
A METHODOLOGY FOR INTEGRATING NEW SCIENTIFIC
DOMAINS AND APPLICATIONS IN A VIRTUAL LABORATORY
ENVIRONMENT
E. C. Kaletas, H. Afsarmanesh, L. O. Hertzberger
University of Amsterdam, Informatics Institute
Kruislaan 403, 1098 SJ Amsterdam, The Netherlands
Keywords:
Virtual laboratory, scientific domain integration, scientific application integration, integration methodology
Abstract:
Emergence of advanced complex experiments in experimental sciences resulted in a change in the way of
experimentation. Several solutions have been proposed to support scientists with their complex experimen-
tations, ranging from simple data portals to virtual laboratories. These solutions offer a variety of facilities
to scientists, such as management of experiments and experiment-related information, and management of
resources. However, issues related to adding new types of experiments to proposed support environments still
remain untouched, causing inefficient utilization of efforts and inadequate transfer of expertise. The main topic
of this paper is to present a methodology for integrating new scientific domains and applications in a multi-
disciplinary virtual laboratory environment. In order to complement the methodology with the right context,
the paper also presents an experiment model that uniformly represents scientific experiments, data models for
modelling experiment-related information, and mechanisms for the management of this information.
1 INTRODUCTION
In recent years, technological advances and system-
atic growth of research efforts in experimental science
domains, such as life sciences, have achieved break-
throughs that go beyond the imagination of mankind
even a decade ago. Advances in the laboratory tech-
niques and the introduction of highly automated lab-
oratory instruments have caused both the automation
of many steps in scientific experiments and the gen-
eration of very large amounts of data, leading to a
change in the way that scientists perform their sci-
entific research. However, new ways of experimenta-
tion pose many requirements and challenges to both
scientists and developers of supporting tools and in-
frastructures. The following can be mentioned among
these challenges, which create a burden on the shoul-
ders of scientists and developers of supporting tools:
• Increasing number and complexity of laboratory
instruments and procedures require significant ef-
fort to perform an experiment. For example, clone
preparation phase of a microarray experiment con-
tains up to 51 steps.
• Large amounts of data are generated during experi-
ments. For instance, material analysis experiments
for complex surfaces typically generate 20 images
per day where data size goes up to 100 MB per im-
age (Frenkel et al., 2001).
• Heterogeneity causes difficulties in modelling and
storage of experimental information. For instance,
281 biological database resources in 18 different
categories were listed in 2001 (Baxevanis, 2001).
• Lack of standards for modelling and representation
of information causes wasted efforts for developing
specific solutions to overcome common problems
and difficulties in comparing the results obtained
in multiple experiments.
New solution methods and problem solving tech-
niques are needed to address the challenges. How-
ever, addressing these challenges require long-term
multi-disciplinary efforts between scientists from
both experimental science domains and computer sci-
ence. In experimental science domains, scientists in-
volved in these experimentations must both advance
their models and mechanisms supporting the analy-
sis of resulted information, and design innovative in-
vestigation approaches leading to discoveries. From
the computer science point of view, however, the
complexity of emerging experimentations necessi-
tates that the support environment be generic and con-
stitute a horizontal base infrastructure, on top which
265
C. Kaletas E., Afsarmanesh H. and O. Hertzberger L. (2004).
A METHODOLOGY FOR INTEGRATING NEW SCIENTIFIC DOMAINS AND APPLICATIONS IN A VIRTUAL LABORATORY ENVIRONMENT.
In Proceedings of the Sixth International Conference on Enterprise Information Systems, pages 265-272
DOI: 10.5220/0002634802650272
Copyright
c
SciTePress

vertical software tools can be developed, each provid-
ing a specific service for the experimentations.
Several solutions have been proposed to sup-
port scientific experimentations, ranging from simple
Web-based query interfaces for scientific databases
to complex problem solving environments and vir-
tual laboratories. Some of these environments target
specific applications while others are offering more
generic support. However, issues such as extension of
support environments with facilities for emerging sci-
entific experiments (i.e. integration of vertical soft-
ware tools in the horizontal support environments)
still remain untouched. More precisely, a method-
ology for integrating new scientific domains and ap-
plications in a support environment is still lacking;
causing inefficient utilization of efforts and inade-
quate transfer of expertise. Yet, a further challenge
would be to provide a methodology for integrating di-
verse heterogeneous scientific applications in a multi-
disciplinary support environment.
The main topic of this paper is to present a method-
ology for integrating new scientific domains and ap-
plications in a multi-disciplinary virtual laboratory
environment. Nevertheless, integration of new do-
mains and applications in a complex environment like
virtual laboratory requires a common understanding
of scientific experiments, availability of both well-
defined data models for modelling experiment-related
information and mechanisms for the management of
this information. The remaining of this paper is struc-
tured as follows: First an overview of existing and
emerging support environments is provided in Sec-
tion 2. In Section 3, general structure of scientific
experiments is presented, followed by a description
of an experiment model. Then ‘process data flows’
are introduced in this section as a tool for modelling
experiments. Section 4 focuses on a specific support
environment, namely the VLAM-G virtual laboratory
environment. Section 5 further focuses on VIMCO,
the information management platform of VLAM-G,
and addresses issues related to modelling and man-
agement of information related to scientific experi-
ments. The proposed methodology is described in
Section 6. Finally, Section 7 concludes.
2 SUPPORT ENVIRONMENTS
This section describes different types of support envi-
ronments for scientific experimentations. The remain-
ing of this paper, however, focuses on virtual labora-
tories.
Science Portals. Science Portals (Ashby et al.,
2001), (Pierce et al., 2002) are emerging as conve-
nient mechanisms for providing a single point of ac-
cess with familiar and simplified interfaces to a spe-
cific set of resources that are of importance to a spe-
cific scientific community. Resources can be, for in-
stance, computational, storage, networking resources,
electronic whiteboards, or a digital library. These re-
sources are usually made available to users as ser-
vices. Users can make use of these services either
in a stand-alone manner using only one service at a
time, or in a collective manner using a number of ser-
vices simultaneously. Science portals only provide a
uniform means for accessing resources. Users them-
selves are responsible for the correct and efficient us-
age of the available resources.
Problem Solving Environments. A Problem Solv-
ing Environment (PSE) (Allen et al., 2001),
(Schuchardt et al., 2002) is a computer system that
provides all the computational facilities needed to
solve a target class of problems (Gallopoulos et al.,
1994). Common characteristics of PSEs include
among others a target class of science or engineer-
ing problems, natural appearance and ease of use,
provision of multiple solution paths or algorithms in
the target areas, and availability of parameterized al-
gorithms. Users can be assisted through automatic
and/or semiautomatic selection of a proper solution
method from the available set of solution methods, or
by providing ways to easily incorporate new solution
methods. Although some examples of generic PSEs
do exist such as Cactus (Allen et al., 2001), most of
the current PSEs target a specific class of problems.
Virtual Laboratories. A Virtual Laboratory (VL)
(Afsarmanesh et al., 2002), (Messina, 2003) provides
an electronic workspace for distributed collaboration
and experimentation in research, to generate and de-
liver results using distributed information and com-
munication technologies (Vary, 2000). It supports
an aggregation of people who pursue a related set
of research activities and share resources, where the
resources including the people may be geographi-
cally distributed and associated with different institu-
tions. Therefore, virtual laboratories bring together
best combination of skills, expertise, and tools to
carry out the same type of research that is done in
a single real laboratory (Messina, 2003).
At present, the concept of ‘virtual laboratory’ has
been associated with several different meanings, de-
pending on the wide variety of application interests
and fields (e.g. education, games, science, engineer-
ing, manufacturing, etc.). On the other hand, there are
some recent VL projects that target the provision of
generic solutions and offer a wide variety of function-
ality, and that are applicable to a wider set of problems
(Afsarmanesh et al., 2002), (Catlin et al., 2000). One
such virtual laboratory that offers generic solutions to
ICEIS 2004 - INFORMATION SYSTEMS ANALYSIS AND SPECIFICATION
266

scientists is the Grid-based Virtual Laboratory Ams-
terdam (VLAM-G), which is described in Section 4.
3 MODELLING EXPERIMENTS
In a VL environment, it is natural that all activities are
centered around experiments. The experiment model
underlying a VL outlines the overall approach that
the VL follows for supporting scientific experiments.
Furthermore, the experiment model allows a method-
ological definition of complex experiments in a prob-
lem domain. Thus, the experiment model lies in the
core of a VL. This section focuses on the modelling
of scientific experiments and describes an experiment
model, by first presenting high-level general structure
of scientific experiments, then providing a description
of the experiment model. This section then introduces
‘process data flows’ as a tool for modelling experi-
ments.
3.1 General Structure of
Experiments
A scientific experiment consists of a number of ex-
periment components. Components of an experiment
correspond to activities, physical entities, or data ele-
ments involved in that experiment. Every experiment
is part of a project, which may contain one or more
experiments that are related to each other (Figure 1).
Project
*
Experiment
*
Experiment Component
Figure 1: General structure of scientific experiments
A project is a grouping of related experiments. Ex-
periments in a project may follow an order (e.g. time
course experiments), or may be unordered (e.g. com-
parison experiments).
Experiments are composed of three kinds of exper-
iment components (see Figure 2 for experiment com-
ponents involved in a typical microarray experiment).
Physical entities are used during laboratory activities
or during instrumentation. Activities may represent
laboratory activities, instrumentations, or computa-
tional processes. Data elements in turn correspond
to both raw data generated by instruments and input
data to computational processes. The generic descrip-
tive elements represent the components that are com-
mon to all experiments and provide descriptive infor-
mation about them, such as organism, gene, or infor-
mation about hardware and software tools (e.g. scan-
ner). Such descriptive information does not change
from one experiment to another.
HYBRIDIZATION CLONE
HYBRIDIZED
ARRAY
ARRAY IMAGEMICROARRAY
ARRAY
MEASUREMENT
ARRAY
SCANNING
ARRAY IMAGE
ANALYSIS
GENE
ORGANISM
IMAGE
ANALYSIS
PROGRAM
SCANNER
NAME
NAME
NAME
NAME
Notation:
Physical Entity
Activity
Data Element
Generic Descriptive
Element
SAMPLE
MRNA PROBE
Figure 2: Components for microarray experiments
3.2 The Experiment Model
An experiment model is developed to uniformly rep-
resent scientific experiments from different domains.
A brief summary of the experiment model is pro-
vided below. For more information on the experiment
model, please refer to (Kaletas et al., 2003).
The experiment model consists of two main com-
ponents, namely experiment procedure and experi-
ment context (Figure 3).
Experiment Procedure
Experiment Context
Experiment
Figure 3: The experiment model
An experiment procedure defines the approach
taken to solve a particular scientific problem, by
defining the experiment components that are typically
involved in the experiments of the same type. Thus,
an experiment procedure standardizes the experimen-
tal approach for experiments of the same type.
An experiment context, on the other hand, de-
scribes the solution. An experiment context is an in-
stantiation of an experiment procedure. It describes
the accomplishment of a particular experiment, by
providing descriptions of each experiment component
involved in the experiment. Thereby, it provides the
context for that particular experiment.
Processing and analysis of large data sets consti-
tute an important part of scientific experiments, and
hence require special attention. In an experiment, dur-
ing its processing and analysis, data flows from one
computational process to another. This data flow is
represented by a directed graph. Nodes in the graph
are computational processes, while the connecting
arcs are the data flowing through the processes. This
A METHODOLOGY FOR INTEGRATING NEW SCIENTIFIC DOMAINS AND APPLICATIONS IN A VIRTUAL
LABORATORY ENVIRONMENT
267
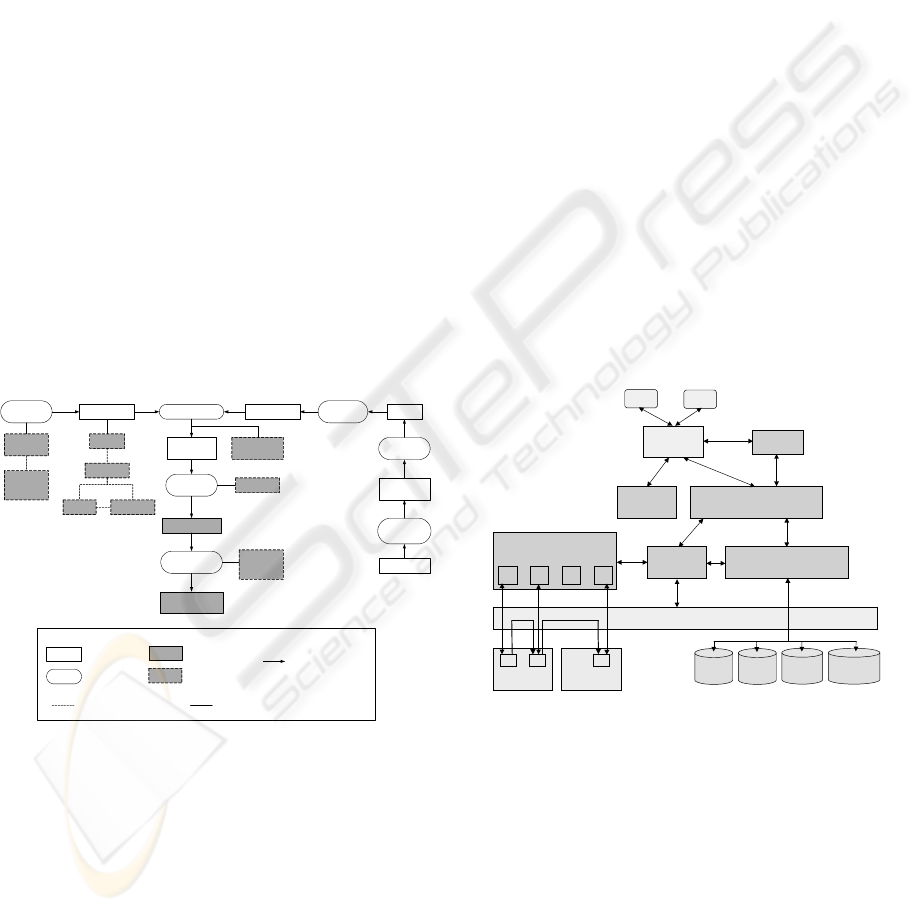
data flow graph is called as computational processing.
An experiment procedure may contain computational
processes. Consequently, particular experiments that
are instantiating this procedure contain descriptions
of these processes in their experiment contexts.
3.3 Process Data Flows
Given a scientific domain, the first step in modelling
the experiment-related information is to perform a de-
tailed analysis of the experiment components for each
experiment in that domain. Result of the analysis is a
conceptual model, which includes a comprehensive
component-by-component definition of experiments
in the domain.
In this conceptual model, every component in-
volved in an experiment, the properties of each com-
ponent, and the relationships among the components
are defined. Such a detailed and comprehensive def-
inition of an experiment is referred to as the Process
Data Flow (PDF). Figure 4 presents the microarray
PDF as an example using the same notation as Fig-
ure 2 for experiment components. This example mi-
croarray PDF depicts the order of components in the
experiment (i.e. the experiment flow) and also the re-
lationships between components and generic descrip-
tive elements in microarray experiments.
HYBRIDIZATION
CLONE
HYBRIDIZED
ARRAY
ARRAY IMAGE
MICROARRAY
ARRAY
MEASUREMENT
ARRAY
SCANNING
ARRAY IMAGE
ANALYSIS
GENE ORGANISM
IMAGE
ANALYSIS
PROGRAM
SCANNER
NAME
NAME
NAME
NAME
Notation:
ARRAY
SPOTTING
ARRAY
SPOTTER
ARRAY
SPOTTER
PROGRAM
MRNA PROBE
MRNA
LABELING
MRNA
MRNA
EXTRACTION
TREATED
CELL SAMPLE
CELL SAMPLE
TREATMENT
CELL SAMPLE
SLIDE
PROCESSOR
SPOT
Physical Entity
Activity
Data Element
Generic Descriptive
Element
Experiment flow /
Relationship between
experiment components
Relationship between an experiment
component and a generic descriptive element
Relationship between two
generic descriptive elements
Figure 4: Process-Data Flow for microarray experiments
4 VLAM-G
The Dutch VLAM-G (Grid-based Virtual Laboratory
Amsterdam) project (Afsarmanesh et al., 2002), (Af-
sarmanesh et al., 2001) provides the main context
for the work presented in this paper. VLAM-G is
a multi-disciplinary virtual laboratory environment
that provides the required generic environment for
multi-disciplinary research in experimental science
domains.
The two initial application cases of VLAM-G were
the DNA microarray application from the life sci-
ences domain and the material analysis for complex
surfaces application (MACS) (Frenkel et al., 2001)
from the physics domain. The work described in this
paper has been initiated and motivated by the need
to support the requirements of both these initial ap-
plication cases and any future applications within the
VLAM-G.
VLAM-G allows its users to:
• perform multi-disciplinary, collaborative experi-
ments in a uniform, integrated environment,
• complement their in-vitro experiments with in-
silico experiments,
• define customized experimental procedures and
analysis flows,
• reuse generic software components, and
• share hardware, software, storage, networking re-
sources as well as knowledge and experience.
The architecture of the VLAM-G and interaction
among its components are shown in Figure 5. Main
components of the architecture are described below.
user
user
Front-End
Collaboration
Session Manager
RTS
Assistant
VIMCO
Grid / Globus services
M1 M2 M3 M4
Module Repository
Resource A Resource B
…
RTS
DB
Project
DB
VIMCO
DB
…
…
Application
DBs
Figure 5: VLAM-G architecture
Front-End is the user environment of the VLAM-
G. Users interact with the VLAM-G only through the
Front-End. The Front-End consists of a number of
graphical user interfaces (GUIs), which present the
VLAM-G functionality to its users in a uniform way.
Session Manager acts as a gateway to the VLAM-
G server side. It manages the active user sessions, and
is responsible for coordinating the interactions among
the VLAM-G components.
The distributed computing and networking re-
sources on the Grid are made available to VLAM-
G users through the Run Time System. Generic
ICEIS 2004 - INFORMATION SYSTEMS ANALYSIS AND SPECIFICATION
268

VLAM-G processing functionality and application-
specific problem solvers are presented to its users as
self-contained software entities. RTS provides an API
which encapsulates the Grid computing code within a
simple interface. Users compose their computational
processings by attaching a number of software entities
to each other, which are then executed by the RTS on
the Grid environment.
Collaboration component of the VLAM-G en-
ables simultaneous collaborative design and execu-
tion of experiments. The Assistant assists users dur-
ing the design of an experiment, for instance, by sug-
gesting the most efficient software to perform a spe-
cific task. Implementation of these two components
is planned for future work.
Module Repository is a persistent storage for bi-
naries of software entities.
The Virtual Laboratory Information Management
for Cooperation (VIMCO) is the information man-
agement platform of VLAM-G. VIMCO is described
in details in the following section.
5 VIMCO
This section describes the VIMCO facilities for mod-
elling and manipulation of information related to sci-
entific experiments in the VLAM-G environment.
5.1 Modelling Information Related
to Scientific Experiments
Modelling experiment-related information handled in
a VL is important, since the employed data models
have a direct influence on the architectural compo-
nents of the VL and their functionalities. Information
related to scientific experiments include experiment
information (e.g. experiment procedures, experiment
contexts, computational processings), user informa-
tion (e.g. information about users and user roles, ac-
cess rights definitions), and VL-related information
(e.g. session information, information about the ser-
vices provided by the VL). This section builds on the
experiment model presented earlier in this paper, and
describes the modelling of information related to sci-
entific experiments in VL.
5.1.1 Experiment Information
In this subsection, data models developed for the
modelling of different types of information about sci-
entific experiments are described. Note that all data
models described below are object-oriented. Due to
space limitations, however, UML diagrams and de-
tailed descriptions of the data models are omitted. De-
tails of the data models presented here can be found
in (Kaletas et al., 2003).
Modelling Experiment Procedures. The
Procedure Data Model facilitates the rep-
resentation and storage of experiment pro-
cedures, and consists of the following
classes: Procedure, ProcedureElement,
ProcedureConnection, and ProcedureGUI.
An instance of the Procedure class contains the
general information about a procedure, including its
version. Multiple versions of the same procedure can
exist. Every Procedure object consists of a set of
ProcedureElements, which correspond to exper-
iment components. For each procedure element (and
hence experiment component), there is a correspond-
ing data type in the application database schema to
hold the information about that experiment compo-
nent. The corresponding data type in the database
schema for a procedure element is represented by the
className attribute. isProcessing attribute of
a procedure element indicates whether the element
actually corresponds to a processing. Instances of
ProcedureConnection class correspond to
the relationships between classes in the database
schema, indicated by the relName attribute. Finally,
ProcedureGUI objects keep information about the
graphical properties of a procedure for displaying
it to users. Procedure definitions are stored in the
application databases.
Modelling Experiment Contexts. A context is an
instance of a procedure, and the procedure elements
correspond to data types in an application database.
This means, a context actually consists of a number of
objects in the application database, hence, modelling
the application database schema for the domain of an
experiment is sufficient for modelling the contexts of
that experiment.
Using the general structure of scientific experi-
ments, a base data model for experimental infor-
mation called Experimentation Environment Data
Model (EEDM) is designed. EEDM covers the com-
mon aspects of experiments; namely scientist owning
the experiment, input, activities applied on the input,
output obtained from an activity, hardware and soft-
ware used during the experiment, conditions and pa-
rameters for the activities, devices and software, and a
recursive flow of processes and data where a specific
order is followed during an experiment.
EEDM provides generic constructs for these com-
mon aspects of scientific experiments, which are ex-
tended (sub-typed) in application databases for mod-
elling experiment-specific information.
A METHODOLOGY FOR INTEGRATING NEW SCIENTIFIC DOMAINS AND APPLICATIONS IN A VIRTUAL
LABORATORY ENVIRONMENT
269

Modelling Software Entities and Computational
Processings. A computational processing covers
both descriptions of the executables used (i.e. soft-
ware entities) and run-time information for the pro-
cessing. Software entities represent self-contained ex-
ecutable programs. They are mainly characterized by
the tasks that they perform, their input and output data
types, parameters, and run-time requirements. For ev-
ery executable, its developer specifies the run-time re-
quirements to help a user to decide which software
entities to use in his/her processing. A user designs
a computational processing by attaching a number of
software entities to each other through their input and
output ports. This design is stored in RTS DB for fu-
ture references and for possible re-runs.
5.1.2 User Information
User information is used in a VL for security and
access control, and for administration purposes. In
addition, application databases contain user informa-
tion to capture, for instance, the owner of a physical
entity or data element (e.g. microarray or array im-
age), or operator of an instrument (e.g. array scan-
ner). As such, information about VL users is defined
and stored in all VL databases.
Every VL user has a unique username, and assumes
a role. Roles are used to define and enforce access
rights for groups of users. Access to information in
the VL is controlled through a set of restrictions. It
is considered that by default every user has all privi-
leges on all available information. Depending on the
role that a user assumes, these unlimited privileges are
restricted by a set of ‘restrictions’. Below, the specific
roles defined for VL users are summarized.
• Application users are the actual users of the VL.
An application user is typically associated with a
scientific domain (e.g. molecular biology), and can
only access the application database for this do-
main. S/he has full control on her/his contexts and
processings but only read access to procedures.
• Domain experts are application users who have
extensive knowledge and experience in a given do-
main and on the experiments being performed in
that domain. They are responsible for modelling
application databases (together with Information
Management Admin), designing experiment proce-
dures, defining laboratory protocols, and register-
ing descriptions of software entities.
• Tool developers are the developers of software en-
tities (i.e. vertical software tools).
• Administrators are responsible for the proper
management and operation of the VL. Informa-
tion Management Admin is responsible for mod-
elling application database schemas and for main-
taining these databases. VL Admin is responsi-
ble for resource management, infrastructure main-
tenance, and user management.
5.1.3 VL-Related Information
During its operation, the VL itself manages and ma-
nipulates certain types of information, such as infor-
mation about active sessions and information about
the services it provides.
Session management is an important functionality
in a VL for controlling and coordinating user activi-
ties. All user activities occur within a session. For re-
covery reasons, VIMCO provides persistence to ses-
sion information (in VIMCO DB). A Session class
is defined to store the session information, which con-
tains owner of the session, observers in case of a col-
laborative session, and the procedures, contexts, and
processings being used in the session.
The services that are made available by the VL
to its users are highly dependent on the existing
experiment-related information in the VL. For in-
stance, availability of a ‘microarray experiment pro-
cedure’ allows a VL to provide ’making microarray
experiments’ as a service to its users. In order to pro-
vide a list of services to users, VL makes use of the
experiment-related information stored and managed
by VIMCO. The list of available services is generated
by VIMCO by extracting the necessary information
from what is available in the databases.
5.2 Managing Information Related
to Scientific Experiments
VIMCO provides the necessary functionality and
mechanisms for the manipulation of different types
of information that it manages. Very briefly, VIMCO
supports the following functionality:
• managing experiment procedures, contexts, and
computational processings;
• defining and enforcing usage policies for experi-
ment contexts;
• managing user information;
• defining and enforcing access rights;
• managing VL-related information;
• uniformly accessing multiple, heterogeneous appli-
cation databases; and
• sharing and exchanging information among collab-
orating partners.
ICEIS 2004 - INFORMATION SYSTEMS ANALYSIS AND SPECIFICATION
270

6 THE PROPOSED
METHODOLOGY
Lack of a methodology for supporting new scien-
tific domains and applications in a virtual laboratory
causes inadequate transfer of expertise and hence in-
efficient utilization of efforts. In this paper, sev-
eral different aspects of a virtual laboratory environ-
ment were addressed with a focus on its informa-
tion management component. In specific, an exper-
iment model is presented for uniformly representing
diverse scientific experiments. Furthermore, several
data models for modelling different types of informa-
tion related to scientific experiments, and function-
ality for the actual management of this information
are provided. The results obtained and the experience
gained during the VLAM-G project, and especially
during the two application cases, are used for defining
a methodology for integrating new scientific domains
and applications in VL. The methodology provides
step-by-step guidance to domain experts, tool devel-
opers and administrators during the process of adding
new domains, applications and resources to VL.
The methodology provided in this section is
generic and reusable; it can be uniformly applied to
different scientific domains and to different scientific
applications. This is partially due to the genericness
and reusability of the models described here.
Following subsections describe the methodology.
6.1 Integration of a New Domain
Integration of a new domain in VL covers the devel-
opment of a domain-specific application database, in-
tegrating and registering this database into the VL and
its information management platform, and defining
and registering the domain users (application users).
Steps to take when integrating a new domain in VL
are enumerated below.
1. Study different types of experiments being per-
formed in the domain, to the level of detail of single
components involved in the experiments and the re-
lated information for each component. This step is
realized by the experts in the domain, in close col-
laboration with an Information Management Ad-
min (IM Admin).
2. Generate process data flows (PDFs) for each ex-
periment type. Discuss the PDFs with the experts
from the domain, make any necessary modifica-
tions, and eventually confirm the PDFs. This step
is performed by a Domain Expert in close collabo-
ration with an IM Admin.
3. For each PDF, map the PDF elements to the base
data types in the EEDM, and model each PDF el-
ement by sub-typing one of the base EEDM data
types. The result is the process data flows with ele-
ments that are defined as sub-types of base EEDM
types. These PDFs form the base for the applica-
tion database schema for the domain. This step is
performed by an IM Admin together with a Do-
main Expert.
4. Cross-study different experiment types defined in
terms of extended EEDM constructs, and identify
the elements that are common to different exper-
iments (e.g. organism info). Identify the relation-
ships between components in different experiments
and these common elements. Confirm the result
with experts in the domain. The result is the core
schema for the application database. This step is
performed by a Domain Expert and an IM Admin.
5. Map the core database schema to a data definition
language (e.g. Object Definition Language). Add
any other data models required by the information
management platform (e.g. procedure data model).
This step is performed by an IM Admin.
6. Create the application database by loading the
schema defined in the data definition language.
This step is performed by an IM Admin.
7. Register the database to the information manage-
ment platform by providing the required informa-
tion about the new data source. This step is per-
formed by an IM Admin.
8. Define and register the domain users using the user
management functionality. This step is performed
by a VL Admin.
6.2 Integration of a New Application
Once the new domain is integrated, new applications
from that domain can be integrated. Steps to take
when integrating new applications are given below.
1. Define experiment procedures for different types of
experiments that will be offered to users of this ap-
plication. Graphical editors provided by the VL can
be used for procedure definition. This activity is
performed by a Domain Expert.
2. Develop software entities for application specific
functionality (e.g. analysis tool for microarray
data). This step is performed by a Tool Developer.
3. After the quality control applied to software enti-
ties, store their definitions in the information man-
agement platform, and register them to VL. These
activities are performed by a Domain Expert.
4. Using the access rights management functionali-
ties, define the user roles and restrictions for each
role on the data types of this domain. Assign one
role to each domain user. These activities are per-
formed by a Domain Expert.
A METHODOLOGY FOR INTEGRATING NEW SCIENTIFIC DOMAINS AND APPLICATIONS IN A VIRTUAL
LABORATORY ENVIRONMENT
271

7 CONCLUSIONS
The main subject of this paper was to provide a
methodology for integrating emerging scientific do-
mains and applications in a virtual laboratory envi-
ronment. Integration of new domains and applications
in a complex virtual laboratory environment requires
a common understanding of scientific experiments,
availability of both well-defined data models for mod-
elling experiment-related information and functional-
ity / mechanisms for the management of this informa-
tion. Therefore, in this paper:
1. an experiment model is introduced that is capable
of uniformly representing different aspects of het-
erogeneous scientific experiments;
2. several data models are provided for representing
the experiment-related information, which are uni-
form and reusable (to model information related to
heterogeneous experiments), and open, flexible and
extendible (to improve the developed models in the
future when needed and to model new types of ex-
perimental information);
3. mechanisms for managing the experiment-related
information in a VL are mentioned, that are generic
and reusable (to support uniform management of
heterogenous experimental information); and
4. a methodology is defined for integration of new do-
mains and applications in VL based on the results
obtained and experience gained during the VLAM-
G project presented here, which provides a step-
by-step guidance to domain experts, tool develop-
ers and administrators during the process of adding
new applications / resources to the VL.
The methodology defined in this paper is generic
and can be implemented by different support envi-
ronments in different ways. During the course of the
VLAM-G project, the methodology has been applied
to the two initial application cases (the DNA microar-
ray and MACS applications), and the received feed-
back was used to further improve/refine the method-
ology. Currently, two databases for these applications
together with experiment procedures and some anal-
ysis tools are provided to VLAM-G users. Further-
more, development of a new application for gene se-
quence analysis studies using this methodology is on-
going.
REFERENCES
Afsarmanesh, H., Belleman, R. G., Belloum, A. S. Z., Ben-
abdelkader, A., van den Brand, J. F. J., Eijkel, G. B.,
Frenkel, A., Garita, C., Groep, D. L., Heeren, R.
M. A., Hendrikse, Z. W., Hertzberger, L. O., Kaan-
dorp, J. A., Kaletas, E. C., Korkhov, V., de Laat, C.
T. A. M., Sloot, P. M. A., Vasunin, D., Visser, A., and
Yakali, H. H. (2002). Vlam-g: A grid-based virtual
laboratory. Scientific Programming, 10(2):173–181.
Afsarmanesh, H., Kaletas, E. C., Benabdelkader, A., Garita,
C., and Hertzberger, L. O. (2001). A reference archi-
tecture for scientific virtual laboratories. Future Gen-
eration Computer Systems, 17(8):999–1008.
Allen, G., Benger, W., Dramlitsch, T., Goodale, T., Hege,
H. C., Lanfermann, G., Merzky, A., Radke, T., Seidel,
E., and Shalf, J. (2001). Cactus tools for grid applica-
tions. Cluster Computing, 4(3):179–188.
Ashby, J. V., Bicarregui, J. C., Boyd, D. R. S., van Dam,
K. K., Lambert, S. C., Matthews, B. M., and O’Neill,
K. D. (2001). A multidisicplinary scientific data por-
tal. In Proceedings of the 9th International Confer-
ence and Exhibition on High-Performance Computing
and Networking (HPCN Europe 2001), pages 13–22.
Baxevanis, A. D. (2001). The molecular biology database
collection: An updated compilation of biological
database resources. Nucleic Acids Research, 29(1):1–
10.
Catlin, A. C., Gaitatzes, M., Houstis, E., Ma, Z., Markus, S.,
Rice, J. R., Wang, N.-H., and Weerwarana, S. (2000).
Enabling Technologies for Computational Science -
Frameworks, Middleware and Environments, chap-
ter 24, pages 301–313. Kluwer Academic Publishers.
Frenkel, A., Afsarmanesh, H., Eijkel, G. B., and
Hertzberger, L. O. (2001). Information management
for material science applications in a virtual labora-
tory. In Proceedings of the 12th International Con-
ference on Database and Expert Systems Applications
(DEXA 2001), pages 165–174.
Gallopoulos, E., Houstis, E., and Rice, J. R. (1994). Com-
puter as thinker/doer: Problem-solving environments
for computational science. Computing in Science and
Engineering, 1(2):11–23.
Kaletas, E. C., Afsarmanesh, H., and Hertzberger, L. O.
(2003). Modelling multi-disciplinary scientific exper-
iments and information. In Proceedings of the Eigh-
teenth International Symposium on Computer and In-
formation Sciences (ISCIS03), pages 75–82.
Messina, P. (2003). The emergence of virtual laborato-
ries for science and engineering - igrid2002 presen-
tation. Electronically available at: http://www.
igrid2002.org/ppt/Paul_Messina.ppt.
Pierce, M., Youn, C., and Fox, G. C. (2002). The gateway
computational web portal. Concurrency and Com-
putation: Practice and Experience, 14(13–15):1411–
1426.
Schuchardt, K. L., Myers, J. D., and Stephan, E. G. (2002).
A web-based data architecture for problem solving en-
vironments: Application of distributed authoring and
versioning to the extensible computational chemistry
environment. Cluster Computing, 5(3):287–296.
Vary, J. P. (2000). Report of the expert meeting on virtual
laboratories. Technical Report CII-00/WS/01, United
Nations Educational, Scientific and Cultural Organi-
zation.
ICEIS 2004 - INFORMATION SYSTEMS ANALYSIS AND SPECIFICATION
272
