio of 60:10:30. This setting has
been chosen to be consistent with Habibi et al. (2017).
We use this dataset with the IOBES tagging scheme.
BioSemantics. Similar to the CEMP corpus, the
Biosemantics corpus (Akhondi et al., 2014) has been
constructed from 200 patents taken from the Euro-
pean Patents Office, the United States Patents and
Trademark Office and the World Intellectual Property
Organization. The corpus has been downloaded from
the Biosemantics official webpage
3
. It has been split
into training, validation, and test sets in the ratio of
60:10:30. The identification numbers of the docu-
ments that go into training, validation, and test sets
have been taken from Github
4
. We use this dataset
with the IOBES tagging scheme.
3 EXPERIMENTAL SETUP
In this study we used the Bi-LSTM-CRF network de-
scribed in Reimers and Gurevych (2017). The tag-
ging schemes and data split have been discussed in
section 2.3. We have padded each word to make it 50
characters for CNN based representation. An early
3
https://biosemantics.org/index.php/resources/chemical-
patent-corpus
4
https://github.com/BaderLab/Transfer-Learning-
BNER-Bioinformatics-2018/tree/master/corpora
stopping parameter of value 5 is used to prevent over-
fitting. That is, if the performance does not improve
for five epochs, the network stops training. For all the
runs, the models have been trained for 30 epochs with
the variational dropout (Gal and Ghahramani, 2016)
of (0.5,0.5). Nadam optimizer is used with the default
learning rate in all the experiments.
The network has two LSTM layers with 100 re-
current units in each layer. Since the layers are Bi-
LSTM, each layer has 200 units. The mini-batch size
is 32. We have chosen these parameters after ex-
perimenting with different hyperparameter values and
these values gave us the best results.
4 RESULTS AND DISCUSSION
In this study we used Bi-LSTM-CRF networks in
combination with static and contextual embeddings.
We validated our approach by comparing it with five
previously published studies on four benchmark cor-
pora. We used the F1 metric for evaluation with the
Conference on Natural Language Learning (CoNLL)
scheme. We perform three sets of experiments: a
baseline (without ELMo), with ELMo (general do-
main pre-trained model) and ELMo (pre-trained on
Pubmed). Each experiment was repeated five times
for each dataset and we report the average F1-score
(on the test set) from the epoch that has the best vali-
dation score.
For BC5CDR (shown in Table 2) and
BC4CHEMDNER (Table 3) corpora, our model
ELMo (Pubmed) achieves the highest F1-score when
compared to the previously published methods, that
is 93.02 and 90.80. Additionally, our experiments
also show that ELMo pre-trained on Pubmed results
in better performance as compared to ELMo pre-
trained on the general domain corpus. Our method is
completely entity agnostic and does not rely on any
inductive transfer for the hidden layers as transfer
learning (Giorgi and Bader, 2018) and multi-task
learning (Crichton et al., 2017) do.
Our initial hypothesis, that concatenating ELMo
with word2vec representation will outperform the
baseline Bi-LSTM-CRF, holds for the CEMP and
Biosemantics corpora. The F1 scores for CEMP and
Biosemantics are 81.66 and 76.09 respectively for the
baseline model and improved to 82.37 and 77.70 af-
ter using ELMo. However, the model did not out-
perform the best performing competing systems over
these two corpora. Our results are not directly com-
parable with Chemlistem and LSTMVoter as their
training/validation/test sets use different proportions.
Also, LSTMVoter (Hemati and Mehler, 2019) uses
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
140
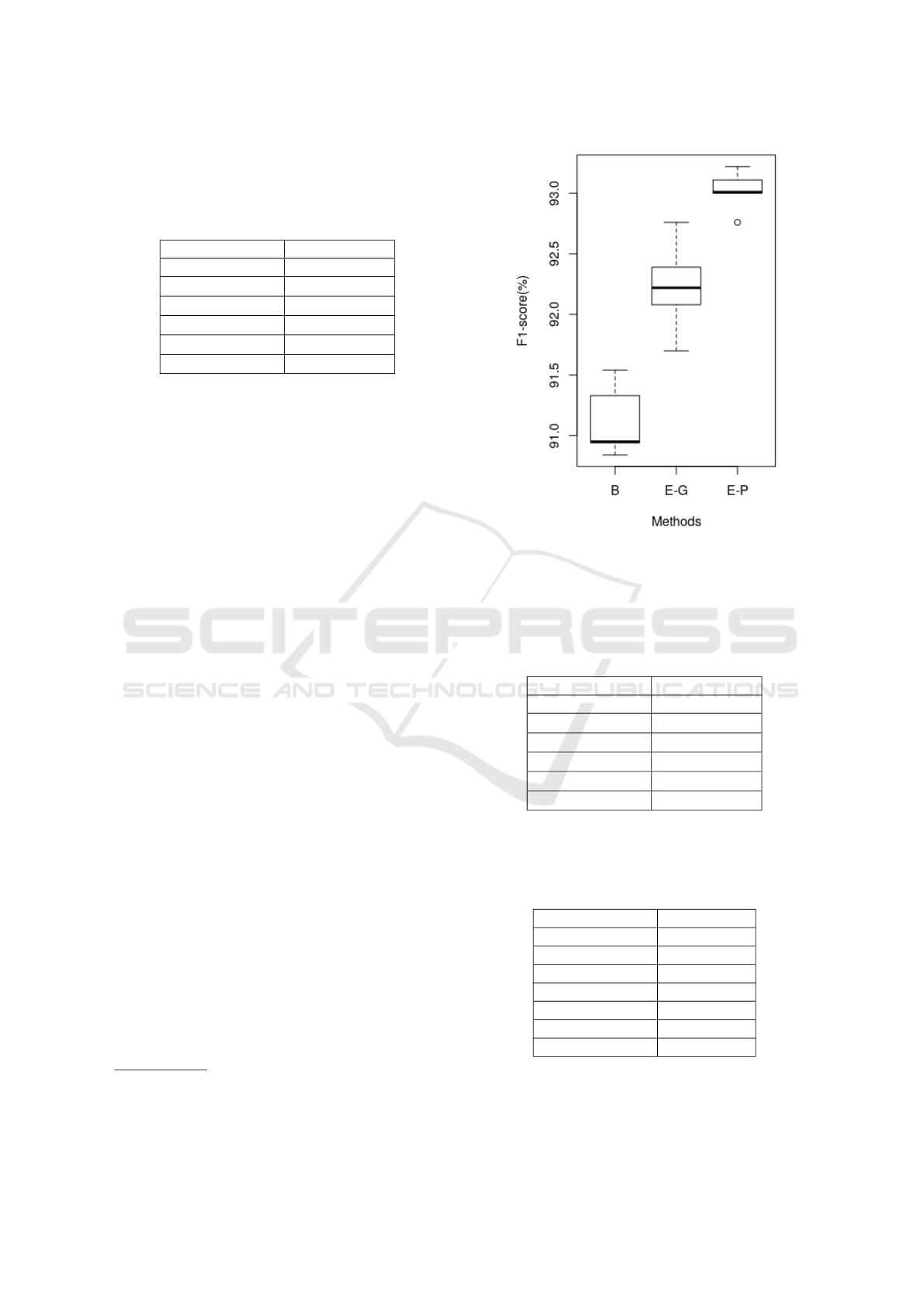
Table 2: F1 score on BC5CDR. Best F1 score in bold and
significantly worse than our model **. First three rows
show our results which are averaged F1 ± SD over five runs
(random seeds). The rest of the results are reported directly
from the respective papers.
Methods F1- score
ELMo (Pubmed) 9
9
93
3
3.
.
.0
0
02
2
2 ±
±
± 0
0
0.
.
.1
1
17
7
7
ELMo (General) 92.23 ± 0.39
**
Baseline 91.02 ± 0.42
**
Habibi 90.63
**
Giorgi TL 91.64
**
Crichton MTL 89.22
**
30,000 patents for the CEMP task, whereas the avail-
able number of patents is only 21,000 (P
´
erez-P
´
erez
et al., 2017) out of which only 14,000 are available
for training purposes. The data used for LSTMVoter
possibly have been combined another corpus with the
original CEMP corpus.
Our results can only be directly compared with the
transfer learning system (Giorgi and Bader, 2018) and
Habibi’s system (Habibi et al., 2017). Our best per-
forming model achieves F1 scores of 82.37 and 77.70
for CEMP, Table 4 and Biosemantics, Table 5. For
these two tasks, our best performing model underper-
forms the transfer learning and Habibi’s systems.
This difference in performance could be attributed
to the hyperparameter tuning or to pre-processing
done to the data. Unfortunately, Giorgi and Bader
(2018) and Habibi et al. (2017) do not make their
pre-processed data available due to the licensing re-
strictions on redistribution of the datasets, so we can-
not conclusively determine the reasons for the differ-
ence in performance. We have downloaded the data
from their respective websites
5
and transformed in
BIO tagging format using a tool
6
. Giorgi and Bader
(2018) use the corpus in brat standoff annotation for-
mat, and we use the BIO tagging scheme. Our method
detected some incorrect B-beginning and I-inside tags
during evaluation and converted them to O-outside
tags. The lower performance of our method for these
two datasets could be due to the incorrect tags af-
ter conversion from brat standoff to BIO encoding or
due to different hyperparameters used in Habibi et al.
(2017) and Giorgi and Bader (2018) systems.
Also, we perform a two-tailed t-test with α val-
ues of 0.01 and 0.05. We consider a model signif-
icantly worse than ELMo (Pubmed) when p ≤ 0.01
(represented by ** in the tables) and if p ≤ 0.05 (rep-
resented by * in the tables). Figures 5, 6, 7 and 8 show
the boxplots for evaluation of corpora on three mod-
5
https://biosemantics.org/index.php/resources/chemical-
patent-corpus
6
https://github.com/spyysalo/standoff2conll
Figure 5: F1-score on BC5CDR for baseline (B), ELMo-
general (E-G) and ELMo-Pubmed (E-P).
Table 3: F1 score on BC4CHEMDNER. Best F1-score in
bold and significantly worse than our model **. First three
rows show our results which are averaged F1 ± SD over
five runs (random seeds). The rest of the results are reported
directly from the respective papers.
Methods F1- score
ELMo (Pubmed) 9
9
90
0
0.
.
.8
8
80
0
0 ±
±
± 0
0
0.
.
.1
1
11
1
1
ELMo (General) 88.36 ± 0.95
**
Baseline 88.75 ± 0.18
**
Habibi 86.62
**
LSTMVoter 90.02
**
Crichton MTL 82.51
**
Table 4: F1 score on CEMP, best F1-score in bold. First
three rows show our results which are averaged F1 ± SD
over five runs (random seeds). The rest of the results are
reported directly from the respective papers.
Methods F1- score
ELMo (Pubmed) 82.37 ±0.31
ELMo (General) 81.77 ± 0.29
Baseline 81.66 ± 0.05
Giorgi TL 86.05
Habibi 85.38
LSTMVoter 89.01
Chemlistem 90.33
els, ELMo (Pubmed), ELMo (General) and baseline.
ELMo (Pubmed) gives the highest score and the base-
line gives the lowest F1 score. These figures show
Chemical Named Entity Recognition with Deep Contextualized Neural Embeddings
141
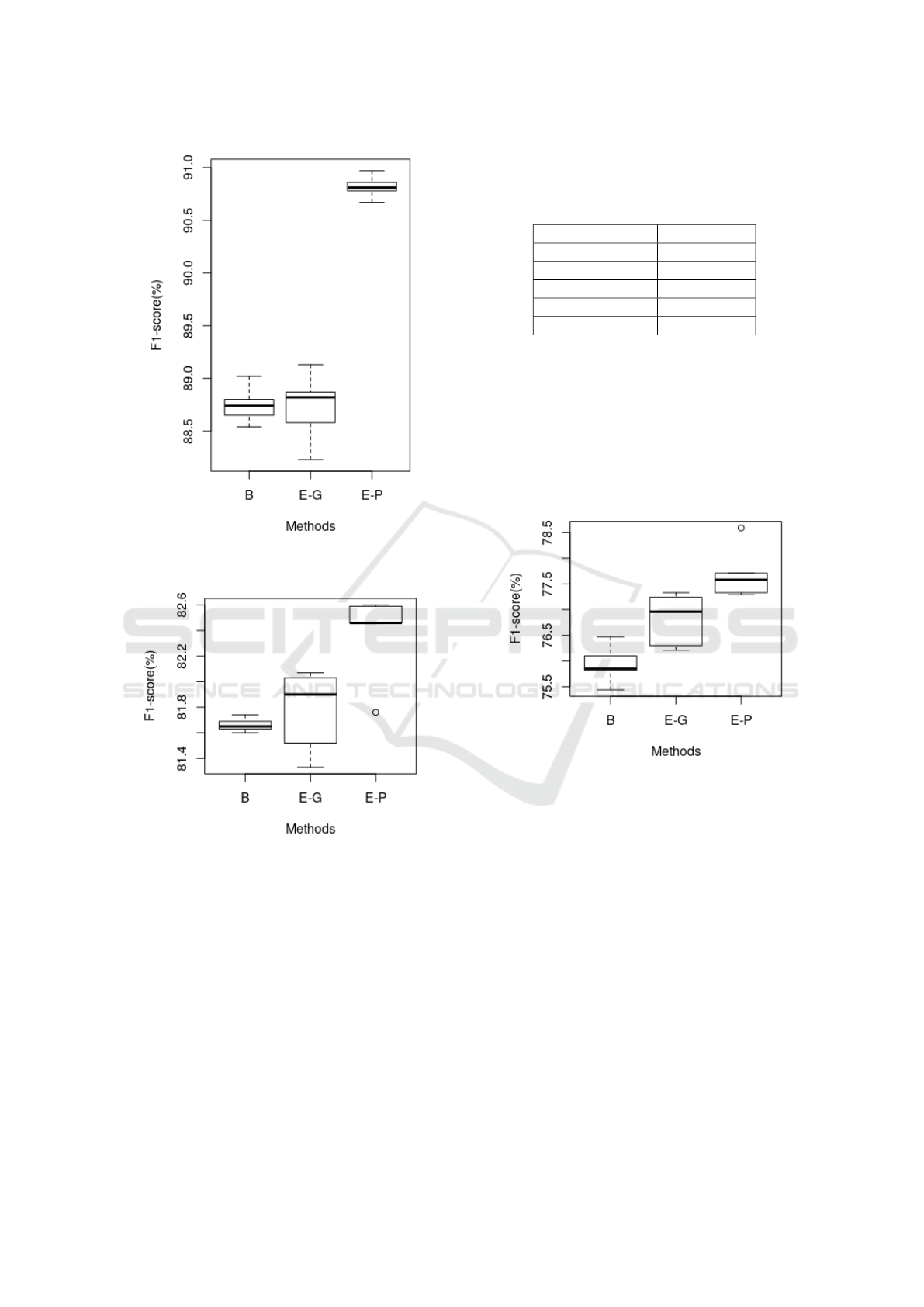
Figure 6: F1 score on BC4CHEMDNER for baseline (B),
ELMo-general (E-G) and ELMo-Pubmed (E-P).
Figure 7: F1 score on CEMP for baseline (B), ELMo-
general (E-G) and ELMo-Pubmed (E-P).
that ELMo (Pubmed) consistently outperforms base-
line and ELMo (General) on all corpora. The perfor-
mance reported is the averaged F1 score over five runs
for the test sets from the epoch that has the highest de-
velopment/validation score.
5 CONCLUSION
In the present study we show that incorporating
ELMo embeddings into the static embeddings for a
Bi-LSTM-CRF network results in a statistically sig-
Table 5: F1 score on Biosemantics, best F1-score in bold.
First three rows show our results which are averaged F1 ±
SD over five runs (random seeds). The rest of the results are
reported directly from the respective papers.
Methods F1- score
ELMo (Pubmed) 77.7 ± 0.47
ELMo (General) 76.80 ± 0.46
Baseline 76.09 ± 0.26
Giorgi TL 86.95
Habibi 81.99
nificant increase in F1 score for all the evaluated cor-
pora. Our results also show that ELMo pre-trained on
Pubmed results in better performance than ELMo pre-
trained on a general domain corpus. The higher per-
formance of ELMo (Pubmed) than ELMo (General)
also shows that transfer learning results in higher F1
score if the source dataset represents the domain of
the target dataset. We confirm our findings on four
benchmark ChemNER corpora.
Figure 8: F1 score on Biosemantics for baseline (B),
ELMo-general (E-G) and ELMo-Pubmed (E-P).
For future work, character-level language mod-
els such as Flair Embeddings (Akbik et al., 2018) or
BERT (Devlin et al., 2018) could be used for the rep-
resentation to see whether they complement or sub-
sume the ELMo representation. Another potential
area of research would be to improve the hyperpa-
rameter tuning using random search, grid search or
Bayesian optimisation methods. Lastly, cross-corpus
evaluation should be performed to measure the gen-
eralizability of the models. All the relevant methods
that we have compared do not evaluate on any exter-
nal corpora. Generalizability is a concern for us as
information extraction methods need to be deployed
at Pubmed scale, and the models should not overfit on
the data that they were trained on. Lastly, multi-task
learning and transfer learning techniques could also
be explored for this task.
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
142

REFERENCES
Akbik, A., Blythe, D., and Vollgraf, R. (2018). Contextual
string embeddings for sequence labeling. In Proceed-
ings of the 27th International Conference on Compu-
tational Linguistics, pages 1638–1649.
Akhondi, S. A., Klenner, A. G., Tyrchan, C., Manchala,
A. K., Boppana, K., Lowe, D., Zimmermann, M., Ja-
garlapudi, S. A., Sayle, R., Kors, J. A., et al. (2014).
Annotated chemical patent corpus: a gold standard for
text mining. PloS one, 9(9):e107477.
Bengio, Y., Simard, P., Frasconi, P., et al. (1994). Learn-
ing long-term dependencies with gradient descent is
difficult. IEEE Transactions on Neural Networks,
5(2):157–166.
Bergstra, J. S., Bardenet, R., Bengio, Y., and K
´
egl, B.
(2011). Algorithms for hyper-parameter optimization.
In Advances in Neural Information Processing Sys-
tems, pages 2546–2554.
Chiu, J. P. and Nichols, E. (2016). Named entity recogni-
tion with bidirectional LSTM-CNNs. Transactions of
the Association for Computational Linguistics, 4:357–
370.
Corbett, P. and Boyle, J. (2018). Chemlistem: chemical
named entity recognition using recurrent neural net-
works. Journal of Cheminformatics, 10(1):59.
Crichton, G., Pyysalo, S., Chiu, B., and Korhonen, A.
(2017). A neural network multi-task learning ap-
proach to biomedical named entity recognition. BMC
Bioinformatics, 18(1):368.
Dernoncourt, F., Lee, J. Y., and Szolovits, P. (2017).
NeuroNER: an easy-to-use program for named-entity
recognition based on neural networks. arXiv preprint
arXiv:1705.05487.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Eltyeb, S. and Salim, N. (2014). Chemical named entities
recognition: a review on approaches and applications.
Journal of Cheminformatics, 6(1):17.
Finkel, J. R., Grenager, T., and Manning, C. (2005). Incor-
porating non-local information into information ex-
traction systems by gibbs sampling. In Proceedings of
the 43rd annual meeting on Association for Compu-
tational Linguistics, pages 363–370. Association for
Computational Linguistics.
Gal, Y. and Ghahramani, Z. (2016). A theoretically
grounded application of dropout in recurrent neural
networks. In Advances in Neural Information Pro-
cessing Systems, pages 1019–1027.
Geyer, K., Greenfield, K., Mensch, A., and Simek, O.
(2016). Named Entity Recognition in 140 Characters
or Less. In # Microposts, pages 78–79.
Giorgi, J. M. and Bader, G. D. (2018). Transfer learning
for biomedical named entity recognition with neural
networks. Bioinformatics, 34(23):4087–4094.
Graves, A. and Schmidhuber, J. (2005). Framewise
phoneme classification with bidirectional LSTM and
other neural network architectures. Neural Networks,
18(5-6):602–610.
Habibi, M., Weber, L., Neves, M., Wiegandt, D. L., and
Leser, U. (2017). Deep learning with word embed-
dings improves biomedical named entity recognition.
Bioinformatics, 33(14):i37–i48.
Hemati, W. and Mehler, A. (2019). LSTMVoter: chemi-
cal named entity recognition using a conglomerate of
sequence labeling tools. Journal of Cheminformatics,
11(1):3.
Hochreiter, S. (1998). The vanishing gradient problem dur-
ing learning recurrent neural nets and problem solu-
tions. International Journal of Uncertainty, Fuzziness
and Knowledge-Based Systems, 6(2):107–116.
Hochreiter, S. and Schmidhuber, J. (1997). Long short-term
memory. Neural Computation, 9(8):1735–1780.
Huang, Z., Xu, W., and Yu, K. (2015). Bidirectional LSTM-
CRF models for sequence tagging. arXiv preprint
arXiv:1508.01991.
Jelier, R., Jenster, G., Dorssers, L. C., van der Eijk, C. C.,
van Mulligen, E. M., Mons, B., and Kors, J. A.
(2005). Co-occurrence based meta-analysis of scien-
tific texts: retrieving biological relationships between
genes. Bioinformatics, 21(9):2049–2058.
Khare, R., Leaman, R., and Lu, Z. (2014). Accessing
biomedical literature in the current information land-
scape. In Biomedical Literature Mining, pages 11–31.
Springer.
Kim, S., Yoon, J., Park, K.-M., and Rim, H.-C. (2005).
Two-phase biomedical named entity recognition using
a hybrid method. In International Conference on Nat-
ural Language Processing, pages 646–657. Springer.
Krallinger, M., Rabal, O., Leitner, F., Vazquez, M., Salgado,
D., Lu, Z., Leaman, R., Lu, Y., Ji, D., Lowe, D. M.,
et al. (2015). The CHEMDNER corpus of chemicals
and drugs and its annotation principles. Journal of
Cheminformatics, 7(1):S2.
Kudo, T. (2010). CRF++: Yet another CRF toolkit
(2005). Available under LGPL from the following
URL: http://crfpp. sourceforge. net.
Lafferty, J. D., McCallum, A., and Pereira, F. C. N. (2001).
Conditional Random Fields: Probabilistic Models for
Segmenting and Labeling Sequence Data. In Proceed-
ings of the Eighteenth International Conference on
Machine Learning, ICML ’01, pages 282–289, San
Francisco, CA, USA. Morgan Kaufmann Publishers
Inc.
Lample, G., Ballesteros, M., Subramanian, S., Kawakami,
K., and Dyer, C. (2016). Neural architectures
for named entity recognition. arXiv preprint
arXiv:1603.01360.
Li, J., Sun, Y., Johnson, R. J., Sciaky, D., Wei, C.-H., Lea-
man, R., Davis, A. P., Mattingly, C. J., Wiegers, T. C.,
and Lu, Z. (2016). BioCreative V CDR task cor-
pus: a resource for chemical disease relation extrac-
tion. Database, 2016:1–10.
Ma, X. and Hovy, E. (2016). End-to-end sequence labeling
via bi-directional LSTMS-CNNs-CRF. arXiv preprint
arXiv:1603.01354.
Chemical Named Entity Recognition with Deep Contextualized Neural Embeddings
143

Mikolov, T., Sutskever, I., Chen, K., Corrado, G. S., and
Dean, J. (2013). Distributed representations of words
and phrases and their compositionality. In Advances in
Neural Information Processing Systems, pages 3111–
3119.
Moen, S. and Ananiadou, T. S. S. (2013). Distributional se-
mantics resources for biomedical text processing. In
Proceedings of the 5th International Symposium on
Languages in Biology and Medicine, Tokyo, Japan,
pages 39–43.
M
¨
uller, T., Schmid, H., and Sch
¨
utze, H. (2013). Efficient
higher-order CRFs for morphological tagging. In Pro-
ceedings of the 2013 Conference on Empirical Meth-
ods in Natural Language Processing, pages 322–332.
Pennington, J., Socher, R., and Manning, C. (2014). Glove:
Global vectors for word representation. In Proceed-
ings of the 2014 Conference on Empirical Methods in
Natural Language Processing (EMNLP), pages 1532–
1543.
P
´
erez-P
´
erez, M., Rabal, O., P
´
erez-Rodr
´
ıguez, G., Vazquez,
M., Fdez-Riverola, F., Oyarzabal, J., Valencia, A.,
Lourenc¸o, A., and Krallinger, M. (2017). Evalua-
tion of chemical and gene/protein entity recognition
systems at BioCreative V. 5: the CEMP and GPRO
patents tracks. Proceedings of the BioCreative V.5
Challenge Evaluation Workshop, pages 11–18.
Peters, M. E., Neumann, M., Iyyer, M., Gardner, M.,
Clark, C., Lee, K., and Zettlemoyer, L. (2018). Deep
contextualized word representations. arXiv preprint
arXiv:1802.05365.
Rebholz-Schuhmann, D., Yepes, A. J. J., Van Mulligen,
E. M., Kang, N., Kors, J., Milward, D., Corbett, P.,
Buyko, E., Beisswanger, E., and Hahn, U. (2010).
CALBC silver standard corpus. Journal of Bioinfor-
matics and Computational Biology, 8(01):163–179.
Reimers, N. and Gurevych, I. (2017). Reporting Score Dis-
tributions Makes a Difference: Performance Study of
LSTM-networks for Sequence Tagging. In Proceed-
ings of the 2017 Conference on Empirical Methods in
Natural Language Processing (EMNLP), pages 338–
348, Copenhagen, Denmark.
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
144
