
FoodOntoMap: Linking Food Concepts across Different Food Ontologies
Gorjan Popovski
1,2 a
, Barbara Korou
ˇ
si
´
c Seljak
3 b
and Tome Eftimov
3,4,5 c
1
Faculty of Computer Science and Engineering, Ss. Cyril and Methodius University, 1000 Skopje, North Macedonia
2
Jo
ˇ
zef Stefan International Postgraduate School, 1000 Ljubljana, Slovenia
3
Computer Systems Department, Jo
ˇ
zef Stefan Institute, 1000 Ljubljana, Slovenia
4
Center for Population Health Sciences, Stanford University, 94305 California, U.S.A.
5
Department of Biomedical Data Science, Stanford University, 94305 California, U.S.A.
Keywords:
Food Data Normalization, Food Data Linking, Food Ontology, Food Semantics.
Abstract:
In the last decade, a great amount of work has been done in predictive modelling in healthcare. All this work is
made possible by the existence of several available biomedical vocabularies and standards, which play a crucial
role in understanding health information. Moreover, there are available systems, such as the Unified Medical
Language System, that bring and link together all these biomedical vocabularies to enable interoperability
between computer systems. However, in 2019, Lancet Planetary Health published that the year 2019 is going
to be the year of nutrition, where the focus will be on the links between food systems, human health, and the
environment. While there is a large number of available resources for the biomedical domain, only a limited
number of resources can be utilized in the food domain. There is still no annotated corpus with food concepts,
and there are only a few rule-based food named-entity recognition systems for food concepts extraction. There
are also several food ontologies that exist, each developed for a specific application scenario. However there
are no links between these ontologies. For this reason, we have created a FoodOntoMap resource that consists
of food concepts extracted from recipes. For each food concept, semantic tags from four food ontologies are
assigned. With this, we have created a resource that provides a link between different food ontologies that can
be further reused to develop applications for understanding the relation between food systems, human health,
and the environment.
1 INTRODUCTION AND
MOTIVATION
Nowadays, the use of predictive modeling in health-
care increases with the large amount of data that is
becoming available. One example of such data are
the electronic health records (EHRs) (Gligic et al.,
2019; Wang et al., 2019), which represent the largest
source of medical data. Analyzing them, the medi-
cal information is presented as natural language text
(i.e. unstructured data) and the key challenge is to ex-
tract terms that are different medical concepts (e.g.,
drugs, diseases, procedures, treatments, etc.). For
this reason, a lot of named-entity recognition meth-
ods (NERs) have been developed (Boag et al., 2015),
which are further used to extract this information for
a
https://orcid.org/0000-0001-9091-4735
b
https://orcid.org/0000-0001-7597-2590
c
https://orcid.org/0000-0001-7597-2590
each patient and then trying to find a patient’s rep-
resentation for some predictive study. Besides the
unstructured data, there are also resources that con-
sists of structured patient medical information. One
such example is the MIMIC-III data (Johnson et al.,
2016), which consists of data relating to patients who
stayed within the intensive care units at Beth Israel
Deaconess Medical Center. The common thing about
the medical data, no matter from where it comes
(unstructured or structured data), is that it is further
used to find a patient’s representation by projecting
the data into a continuous vector space (Beam et al.,
2018; Choi et al., 2016; Miotto et al., 2016). In
this way, medical embeddings are learned in order
to capture the non-linear relationships that exist be-
tween the medical concepts. These representations
are further used with some advanced machine learn-
ing or deep learning methods to perform predictive
studies in healthcare. However, all this happens as
a result of the availability of biomedical vocabularies
Popovski, G., Seljak, B. and Eftimov, T.
FoodOntoMap: Linking Food Concepts across Different Food Ontologies.
DOI: 10.5220/0008353201950202
In Proceedings of the 11th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2019), pages 195-202
ISBN: 978-989-758-382-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
195

and standards that can be used to normalize the med-
ical concepts before learning the embedding space.
One such example is the Unified Medical Language
System (UMLS) that brings and links together several
biomedical vocabularies to enable interoperability be-
tween computer systems (Bodenreider, 2004).
However, in 2019, the Lancet Planetary Health
published that the year 2019 is going to be the year
of nutrition, where the focus will be on the links be-
tween food systems, human health, and the environ-
ment. Contrary to the large number of available re-
sources for the biomedical domain, in the food do-
main there is a limited number of resources that can
be used. There is still no annotated corpus with food
concepts, and there are few rule-based food named-
entity recognition systems that can be used for food
concepts extraction (Eftimov et al., 2017b; Popovski
et al., 2019). Additionally, a number of food ontolo-
gies exist, each developed for a specific application
scenario, but there are no links between them (Bou-
los et al., 2015). In order to move ahead the work for
finding relationships between the human health, food
systems and environment, we should have resources
that will be used for food concepts normalization. For
this reason, we have created a FoodOntoMap resource
that consists of food concepts extracted from recipes
and for each one the semantic tags from four food on-
tologies are assigned. With this, we have created a
resource that provides a link between different food
ontologies that can be further reused to develop em-
bedding space for food concepts and applications for
understanding the relation between food system, hu-
man health, and the environment.
2 RELATED WORK
In this section we provide an overview of the avail-
able food semantic resources that can be used for food
data normalization, followed by the methodologies
that can be used for food information extraction and
normalization.
2.1 Food Semantic Resources
In the domain of food, several food ontologies already
exist, such as:
FoodWiki provides a model of different types of
foods, together with their nutritional information, and
the re-commended daily intake (C¸ elik, 2015).
AGROVOC is a large multilingual thesaurus,
whose terminology is widely used in practice for sub-
ject fields in agriculture,fisheries, forestry, food and
related domains (Caracciolo et al., 2012).
Open Food Facts is an open source global food
database that allows users to learn about a food’s
nutritional information and compare products from
around the world (Boulos et al., 2015). It is also ben-
eficial for the food industry, where it can be used to
track, monitor, and strategically plan food production.
Food Product Ontology describes food products
using common representation, vocabulary and lan-
guage for the food product domain. It is an extended
version of a widely used standardized ontology for
product, price, store, and company data (Kolchin and
Zamula, 2013).
FOODS (Diabetics Edition) is an ontology-driven
system that delivers a web-based food-menu recom-
mendation system for patients with diabetes in Thai-
land (Snae and Br
¨
uckner, 2008).
FoodOn focuses on the human-centric categoriza-
tion and handling of food (Griffiths et al., 2016). Its
main goal is to develop semantics for food safety,
food security, agricultural and animal husbandry prac-
tices linked to food production, culinary, nutritional
and chemical ingredients and processes. It uses parts
from several ontologies covering anatomy, taxonomy,
geography and cultural heritage. Its usage is related
to research and clinical data sets in academia and gov-
ernment.
A detailed review of the aforementioned food on-
tologies was provided by Boulos et. al. (Boulos et al.,
2015).
OntoFood is an ontology with SWRL rules of nu-
trition for diabetic patients and is available in the Bio-
Portal.
SNOMED CT (Systematized Nomenclature of
Medicine - Clinical Terms) is a standardized, multi-
lingual vocabulary of clinical terminology that is used
by physicians and other health care providers for the
electronic health records (Donnelly, 2006). Beside
the medical concepts that are the main focus of this
ontology, there is also a Food concept that can be fur-
ther used for food concept normalization.
The Hansard corpus is a collection of text and
concepts created as a part of the SAMUELS project
(2014-2016). It consists of nearly every speech given
in the British Parliament from 1803-2005. The main
benefit is that it allows semantically-based searches of
these speeches. More details about semantic tags can
be found in (Alexander and Anderson, 2012; Rayson
et al., 2004). The words are organized in 37 higher
level semantic groups, in which one of them is also
Food and Drink (i.e. AG).
Table 1 summarizes the availability of food se-
mantic resources.
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
196

Table 1: Availability of food semantic resources.
Resource name Availability
FoodWiki https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4496660/
AGROVOC http://agroportal.lirmm.fr/ontologies/AGROVOC
Open Food Facts https://vest.agrisemantics.org/content/open-food-facts-food-ontology
Food Product Ontology https://vest.agrisemantics.org/content/food-product-ontology
FoodOn https://foodontology.github.io/foodon/
OntoFood https://bioportal.bioontology.org/ontologies/OF/?p=summary
SNOMED CT https://confluence.ihtsdotools.org/display/DOC/Technical+Resources
Hansard corpus https://www.hansard-corpus.org/
2.2 Food Named-entity Recognition
System
Contrary to the extensive work in named-entity recog-
nition methods for biomedical tasks, the situation in
the food and nutrition science is completely different.
The UCREL Semantic Analysis System (USAS)
is a framework for automatic semantic analysis of
text, which distinguishes between 21 major cate-
gories, one of which is “food and farming” (Rayson
et al., 2004). The USAS can provide additional infor-
mation about the food entity, but the limitation is that
it works on a token level. For example, if in the text
two words (i.e. tokens), like “grilled chicken”, denote
one food entity that needs to be extracted and ana-
lyzed, the semantic tagger would actually parse the
words “grilled” and “chicken” as separate entities and
obtain separate semantic tags.
In (Eftimov et al., 2017b), a rule-based NER used
for information extraction (IE) from evidence-based
dietary recommendation, called drNER, is presented,
where among other entities, food entities were also of
interest. This work was extended into a rule-based
named-entity recognition method for food informa-
tion extraction, called FoodIE (Popovski et al., 2019).
It is a rule engine, where the rules are based on com-
putational linguistics and semantic information that
describe the food entities. Evaluation showed that
FoodIE behaves consistently using different indepen-
dent evaluation data sets and very promising results
have been achieved.
The NCBO Annotator is a Web service that anno-
tates text provided by the user by using relevant ontol-
ogy concepts (Jonquet et al., 2009). It is available as
a part of the BioPortal software services (Noy et al.,
2009). The annotation workflow is based on a highly
efficient syntactic concept recognition (using concept
names and synonyms) engine and on a set of seman-
tic expansion algorithms that leverage the semantics
in ontologies. The methodology leverages ontologies
to create annotations of raw text and returns them us-
ing semantic web standards.
2.3 Food Concepts Normalization
In the last few years, food concepts normalization is
an open research question that is highly researched by
the food and nutrition science community, calling it
food matching. For this reason, StandFood (Eftimov
et al., 2017a) is recently introduced, which a semi-
automatic system for classifying and describing foods
according to a description and classifictaion system,
such as FoodEx2, proposed by the European Food
Safety Agency (EFSA) ((EFSA), 2015). It consists of
three parts. The first involves a machine learning ap-
proach and classifies foods into four categories, with
two for single foods: raw (r) and derivatives (d), and
two for composite foods: simple (s) and aggregated
(c). The second uses a natural language processing
approach and probability theory to perform food con-
cepts normalization. The third combines the result
from the first and the second part by defining post-
processing rules in order to improve the result for the
classification part. However, the food normalization
process was based only on lexical similarity between
the food concepts names, avoiding the semantic simi-
larity between them.
3 FoodOntoMap DESIGN
To provide a data set in which food concepts are
normalized by semantic tags from different food on-
tologies, we selected 22,000 recipes from Allrecipes
(Groves, 2013), which is the largest food-focused so-
cial network where people share their recipes and pro-
vide information about recipes in a non-standardized
manner. The recipes were selected from five recipe
categories: Appetizers and snacks, Breakfast and
Lunch, Dessert, Dinner, and Drinks.
To extract and annotate food concepts using dif-
ferent food ontologies we used two approaches.
FoodOntoMap: Linking Food Concepts across Different Food Ontologies
197

First, we used the recently proposed rule-based food-
named entity recognition method called FoodIE. Us-
ing FoodIE, the extracted food concepts are addi-
tionally annotated using the Hansard corpus seman-
tic tags. Then, we also used the NCBO annota-
tor together with the food ontologies that are avail-
able in the BioPortal (i.e. FoodOn, OntoFood, and
SNOMED CT). The semantic tags that are assigned
to each food concept are the semantic tags that belong
to the tokens from which the concept is constructed,
so it is a multi-label annotation process. Further in
the paper, we are going to explain each step in more
detail.
After collecting the recipes, FoodIE is used to
extract the food concepts. Further, we addition-
ally annotated the food concepts by assigning the
semantic tags from the Hansard corpus. For ex-
ample, if the food concept is ”grilled chicken”, it
obtains the semantic tags AG.01.t.07[Cooking] and
AG.01.d.06[Fowls]. After assigning the semantic tags
to each food concept, the results were organized in the
BioC format, which is a simple format to share text
data and annotations, with the goals of simplicity, in-
teroperability, and broad use and reuse (Comeau et al.,
2013). The BioC format for one recipe and its anno-
tations are presented in Figure 1. From it, it is evident
that each recipe is presented as a document for which
the category, description (text), and food annotations
are included. Each annotation consists of the food
concept that is extracted, the semantic tags from the
Hansard corpus that are assigned to it, and the offset
that points the token position from the beginning of
the text where the food concept starts and its charac-
ter length. The FoodIE NER method is publicly avail-
able on GitHub (https://github.com/GorjanP/foodie).
The data set organized in the BioC format is also pub-
licly available at http://cs.ijs.si/repository/FoodBase/
foodbase.zip.
Furthermore, the same recipe data set was pro-
cessed using the NCBO annotator with the FoodOn,
OntoFood, and SNOMED CT ontology. For this
reason, we used an R client that uses the An-
notator API available at http://data.bioontology.org/
documentation. The annotator API was used with the
recipe text as input and filters provided by the ontol-
ogy id (i.e FoodOn, OntoFood and SNOMED CT).
When SNOMED CT was used, additionally another
filter was applied based on a semantic type for which
food was specified (i.e. Food (T168)). An example
of an annotation is presented in Figure 2. Using this
figure, it is evident that for each annotation we have
the semantic tag (i.e. id), the food concept text, and
two numbers indicating the starting and ending posi-
tion of the food concept mention, which are referred
as from and to. It is important to note that the BioC
recipe format uses different metrics to define the po-
sition of the mentioned food concept. The NCBO
annotator uses character based offsets (from and to),
while the BioC format uses two metrics referred to as
offest and length. The offset indicates at which token
from the beginning of the recipe text the mentioned
food concept occurs, while the length indicates the
character length of the food concept. Due to this dif-
ference, these numbers were converted between the
two types of location description while the mapping
was being performed. Using these location metrics it
is determined which food concept mentions extracted
by the NCBO annotator refer to the same food con-
cept. More specifically, if some food concept mention
is a subset of another mention, their information is ag-
gregated and represented as the superset food concept
mention. With this duplicates, synonyms and multiple
labels are resolved into a more complete food entity.
The FoodOntoMap data set consists of food con-
cepts extracted from the recipes and normalized to
different food ontologies. It also provides a link be-
tween the food ontologies. For this reason, the food
concepts are matched and for each food concept the
semantic information from each dataset is assigned.
The concept matching is done by iterating through
each food concept that is extracted by the NER
method FoodIE. If the concept is also recognized
wholly or partially by the NCBO annotator in com-
bination with any of the selected ontologies, the se-
mantic tags from that ontology are also assigned to
the food concept. However, it is not uncommon while
using the NCBO annotator for it to provide semantic
tags on a token level instead of on a concept level.
Such an example would be when an ontology returns
two outputs for the food concept “salad dressing”, in-
stead of classifying it as a single food concept con-
sisting of two tokens. In these cases, each incom-
plete food concept extracted by the NCBO needs to
be matched to its corresponding superset food con-
cept concept extracted by FoodIE. This was done by
checking if the location metrics from the NCBO an-
notator are in accordance (more specifically, whether
they are a subrange) with the location metrics of a
food concept provided by FoodIE. If such a match ex-
ists, the NCBO food concept is added to the mapping
set of the FoodIE concept. By doing this, the map-
ping sets aggregate all the corresponding food con-
cept mentions that map to a food concept, along with
their semantic information, even if the NCBO anno-
tator does not classify some food concepts wholly.
To illustrate this with an example, on Figure 1
it is evident that we have multiple food concept
mentions that overlap. Such mentions are “SALAD
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
198
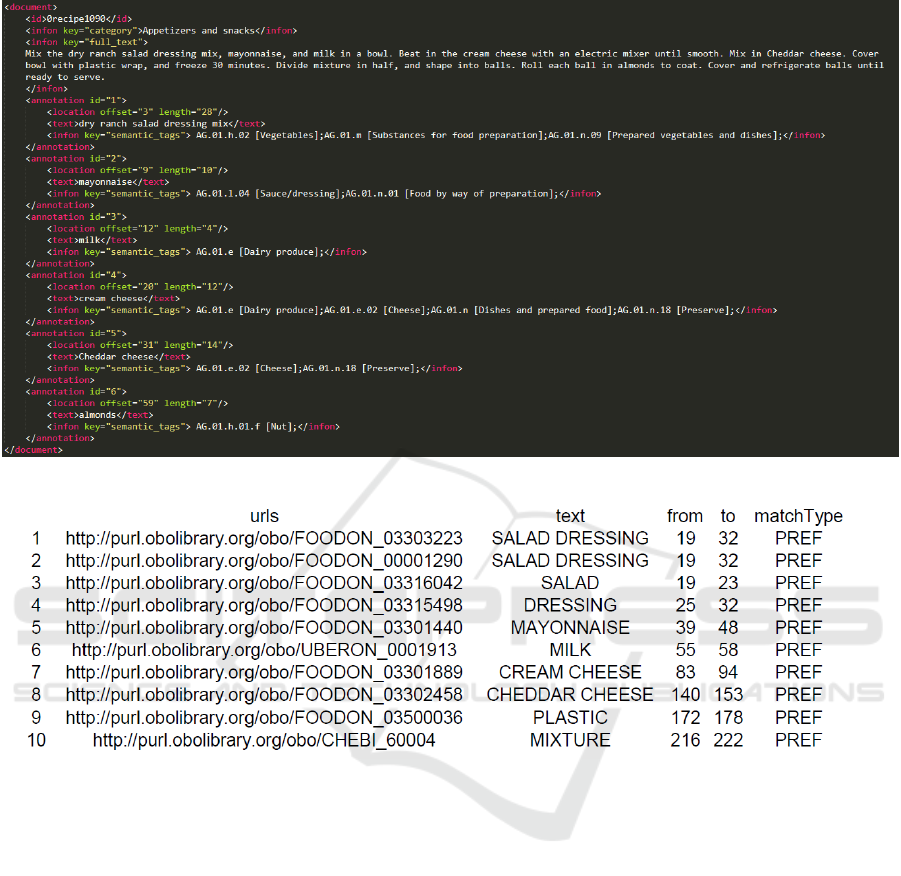
Figure 1: BioC format for a single recipe.
Figure 2: NCBO tagger output for a single recipe, using the ontology FoodOn.
DRESSING”, starting at 19 and ending at 32 and
appearing twice with different semantic information;
“SALAD”, starting at 19 and ending at 23; and
“DRESSING”, starting at 25 and ending at 32. Dur-
ing the matching step that is mentioned above, all
these are aggregated into a single food concept men-
tion that holds all the semantic information of its com-
ponent food concepts. The sole food concept mention
becomes “SALAD DRESSING”, starting at 19 and
ending at 32 along with four different semantic tags,
one from each component mention. Then, after con-
verting the location metrics to be compatible with the
BioC recipe format, the food concept is matched to
its corresponding food concept from the BioC recipe
dataset. This means that the resulting mapping is “dry
ranch salad dressing mix” to “SALAD DRESSING”.
Notice that not all tokens are caught by the NCBO
annotator using FoodOn. With this, the normaliza-
tion is complete, the code mapping being A000046 to
B000027.
In instances where one code from a dataset maps
to two separate codes from another dataset, the NCBO
tagger failed to produce one food concept mention
which contains the full semantic information. Instead,
it produced only a token-level mentions, and as such
they were not aggregated into one larger food concept
mention. Such an example is “SALTED WATER”,
which maps to “WATER” and “SALTED” separately,
the codes being A000123, B000012 and B000065, re-
spectively.
The flowchart of the FoodOntoMap design is pre-
sented in Figure 3.
The availability of the resources that are used to
create the FoodOntoMap is presented in Table 2.
The results from the FoodOntoMap are four dif-
ferent datasets and one mapping. Each dataset con-
sists of an artificial id for each unique food concept
that is extracted using each approach, the name of the
FoodOntoMap: Linking Food Concepts across Different Food Ontologies
199
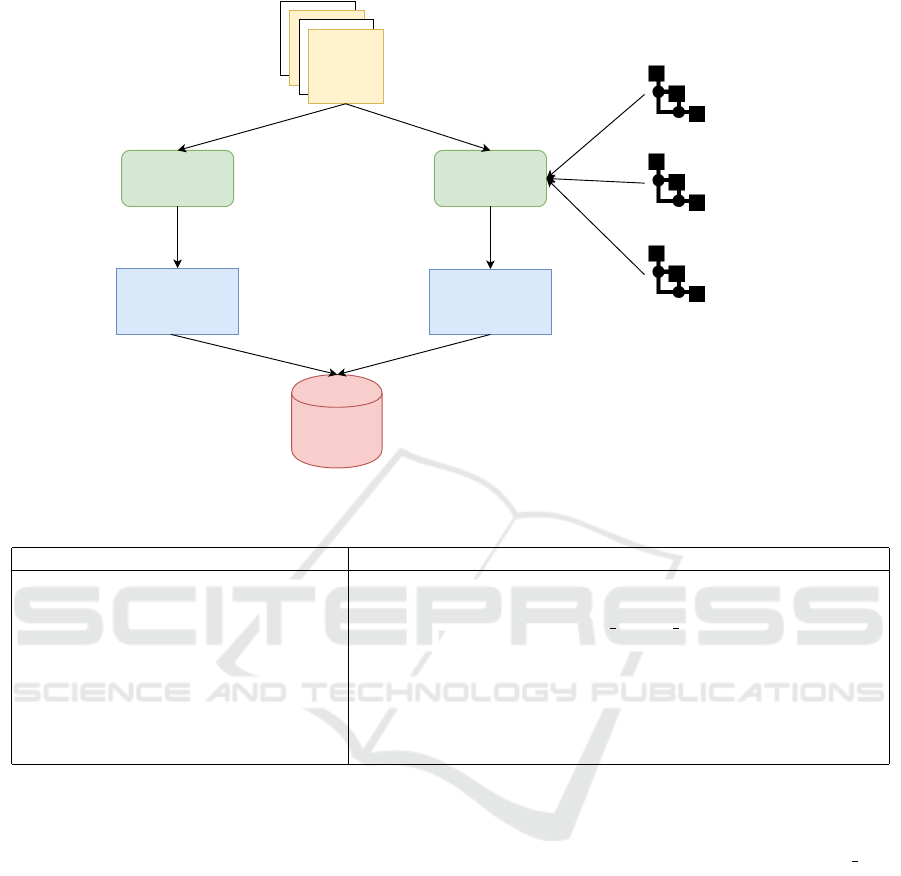
Recipes
FoodIE NCBO annotator
FoodON
OntoFood
SNOMEDCT
Food concepts
annotated using the
Hansard corpus
Food concepts
annotated using the
selected
ontologies
FoodOntoMap
Figure 3: FoodOntoMap design.
Table 2: Availability of resources used to create FoodOntoMap.
Resource name Availability
FoodIE https://github.com/GorjanP/foodie
BioC format recipe set http://cs.ijs.si/repository/FoodBase/foodbase.zip
Mapper client and NCBO annotator client https://github.com/GorjanP/FOM mapper client
Hansard corpus https://www.hansard-corpus.org/
NCBO annotator http://bioportal.bioontology.org/annotator
NCBO annotator REST API http://data.bioontology.org/documentation
FoodOn https://foodontology.github.io/foodon/
OntoFood https://bioportal.bioontology.org/ontologies/OF/?p=summary
SNOMED CT https://confluence.ihtsdotools.org/display/DOC/Technical+Resources
extracted food concept, and the semantic information
assigned to it. Each dataset corresponds to one of the
four semantic resources: Hansard corpus, FoodOn,
OntoFood, and SNOMED CT. At the end there is one
data set mapping, called FoodOntoMap, which, for
each concept that appears at least in two datasets, pro-
vides the links between them by listing the artificial id
of the concepts from each of the datasets in which it
is mentioned.
An example for one instance from FoodOntoMap
is A000016, B000011, C000012, D000002. This
means that this food concept is mapped from the
Hansard corpus to the respective ontologies, i.e.
FoodOn, SNOMED CT and OntoFood. The provided
codes are artificial unique identifiers that are assigned
to the food concept using each of the aforementioned
food semantic resources, respectively. If we look at
the separate datasets, we can see that A000016 cor-
responds to “garlic” with semantic tag AG.01.h.02.e
[Onion/leek/garlic] from the Hansard corpus,
B000011 corresponds to “garlic” with semantic
tag http://purl.obolibrary.org/obo/NCBITaxon 4682
from the FoodOn, C000012 is for “garlic” with the
semantic tag http://purl.bioontology.org/ontology/
SNOMEDCT/735030001 from the SNOMED
CT, and D000002 corresponds to enquotegarlic
with the semantic tag http://www.owl-ontologies.
com/Ontology1435740495.owl#Garlic from the
OntoFood.
The datasets consist of 13,205; 1,069; 111; and
582 unique food concepts, obtained using Hansard
corpus, FoodOn, OntoFood, and SNOMED CT, re-
spectively. The FoodOntoMap data set consists of
1,459 food concepts that are found in at least two food
semantic resources.
From the results, we can conclude that FoodIE
with the Hansard corpus gives the most promising re-
sults because it can extracted larger number of food
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
200

concepts compared with the NCBO annotator in com-
bination with some of the selected ontologies. More-
over, the results prove that the three food ontologies
(i.e. SNOMED CT, FoodOn, OntoFood) do not rep-
resent the food domain well, as many food concepts
cannot be extracted using the NCBO annotator, which
indicates that they do not exist in the food ontologies
themselves.
4 DISCUSSION
4.1 Impact
To the best of our knowledge, FoodOntoMap is the
first resource that provides normalization of food con-
cepts to different food ontologies, additionally pro-
viding a link between them. The motivation for build-
ing such a resource in the food domain comes from
the existence of the UMLS, which is extensively used
in the biomedical domain. For example, the MR-
CONSO.RRF table that is a part of the UMLS is
used in a lot of semantic web applications because
it can map the medical concepts to different biomed-
ical standards and vocabularies. To make progress in
analysing the large amount of data that is available in
order to find these relations, resources for food con-
cepts normalization are extremely valuable and wel-
come.
The main benefit of using FoodOntoMap is that
the food concepts can be normalzied by mapping
them to a unified system. Furthermore, the seman-
tic tags can be reused to find the non-linear relations
that exist between the concepts in the vector space,
by learning the embedding space. This can also be
done together with some medical and environment
concepts. Once the embedding space is learned, the
embeddings can be used for predictive studies in order
to explain the relations between human health, food
systems, and the environment.
4.2 Reusability
FoodOntoMap can be used as a resource that repre-
sents a normalized dataset of food concepts. Addi-
tionally, users can also follow the described pipeline
of steps used to create the FoodOntoMap mapping
in order to create their own new resource where the
food concepts will be normalized. Both approaches,
FoodIE and NCBO, used for food concepts extrac-
tion have already been well documented and evalu-
ated. The ontologies that are used by the NCBO an-
notator have also been well documented and are easy
to utilize. Furthermore, FoodOntoMap can be easily
extended on additional recipes, as well as a wide vari-
ety of different ontologies. With this it can provide an
ever wider coverage of food concepts. Additionally,
the FoodOntoMap pipeline can be used to normalize
food concepts that exist in food consumption and food
composition databases.
4.3 Availability
The resource FoodOntoMap is published and pub-
licly available for download at https://doi.org/10.
5281/zenodo.2635437. under a Creative Commons
Attribution-NonCommercial-ShareAlike 4.0 Interna-
tional (CC BY-NC-SA 4.0) licence. With this, we
encourage users to further contribute to this resource
and modify it as need be. Zenodo was the platform of
choice, as it provided all the framework tools needed
for such a dataset. Hosted along with the resource’s
datasets is a DCAT specification file, which briefly de-
scribes the structure of the resource. Furthermore, the
resource will be actively maintained and extended as
new ontologies, annotators, NER methods and NLP
methods become available. The goal is to keep the re-
source relevant and contemporary while also improv-
ing its domain coverage with the ever improving NLP
tools.
5 CONCLUSION
The resource that is presented in this paper represents
a data normalization pipeline. FoodOntoMap targets
the domain of food and nutrition science, normaliz-
ing food concepts across three different ontologies
and one semantic dataset. Additionally, it provides
a set with semantic information for each food concept
present in the dataset. This was made possible with
the use of NER methods for information extraction
from unstructured textual data. FoodOntoMap rep-
resents a first of its kind in the domain of Food and
Nutrition, with no other such resources being readily
available to the best of our knowledge. As such, it
is crucial in building tools that improve knowledge in
this domains and the public health effects it implies,
as well as building data models that can be utilized
to further discover relations and links in the food and
nutrition domain.
FoodOntoMap: Linking Food Concepts across Different Food Ontologies
201

ACKNOWLEDGEMENT
This work was supported by the Slovenian Research
Agency Program P2-0098 and the European Union’s
Horizon 2020 research and innovation programme
[grant agreement No 769661].
REFERENCES
Alexander, M. and Anderson, J. (2012). The hansard cor-
pus, 1803-2003.
Beam, A. L., Kompa, B., Fried, I., Palmer, N. P., Shi, X.,
Cai, T., and Kohane, I. S. (2018). Clinical concept
embeddings learned from massive sources of medical
data. arXiv preprint arXiv:1804.01486.
Boag, W., Wacome, K., Naumann, T., and Rumshisky, A.
(2015). Cliner: A lightweight tool for clinical named
entity recognition. AMIA Joint Summits on Clinical
Research Informatics (poster).
Bodenreider, O. (2004). The unified medical language sys-
tem (umls): integrating biomedical terminology. Nu-
cleic acids research, 32(suppl 1):D267–D270.
Boulos, M. N. K., Yassine, A., Shirmohammadi, S., Nama-
hoot, C. S., and Br
¨
uckner, M. (2015). Towards an
“internet of food”: Food ontologies for the internet of
things. Future Internet, 7(4):372–392.
Caracciolo, C., Stellato, A., Rajbahndari, S., Morshed,
A., Johannsen, G., Jaques, Y., and Keizer, J. (2012).
Thesaurus maintenance, alignment and publication as
linked data: the agrovoc use case. International Jour-
nal of Metadata, Semantics and Ontologies, 7(1):65–
75.
C¸ elik, D. (2015). Foodwiki: Ontology-driven mobile safe
food consumption system. The Scientific World Jour-
nal, 2015.
Choi, E., Bahadori, M. T., Searles, E., Coffey, C., Thomp-
son, M., Bost, J., Tejedor-Sojo, J., and Sun, J. (2016).
Multi-layer representation learning for medical con-
cepts. In Proceedings of the 22nd ACM SIGKDD In-
ternational Conference on Knowledge Discovery and
Data Mining, pages 1495–1504. ACM.
Comeau, D. C., Do
˘
gan, R. I., Ciccarese, P., Cohen, K. B.,
Krallinger, M., Leitner, F., Lu, Z., Peng, Y., Rinaldi,
F., Torii, M., et al. (2013). Bioc: a minimalist ap-
proach to interoperability for biomedical text process-
ing. Database, 2013:bat064.
Donnelly, K. (2006). Snomed-ct: The advanced terminol-
ogy and coding system for ehealth. Studies in health
technology and informatics, 121:279.
(EFSA), E. F. S. A. (2015). The food classification and
description system foodex 2 (revision 2). EFSA Sup-
porting Publications, 12(5):804E.
Eftimov, T., Koro
ˇ
sec, P., and Korou
ˇ
si
´
c Seljak, B. (2017a).
Standfood: Standardization of foods using a semi-
automatic system for classifying and describing foods
according to FoodEx2. Nutrients, 9(6):542.
Eftimov, T., Korou
ˇ
si
´
c Seljak, B., and Koro
ˇ
sec, P. (2017b).
A rule-based named-entity recognition method for
knowledge extraction of evidence-based dietary rec-
ommendations. PloS One, 12(6):e0179488.
Gligic, L., Kormilitzin, A., Goldberg, P., and Nevado-
Holgado, A. (2019). Named entity recognition
in electronic health records using transfer learn-
ing bootstrapped neural networks. arXiv preprint
arXiv:1901.01592.
Griffiths, E. J., Dooley, D. M., Buttigieg, P. L., Hoehn-
dorf, R., Brinkman, F. S., and Hsiao, W. W. (2016).
Foodon: A global farm-to-fork food ontology. In
ICBO/BioCreative.
Groves, S. (2013). How allrecipes. com became the worlds
largest food/recipe site. roi of social media (blog).
Johnson, A. E., Pollard, T. J., Shen, L., Li-wei, H. L.,
Feng, M., Ghassemi, M., Moody, B., Szolovits, P.,
Celi, L. A., and Mark, R. G. (2016). Mimic-iii, a
freely accessible critical care database. Scientific data,
3:160035.
Jonquet, C., Shah, N., Youn, C., Callendar, C., Storey, M.-
A., and Musen, M. (2009). Ncbo annotator: seman-
tic annotation of biomedical data. In International
Semantic Web Conference, Poster and Demo session,
volume 110.
Kolchin, M. and Zamula, D. (2013). Food product ontol-
ogy: Initial implementation of a vocabulary for de-
scribing food products. In Proceeding of the 14th
Conference of Open Innovations Association FRUCT,
Helsinki, Finland, pages 11–15.
Miotto, R., Li, L., Kidd, B. A., and Dudley, J. T. (2016).
Deep patient: an unsupervised representation to pre-
dict the future of patients from the electronic health
records. Scientific reports, 6:26094.
Noy, N. F., Shah, N. H., Whetzel, P. L., Dai, B., Dorf, M.,
Griffith, N., Jonquet, C., Rubin, D. L., Storey, M.-A.,
Chute, C. G., et al. (2009). Bioportal: ontologies and
integrated data resources at the click of a mouse. Nu-
cleic acids research, 37(suppl 2):W170–W173.
Popovski, G., Kochev, S., Korou
ˇ
si
´
c Seljak, B., and Efti-
mov, T. (2019). Foodie: A rule-based named-entity
recognition method for food information extraction.
In Proceedings of the 8th International Conference
on Pattern Recognition Applications and Methods,
(ICPRAM 2019), pages 915–922.
Rayson, P., Archer, D., Piao, S., and McEnery, A. (2004).
The ucrel semantic analysis system.
Snae, C. and Br
¨
uckner, M. (2008). Foods: a food-oriented
ontology-driven system. In Digital Ecosystems and
Technologies, 2008. DEST 2008. 2nd IEEE Interna-
tional Conference on, pages 168–176. IEEE.
Wang, Q., Zhou, Y., Ruan, T., Gao, D., Xia, Y., and He,
P. (2019). Incorporating dictionaries into deep neu-
ral networks for the chinese clinical named entity
recognition. Journal of biomedical informatics, page
103133.
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
202
