
Data Mining Techniques for Early Detection of Breast Cancer
Maria Inês Cruz
1
and Jorge Bernardino
1,2 a
1
Polytechnic of Coimbra – ISEC, Rua Pedro Nunes, Quinta da Nora, 3030-199 Coimbra, Portugal
2
CISUC – Centre of Informatics and Systems of University of Coimbra, Pinhal de Marrocos, 3030-290 Coimbra, Portugal
Keywords: Data Mining, Cancer, Breast Cancer, Biomarkers, Ensemble.
Abstract: Nowadays, millions of people around the world are living with the diagnosis of cancer, so it is very important
to investigate some forms of detection and prevention of this disease. In this paper, we will use an ensemble
technique with some data mining algorithms applied to a dataset related to the diagnosis of breast cancer using
biological markers found in routine blood tests, in order to diagnose this disease. From the results obtained,
it can be verified that the model got an AUC of 95% and a precision of 87%. Thus, through this model it is
possible to create new screening tools to assist doctors and prevent healthy patients from having to undergo
invasive examinations.
1 INTRODUCTION
Cancer is a disease where the cells of our body divide
without control due to the fact that they have
undergone mutations in their DNA, and because of
this, cells acquire properties during this division
process (CUF, 2017).
Today, millions of people around the world living
with the diagnosis of cancer. In Portugal, in 2018
were recorded about 58.199 new cases of cancer in
which about 28.960 of these cases don’t survive
(Global Cancer Observatory, 2018). The constant
investigation on this area is extremely necessary.
Some types of cancer can be detected before they
cause problems, and so it is very important to do
screening tests.
One of these types is the breast cancer, a cancer
that forms in tissues of the breast. The most common
type of breast cancer is ductal carcinoma, which
begins in the lining of the milk ducts (thin tubes that
carry milk from the lobules of the breast to the
nipple). Another type of this cancer is lobular
carcinoma, which begins in the lobules (milk glands)
of the breast. Invasive breast cancer is a cancer that
has spread from where it began to surround normal
tissue. Breast cancer can occur in both men and
women, although male breast cancer is rare. It is the
most common no cutaneous cancer in United States
women, with an estimated 62,930 cases of local
a
https://orcid.org/0000-0001-9660-2011
disease and 268,600 cases of invasive disease in
2019. Clinical trials have established that screening
asymptomatic women using mammography, with or
without clinical breast examination, decreases breast
cancer mortality (National Cancer Institute, n.d.).
The early detection of cancer is one of the most
efficient methods for the diagnosis of this disease.
“The cancer kills us because we give time to do it”
writes researcher Patrizia Paterlini-Bréchot in her
book “Kill the Cancer”. This researcher discovered a
blood test that allows visualizing the presence of
cancerous cells, of any type of cancer except
leukaemia and lymphomas, “often before the cancer
can be detected”. It further considers that to “kill the
cancer” it is necessary “extend the methods of early
detection” and that “very early diagnosis is the way
to save millions of lives” (Agência Lusa, 2018).
Therefore, the computational tools of data mining
become very important to analyse all of data that
coming of several medical exams. These can be used
in extracted data from blood tests, thus making an
important contribution to the experts, offering more
screening tools.
The purpose of this paper is to apply many
techniques of data mining to a dataset with some
features found in routine blood tests in order to
predict the presence of breast cancer.
In this paper will be made a univariate and
multivariate descriptive analysis for the data pre-
434
Cruz, M. and Bernardino, J.
Data Mining Techniques for Early Detection of Breast Cancer.
DOI: 10.5220/0008346504340441
In Proceedings of the 11th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2019), pages 434-441
ISBN: 978-989-758-382-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

processing. We will build a model based on ensemble
techniques and use the Stacking Ensemble learning
technique which will be explained in the next section.
The algorithms that we will use in our model are
Logistic Regression, Random Forest, Naive Bayes
and Support Vector Machine. To train and validate
the model will be used Validation Set and Cross-
Validation methods. The aim is to evaluate the
performance of the model in terms of accuracy,
precision, recall, false negatives rate and AUC (Area
Under the ROC Curve).
There are some studies regarding the application
of DM techniques to breast cancer diagnostic
datasets. In 2018, a study for create and analyse the
dataset that will be used in this paper was done
(Patrício et al., 2018).
In this study, a univariate analysis was elaborated
where each variable was evaluated as to normality
using some normalization tests. In the end was using
the ROC curve to evaluate each parameter. In
multivariate analysis the Gini coefficient was used,
on average, in all trees of a Random Forest. The
predictive models used logistic regression, support
vector machines and random forest algorithms. The
Monte Carlo Cross-Validation was adopted in the
training set and the models was evaluated in relation
to AUC, specificity and recall. The SVM using
Glucoses, Resistin, Age and IMC as predictors got a
recall between 82% and 88% and a specificity
between 85% and 90%. The confidence interval of
95% to the AUC was [0.87;0.91].
This paper is organized as follows. In section 2 are
introduced some fundamental concepts. In section 3
the dataset is explored in order to understand the data.
In section 4 the model is created and analysed. In
section 5 the results are discussed and evaluated
according to the metrics. Finally, section 6 presents
the conclusions and some ideas for future work.
2 FUNDAMENTAL CONCEPTS
This section describes some of the fundamental
theoretical concepts to understand the study that will
be performed. We explain the data mining concept, as
well as the various steps of this process.
2.1 Data Mining
Data Mining can be considered as the synonymous of
the term Knowledge Discovery from Data, or KDD,
or as merely an essential step in knowledge discovery
process. This process of discovery is a sequence of
the following steps:
Data Cleaning, to remove the noise and
inconclusive data;
Data Integration, where many data sources can
be integrated;
Data Selection, where the relevant data for the
analysis are extracted from data base;
Data Transformation, where the data are
transformed and consolidated properly to make
the analysis performing summary and aggregation
operations.
Data Mining, the essential process where is used
methods to extract patterns and correlations of the
data;
Patterns Evaluation, to identify the real interest
of the patterns that represent knowledge based on
“interest” metrics;
Knowledge Presentation, where techniques of
visualization and representation of knowledge are
used to present knowledge to users.
This approach shows data mining as a step of the
knowledge discovery process, although essential
because it reveals patterns that are hidden for
evaluation. However, in industry and investigation
the term is frequently used to define all process of
knowledge discovery (Borges, Marques, and
Bernardino, 2013). Therefore, a broad view of data
mining was adopted as the process of discovering
interesting patterns and knowledge from large
amounts of data.
2.2 Data Pre-processing
Typically, the daily data is redundant and
inconsistent, also containing missing values. On the
other hand, there is also the problem of having a large
amount of data or, conversely, a small amount of data.
In order to perform a good analysis of the data, it
is necessary to prepare the data. This process involves
a more in-depth analysis of the attributes and values
of the data.
The starting point for this pre-processing will be
to obtain a statistical description of the data,
identifying its attributes and performing a univariate
and multivariate analysis.
Univariate descriptive analysis involves
describing the central tendency and dispersion of an
attribute. Some measures of the central tendency are
the mean (average number of all values), mode (most
frequent value) and median (number that is in the
middle of the list). The dispersion can be measured
by variance or standard deviation, range of values
(minimum and maximum value), percentiles,
quartiles, and the five-number summary (it involves
Data Mining Techniques for Early Detection of Breast Cancer
435

the minimum, the first quartile, the median, the third
quartile and the maximum).
Multivariate descriptive analysis involves
analysing the correlation between attribute pairs
through scatter plots. After these analyses are
followed the cleaning, transformation and reduction
of the data. This stage is where the outliers and
missing values (attribute values that are missing in
some examples) are treated.
Having thus the data already pre-processed and
prepared for analysis it is possible to move to the
stage of construction of the data mining models.
2.3 Types of Learning
At the stage of model construction, the purpose of the
analysis is to learn to recognize complex patterns and
make intelligent and data-driven decisions. There are
then two types of learning (Kaufmann, Han and
Kamber, 2006): supervised and unsupervised
learning. In this case study, the dataset examples have
an attribute that classifies them, whether the patient
has cancer or not. So, this study will focus on
supervised learning.
In Supervised Learning we find Classification
problems, where the output variable is qualitative (a
class, category or diagnosis), such as the prediction of
a person having or not having a particular disease.
For this type of learning, in the construction of the
model it is necessary to have a training to teach our
method to estimate the model using the available data
examples (Kaufmann, Han and Kamber, 2006). This
training is performed using a learning algorithm, in
this case study will be used Logistic Regression,
Random Forest, Naive Bayes and Support Vector
Machine.
It is then necessary to evaluate the quality of the
model created (Kaufmann, Han and Kamber, 2006),
that is, if the estimate corresponds to the observations.
The goal is for the method to obtain generalization
capability, that is, to be precise in situations that are
not found in the training and not to memorize the
examples. For this evaluation a test set is created with
different examples from those used for training. If
there are examples for testing available, these
examples are used to evaluate the model, if no
examples are available the training set is divided into
two parts, training with one and testing with the other.
For this approach there are two methods of validation:
Validation Set: this method divides 70% of the
data for training and 30% of the data for test;
Cross-validation: there are two techniques for
this method, one of which is k-fold Cross-
validation in which the initial data are randomly
divided into k exclusive subsets, each
approximately of the same size. The training and
the test are done k times. In the first iteration,
subset 1 is used for testing and the rest for
training, and so on. Another technique is the
Leave-one-out Cross-validation which is a
special case of k-fold cross-validation where one
example is taken at a time for testing and the rest
are used for training.
2.4 Ensemble Methods
Ensemble methods is a data mining technique that
combines several base models in order to produce one
optical predictive model. These methods can be
divided in two groups:
Sequential: where the base learners are generated
sequentially (e.g. AdaBoost). The motivation of
these methods is to exploit the dependence
between the base learners. The overall
performance can be boosted by weighing
previously mislabelled examples with higher
weight;
Parallel: where the base learners are generated in
parallel (e.g. Random Forest). The motivation of
these methods is to exploit independence between
the base learners since the error can be reduced
significantly by averaging.
There are three ways for using ensemble methods,
that are bagging, boosting and stacking. In this study
the stacking method described below is used.
2.4.1 Stacking
Stacking is an ensemble learning technique that
combines multiple classification or regression models
via a meta-classifier or a meta-regressor. The base
level models are trained based on a complete training
set, then the meta-model is trained on the outputs of
the base level model as features (Smolyakov, 2017).
In this study we will use the Stacking method with
Random Forest, Naive Bayes and Logistic
Regression as base algorithms and the Support Vector
Machine as meta-classifier.
2.5 Learning Algorithms
In this section we briefly describe how the learning
algorithms used for this study works.
2.5.1 Bayesian Algorithms (Naive Bayes)
Bayesian algorithms follow probabilistic approaches
that create strong assumptions about how data is
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
436

generated and construct a probabilistic model that
incorporates these assumptions. They use a set of
classified training examples to estimate the model
parameters. Classification in the new examples is
done with the Bayes rule by selecting the class that is
most likely to have generated that example
(McCallum and Nigam, 1998).
The Naive Bayes is a probabilistic algorithm
based on Bayes’ theorem and is the simplest classifier
of these algorithms since it is assumed that all
attributes are independent given the class context.
Although this assumption is false in most real-world
data, this classifier performs well most of the time.
Thanks to this assumption, the parameters for each
attribute can be learned separately, and thus there is a
simplification of learning, especially with many
attributes.
This method works with several probabilities for
each class. These probabilities are reflected in the
conditioned probability of each value of the attribute
given to the class, as well as the probability of the
class (Langley, Iba, and Thompson, 1992).
2.5.2 Random Forest
Random Forest is a supervised learning algorithm,
and as the name implies it creates a forest and makes
it somehow random. The “forest” is an ensemble of
Decision Trees (Loh and Shin, 1997), most of the
time trained with the “bagging” method. The general
idea of this method is that a combination of learning
models increases the overall result. In a simple way,
this algorithm builds multiple decision trees and
merges them together to get a more accurate and
stable prediction.
This method adds additional randomness to the
model, while growing trees. Instead of searching for
the most import feature while splitting a node, it
searches for the best feature among a random subset
of features. This results in a wide diversity that
generally results in a better model.
This algorithm is a collection of Decision Trees
but exist some differences. If we input a training
dataset with features and labels into a decision tree, it
will formulate some rules, which will be used to make
the predictions. In comparison, the Random Forest
randomly selects observations and features to build
several decision trees and then averages the results.
One of the vantages of this algorithm is that it
prevents overfitting (in a simple way, it is when a
model learns too much noise) (Technopedia, n.d.)
most of the time, by creating random subsets of the
features and building smaller trees using them.
Afterwards, it combines the subtrees. With decision
trees, the more we increase the depth of the tree the
more likely there is to be overfitting (Donges, 2018).
2.5.3 Logistic Regression
Logistic regression is used in classification problems
in which the attributes are numerical, it is an
adaptation of linear regression methods. Considering
a dataset where the target is a binary categorical
variable, the value of 0 and 1 is given to each of the
categories respectively, and instead of the regression
executing the response directly, it executes the
probability that the response belongs to a category (0
or 1).
If the model is done following the linear
regression approach, the attributes that have values
close to zero will have a negative probability and if
they have very high values the probability will exceed
the value 1 (James, Witten, Hastie, and Tibshirani,
2013). These predictions are not correct because a
true probability, regardless of the value of the
attribute, must be between 0 and 1. Whenever a
straight line is fitted to a binary response that is coded
as 0 or 1, it always be possible to predict p(X) < 0 and
p(X) > 1 at the outset (unless the X range is limited).
To avoid this problem, one must make the
probability model using a function that provides
outputs between 0 and 1 for all values of X.
2.5.4 Support Vector Machine
The objective of the support vector machine
algorithm is to find a hyperplane (decision boundaries
that help classify the data points) in an N-dimensional
space, when the N is the number of features, that
distinctly classifies the data points. To separate the
two classes of data points, there are many possible
hyperplanes that could be chosen. The main objective
is to find a plane that has the maximum distance
between data points of both classes. Therefore, it is
possible to provide some reinforcement so that future
data points can be classified with more confidence.
Support Vectors are data points that are closer to
the hyperplane and influence the position and
orientation of the hyperplane, using this support
vectors it is possible to maximize the margin of the
classifier. Hyperplanes and support vectors are the
core for building an SVM algorithm (Gandhi, 2018).
2.6 Evaluation Metrics
Finally, it is necessary to evaluate the performance of
the model created. For this, there are several
evaluation metrics (
Sunasra, 2017):
Data Mining Techniques for Early Detection of Breast Cancer
437

Accuracy: is the degree of proximity of an
amount with the true value of that quantity, that is,
the model hit rate, the number of times the model
hit the forecasts;
Precision: is the degree to which repeated
measurements under unchanged conditions show
the same results, that is, the generalization ability
of the model;
Recall: is the rate of values that the model
predicted positive and it is positive in dataset;
Specificity: is the rate of values that the model
predicted negative and it is negative in dataset;
False Negatives Rate: is the rate of values that
the model missed, classifying as negatives the
positive values.
Another metric of evaluation is the ROC curve, which
consists of the graphical representation of the pairs,
recall and specificity in all limits of classification
(thresholds) (Google Developers, 2019). This curve
allows you to achieve the AUC (Area Under the
ROC Curve) measurement that measures the entire
area under the ROC curve. The higher the AUC the
better is the model used as it is performing the
predictions correctly. The AUC ranges from 0 to 1. If
a model obtains 100% of missed predictions, will
have an AUC of 0 and vice versa.
3 DATA EXPLORATION
The dataset used for the analysis is called Breast
Cancer Coimbra Dataset and can be consulted
publicly in (Machine Learning Repository, 2018).
This dataset was used for a study at the University of
Coimbra with the objective of constructing a
predictive model that could potentially be used as a
bio marker for breast cancer.
It was created in May of 2018 and contains 10
quantitative attributes and one categorical variable
which indicates the presence or not of breast cancer.
The attributes are anthropometric data and parameters
that can be collected in routine blood tests.
Were collected data of 64 sick women and 52
healthy women. So, the dataset contains 116
examples. The patient data were collected before the
surgery and the treatments (Patrício et al., 2018).
The categorical variable indicates the values 1 and
2, that corresponding, respectively to women healthy
and sick. The dataset is complete, not containing
missing values.
A description of the dataset is given below
(Patrício et al., 2018) (Frazão, 2018):
Age: Age of the patient (24 to 89);
BMI: Body Mass Index (18,37 to 38,58 kg/m
);
Glucose: Quantity of sugar in blood (60 to 201
mg/dL);
Insulin: Hormone produced by pancreas to
reduce the rate of glucose in blood (2,432 to 58,46
U/mL);
HOMA: Homeostatic Model Assessment, is a
method used to quantify the insulin resistance
(Lemos, 2018) (0,467 to 25,05);
Leptin: Protein responsible for the control of food
ingested, send information to the brain (Gunnars,
2018) (4,3 to 90,3 ng/mL);
Adiponectin: Protein responsible for the
regulation of the glucose in blood (1,66 to 38,04
ng/mL);
Resistin: Protein responsible for block the
principal action of the leptin (3,21 to 82,1 ng/mL);
MCP-1: Monocyte Chemoattractant Protein 1,
recruit monocytes and specific cells to spots of
inflammation.
A univariate analysis was performed where the values
of the mean, the standard deviation and the five-
number summary for each attribute were calculated.
The Excel tool was used to perform these
calculations.
Through these values it is possible to create
boxplots. The Orange tool was used to create and
display them. There is greater dispersion of data in the
Age, BMI and Leptin attributes. It is possible to
conclude this by comparing attributes.
Figure 1: Boxplot of attribute Age.
Figure 2: Boxplot of attribute Glucose.
For example, Figure 1 and Figure 2 represent the
boxplots relative to attributes Age and Glucose,
respectively. Note that the interquartile range is
smaller in the Glucose attribute, so the values of this
attribute are mostly close to the mean value.
Therefore, it is concluded that the values of this
attribute are less dispersed compared to age.
In attributes with greater dispersion it becomes
more difficult to find patterns in the data, whereas in
the less dispersed the patterns are found more easily.
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
438

Using the Information Gain method, it is verified
that the attributes most relevant for classification, that
is, for the division of classes are Glucose, HOMA,
Resistin and Insulin.
It is also possible to check the existence of outliers
in all attributes except the Age and BMI attributes by
calculating the upper and lower admissible limits. For
the remaining attributes, it is important to have the
outliers in consideration, since being a medical
dataset, values “out of ordinary” may indicate
important information. It is verified that most of these
values classify diseased patients, which may indicate
that the values have arisen naturally and are important
for the analysis, since they can be a factor of
differentiation in the classification of the problem.
Thus, the same previous analysis was made, but
replacing the outliers with the permissible upper
limit, which showed a single difference in the
dispersion of the data, in which the data became more
dispersed than with the original values. This means
that the outliers do not have great relevance to the
classification of the dataset so they will be kept in the
learning models. We found in Figure 3 that the values
are more dispersed than in Figure 2, with the size
between quartiles increased, which means that the
values are farthest from each other.
Figure 3: Boxplot of attribute Glucose without outliers.
Moving to a multivariate analysis, and through the
visualization of scatter plots, a single correlation
between the HOMA and Insulin attributes is verified,
which is natural since HOMA is a method that
calculates insulin resistance. This correlation is
perceptible because the values form a diagonal line.
The fact that there are two correlated attributes can
mean that it is indifferent whether one exists or not,
since both transmit the same information, and thus
will not interfere in the learning of the model. We
decide to make a prior analysis to verify if the HOMA
attribute when taken from the dataset had a
significance influence on the results and it was
verified that the results did not suffer significant
differences so we will keep all the attributes for the
learning of the model.
4 CONSTRUCTION OF THE
MODEL
In a scenario made previously to this same dataset,
three classification algorithms were analysed:
Decision Tree, Logistic Regression and Naive Bayes.
In this analysis there were no good results, and the
maximum AUC achieved was with logistic regression
with a value of 79% and an accuracy of 74%. The
results are illustrated in Table 1.
Table 1: Results of the individual classifiers.
Algorithm Accuracy Precision Recall AUC Specificity FNR
Logistic
Regression
0.74 0.74 0.73 0.79 0.64 0.28
Decision
Tree
0.72 0.72 0.71 0.73 0.63 0.32
Naive Bayes 0.68 0.68 0.68 0.74 0.58 0.33
Random
Forest
0.66 0.66 0.66 0.70 0.66 0.30
After this, we decided to try to improve these
results with the model proposed below, using
Ensemble methods.
For our model we will then use the ensemble
stacking method, with Logistic Regression, Random
Forest and Naive Bayes as base models and Support
Vector Machine as final meta-classifier. We used the
Orange tool to evaluate the model. The parameters
used in each algorithm were as follows: in Random
Forest 10 trees are created, with 5 attributes at each
split; in Logistic Regression the Tikhonov
regularization was used (Kringstad, 2019) with a cost
strength of 3; in Naive Bayes has no parameters to
adjust; in SVM the cost (penalty term for loss) of the
minimization of the error function is 1, the kernel
function (is a function that transforms attribute space
to a new feature space to fit the maximum-margin
hyperplane) used was polynomial and the permitted
deviation from the expected value was 0,001 and the
limit iterations was 100.
As validation methods are used, first the
Validation Set and then the Cross-Validation, in order
to analyse the differences of the model between both
methods. In the Validation Set on the base models,
the train/test was repeated ten times and in the meta-
classifier was repeated two times.
In the Table 1 it is possible to visualize the values
of precision, recall, accuracy, AUC and false
negatives rate (FNR on Table 2) for the Validation Set
(70-30 on Table 2) method, with 70% of the dataset
Data Mining Techniques for Early Detection of Breast Cancer
439
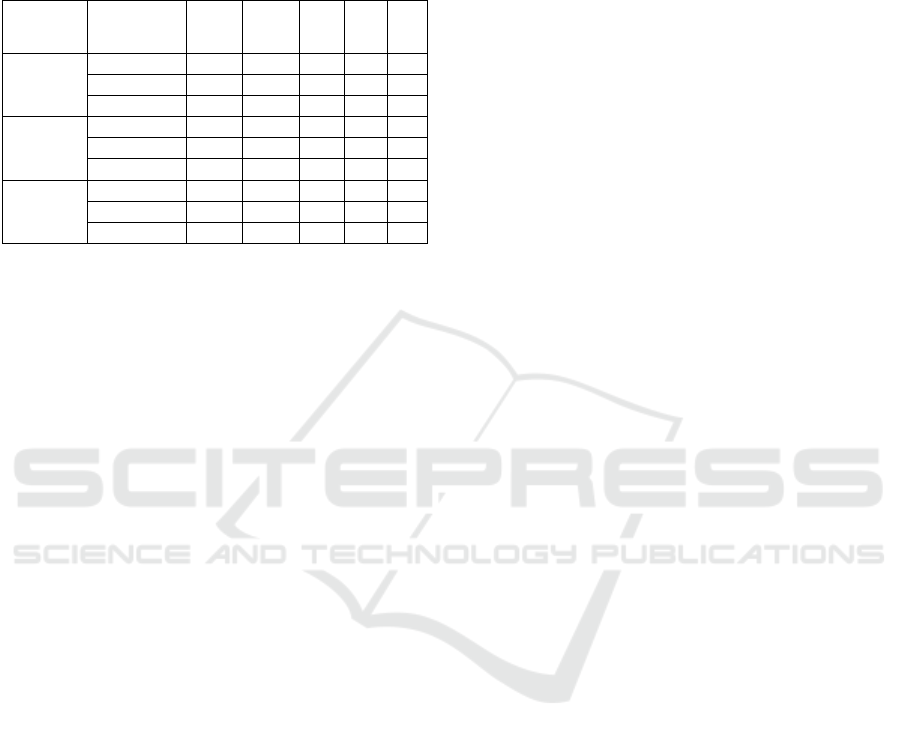
for training and 30% for test, Cross-Validation k-
folds (CV1 in Table 2) and Cross Validation Leave-
one-out (CV2 in Table 2) in base models and meta-
classifier.
Table 2: Results of the model.
Validation
Methods in
Base models
Validation
method in
Meta-Classifier
Accuracy Precision Recall AUC FNR
70-30
70-30 0.86 0.87 0.86 0.95 0.14
CV1 0.73 0.79 0.73 0.96 0.27
CV2 0.62 0.71 0.62 0.84 0.38
CV1
70-30 0.77 0.78 0.76 0.87 0.24
CV1 0.73 0.80 0.73 0.86 0.27
CV2 0.71 0.77 0.71 0.83 0.29
CV2
70-30 0.64 0.79 0.64 0.81 0.36
CV1 0.73 0.80 0.73 0.86 0.27
CV2 0.71 0.77 0.71 0.83 0.29
5 RESULTS DISCUSSION AND
EVALUATION
It is possible to verify through the results that this
model obtains better results than the model made in
the previous study, as would be expected.
We can say that the model presented good results
because all the AUC values are superior to 80%. A
significant difference can be noted when using the
Validation Set method in base models and in meta-
classifier. Thus, achieving an AUC of 95%, a
precision 87% and an FNR of 14%.
In our view, the most important metrics in medical
studies are the ability of the model to adapt to new
cases, that is, the generalization capacity of the model
(precision) and especially the false negative rate,
since the worst case scenario can happen is to
diagnose the person as being healthy (negative
diagnosis) and in fact the person having the disease
(positive diagnosis). It is also very important to have
a good AUC value as it means that the model made
most of the prediction correctly.
Good results with 80% precision and 86% AUC
are also found using Cross-Validation k-folds method
in all algorithms.
Worst values are displayed when we use the
Cross-Validation Leave-one-out in base models and
the Validation Set in meta-classifier.
It is verified through the data exploration of data
that the most relevant features for the distinction
between the classes are Age, BMI, Leptin, Resistin
and Adiponectin.
Due to the small dataset, the results are not very
reliable, so it was interesting made a research with
more subjects to test the model.
6 CONCLUSIONS
The first conclusions were withdrawn in the pre-
processing phase of this study through the
visualization of a decision tree and are that all the
subjects of this dataset aged less than or equal to 74,
Leptin values less than or equal to 31.12, Resistin
higher than 13, Adiponectin higher than 2.2, and BMI
lower than 32 have the disease. There are 26 patients
with these conditions, which makes up 41% of the
patients in the dataset.
With this study it was also possible to conclude
that the ensemble methods significantly improve the
models. In our specific case, using the staking method
it was concluded that the more times we train the base
algorithms the better are the results.
On the other hand, the fact that the best validation
method is the validation set means that many times
(randomly) the examples used for testing are used in
the training, which implies a greater accuracy in the
predictions, but not because the model learned the
results but memorized them.
Despite this, using Cross-Validation k-folds in the
base models and meta-classifier also obtains good
results, so it can be concluded that the model
generally shows good results.
This paper can help other investigators to create
an effective predictive model for detecting cancer
through blood routine exams, before the treatment
becomes more complex and the total elimination of
cancer is more difficult to achieve.
For future work, we intend to improve the results
by testing other algorithms and other ensemble
methods. It was interesting to get new features related
to routine blood test and increase the dataset should
be considered in order to get more efficient results.
REFERENCES
CUF. (2017). O que é o cancro? Retrieved from
https://www.saudecuf.pt/oncologia/o-cancro/o-que-e-
o-cancro/.
Global Cancer Observatory. (2018) International Agency
for Research on Cancer. Retrieved from
https://gco.iarc.fr/today/data/factsheets/populations/62
0-portugal-fact-sheets.pdf/.
National Cancer Institute. (n.d.) NCI Dictionary of Cancer
Terms. Retrieved from https://www.cancer.gov/
publications/dictionaries/cancca-terms/search?contains
=false&q=breast+cancer/.
National Cancer Institute. (n.d.) Breast Cancer Treatment
(PDQ)- Health Professional Version. Retrieved from
https://www.cancer.gov/types/breast/hp/breast-treatme
nt-pqd#_551_toc/.
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
440

Agência Lusa. (2018, April 21). Médica defende teste de
sangue de rotina para detetar células tumorais. [News
Post]. Retrieved from https://www.publico.pt/
2018/04/21/sociedade/noticia/medica-defende-teste-de
-sangue-de-rotina-para-detectar-celulas-tumorais-1811
211/.
Borges, L.C., Marques, V.M. and Bernardino, J. (2013).
Comparison of data mining techniques and tools for
data classification. Proceedings of the International C*
Conference on Computer Science and Software
Engineering (C3S2E’13). ACM, USA, 113-116.
Wikipedia. (2019, March 17). Amplitude Interquartil.
Retrieved from https://pt.wikipedia.org/wiki/
Amplitude_interquartil.
Kaufmann, M., Han, J. and Kamber, M. (2006). General
Approach to Classification. Data Mining: Concepts and
Techniques, 8, 328-330.
Kaufmann, M., Han, J. and Kamber, M. (2006). Model
Evaluation and Selection. Data Mining: Concepts and
Techniques, 8, 364-370.
Smolyakov, V. (2017, August 22). Ensemble Learning to
Improve Machine Learning Results. [Blog Post].
Retrieved from https://blog.statsbot.co/ensemble-
learning-d1dcd548e936/.
McCallum, A., Nigam, K. (1998). A comparison of event
models for Naive Bayes text classification. AAAI-98
Work. on Learning for Text Categorization,752, 41-48.
Langley, P., Iba, W. and Thompson, K. (1992). An analysis
of Bayesian classifiers.
Loh, W., Shin, Y. (1997). Split Selection Methods for
Classification Trees. Statistica Sinica, 815-840.
Technopedia. (n.d.). Overfitting. Retrieved from
https://www.technopedia.com/definition/32512/overfit
ting/.
Donges, N. (2018, February 22). The Random Forest
Algorithm. [Blog Post]. Retrieved from
https://towardsdatascience.com/the-random-forest-
algorithm-d457d499ffcd/.
James, G., Witten, D., Hastie, T. and Tibshirani, R. (2013).
Logistic Regression. An Introduction to Statistical
Learning, 4.3.
Gandhi, R. (2018, June 7). Support Vector Machine-
Introduction to Machine Learning Algorithms. [Blog
Post] Retrieved from https://towardsdatascience.com/
support-vector-machine-introduction-to-machine-lear
ning-algorithms-934a444fca47/.
Sunasra, M. (2017). Performance Metrics for
Classification problems in Machine Learning. Retrieved
from https://medium.com/thalus-ai/performance-metri
cs-for-classification-problems-in-machine-learning-par
t-i-b085d432082b.
Google Developers. (2019). Classification: ROC and AUC.
Retrieved from https://developers.google.com/
machine-learnig/crash-course/classification/roc-and-au
c/.
Machine Learning Repository. (2018). Breast Cancer
Coimbra Data Set. Retrieved from
https://archive.ics.uci.edu/ml/Breast+Cancer+Coimbra
#/.
Patrício, M., Pereira, J., Crisóstomo, J., Matafome, P.,
Gomes, M., Seiça, R., Caramelo, F. (2018). Using
resistin, glucose, age and BMI to predict the presence
of breast cancer. BMC Cancer, 18(1), 1-8.
Frazão, A. (2018). Exame da Glicose: como é feito e valores
de referência. Retrieved from https://www.
tuasaude.com/exame-da-glicose/.
Lemos, M. (2018). Para que serve o índice HOMA.
Retrieved from https://www.tuasaude.com/para-que-
serve-o-indice-homa/.
Gunnars, K. (2018). Leptin and Leptin Resistance:
Everything You Need To Know. Retrieved from
https://www.healthline.com/nutrition/leptin-101/.
Kringstad, A. (2019). Tikhonov regularization. Beyond L2.
Retrieved from https://towardsdatascience.com/
tikhonov-regularization-an-example-other-than-l2-892
2ba512/.
Data Mining Techniques for Early Detection of Breast Cancer
441
